Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy
Abstract
1. Introduction
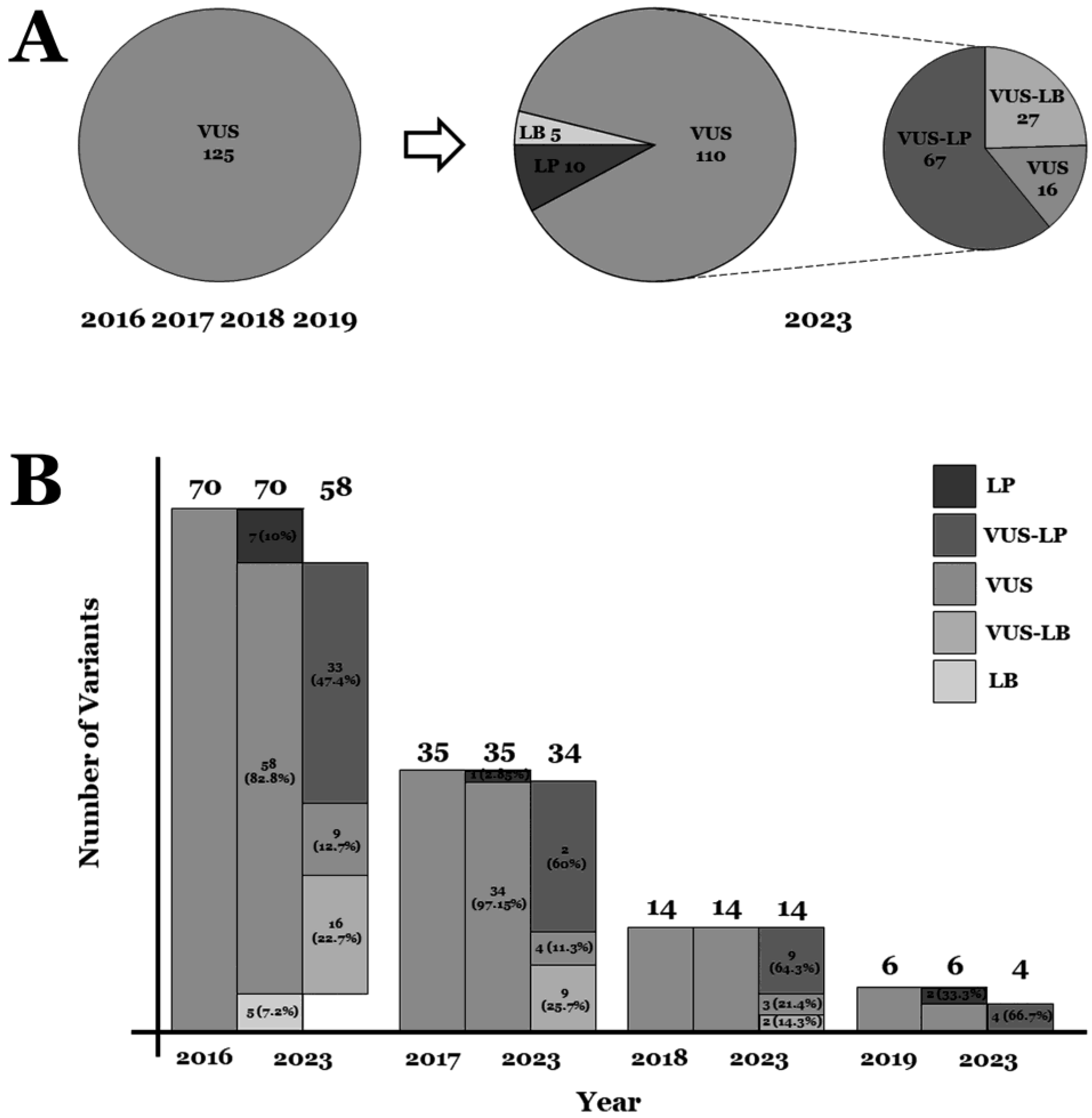
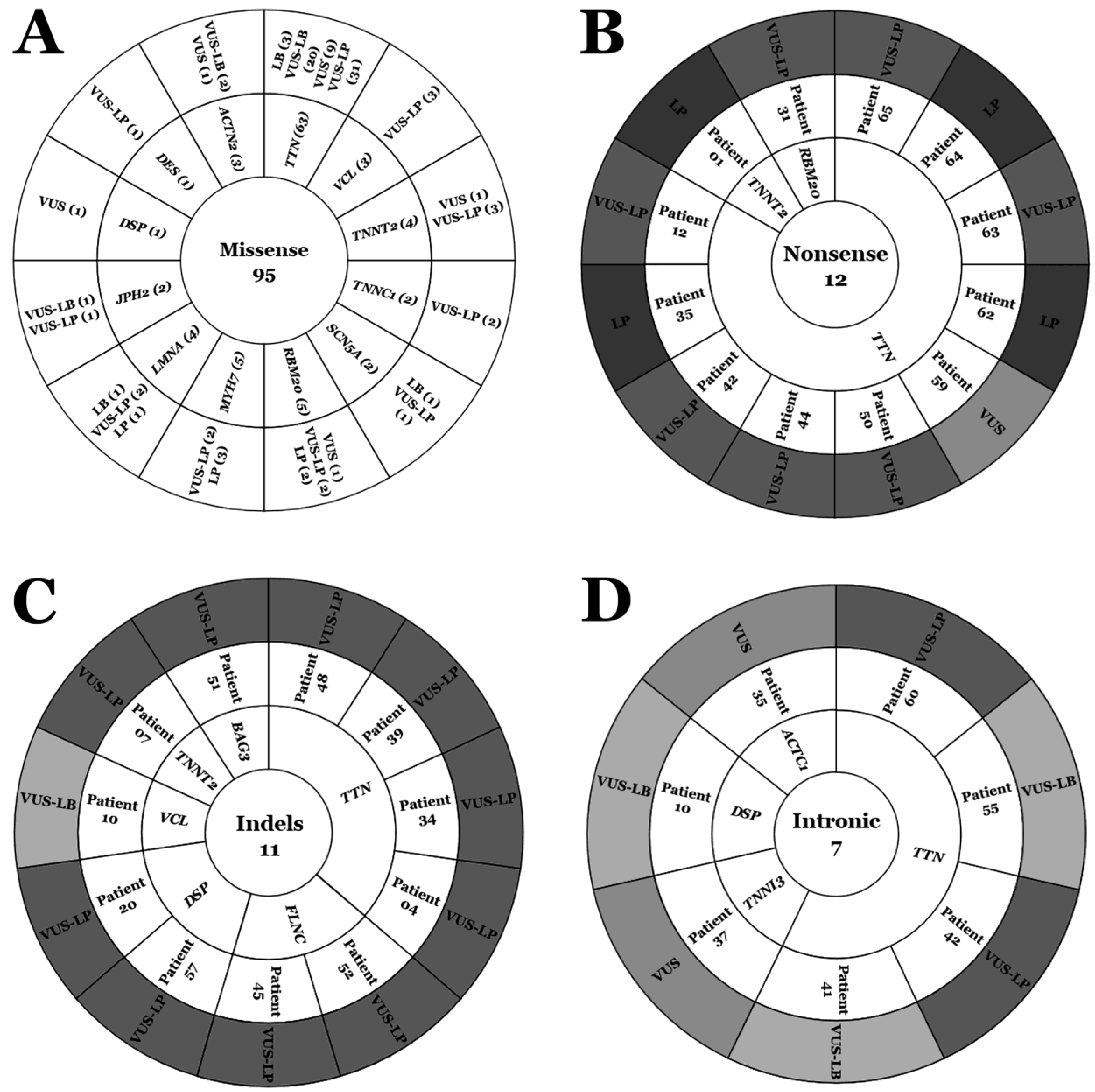
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Genetic Analysis
4.3. Data Sources
4.4. Classification/Interpretation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arbelo, E.; Protonotarios, A.; Gimeno, J.R.; Arbustini, E.; Barriales-Villa, R.; Basso, C.; Bezzina, C.R.; Biagini, E.; A Blom, N.; A de Boer, R.; et al. 2023 ESC Guidelines for the management of cardiomyopathies. Eur. Heart J. 2023, 44, 3503–3626. [Google Scholar] [CrossRef] [PubMed]
- Wilde, A.A.M.; Semsarian, C.; Marquez, M.F.; Shamloo, A.S.; Ackerman, M.J.; Ashley, E.A.; Sternick, E.B.; Barajas-Martinez, H.; Behr, E.R.; Bezzina, C.R.; et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the State of Genetic Testing for Cardiac Diseases. Europace 2022, 24, 1307–1367. [Google Scholar] [CrossRef]
- Muller, R.D.; McDonald, T.; Pope, K.; Cragun, D. Evaluation of Clinical Practices Related to Variants of Uncertain Significance Results in Inherited Cardiac Arrhythmia and Inherited Cardiomyopathy Genes. Circ. Genom. Precis. Med. 2020, 13, e002789. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.; Peterson, L.; Ai, T.; Asatryan, B.; Bronicki, L.; Brown, E.; Celeghin, R.; Edwards, M.; Fan, J.; Ingles, J.; et al. An Evidence-Based Assessment of Genes in Dilated Cardiomyopathy. Circulation 2021, 144, 7–19. [Google Scholar] [CrossRef]
- Landstrom, A.P.; Chahal, A.A.; Ackerman, M.J.; Cresci, S.; Milewicz, D.M.; Morris, A.A.; Sarquella-Brugada, G.; Semsarian, C.; Shah, S.H.; Sturm, A.C.; et al. Interpreting Incidentally Identified Variants in Genes Associated with Heritable Cardiovascular Disease: A Scientific Statement from the American Heart Association. Circ. Genom. Precis. Med. 2023, 16, e000092. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Asatryan, B.; Shah, R.A.; Sharaf Dabbagh, G.; Landstrom, A.P.; Darbar, D.; Khanji, M.Y.; Lopes, L.R.; van Duijvenboden, S.; Muser, D.; Lee, A.M.; et al. Predicted Deleterious Variants in Cardiomyopathy Genes Prognosticate Mortality and Composite Outcomes in UK Biobank. JACC Heart Fail. 2023. ahead of print. [Google Scholar] [CrossRef]
- Hofmeyer, M.; Haas, G.J.; Jordan, E.; Cao, J.; Kransdorf, E.; Ewald, G.A.; Morris, A.A.; Owens, A.; Lowes, B.; Stoller, D.; et al. Rare Variant Genetics and Dilated Cardiomyopathy Severity: The DCM Precision Medicine Study. Circulation 2023, 148, 872–881. [Google Scholar] [CrossRef]
- Stroeks, S.; Verdonschot, J.A.J. The next step toward personalized recommendations for genetic cardiomyopathies. Eur. J. Hum. Genet. 2023, 31, 1201–1203. [Google Scholar] [CrossRef]
- Campuzano, O.; Sarquella-Brugada, G.; Fernandez-Falgueras, A.; Coll, M.; Iglesias, A.; Ferrer-Costa, C.; Cesar, S.; Arbelo, E.; García-Álvarez, A.; Jordà, P.; et al. Reanalysis and reclassification of rare genetic variants associated with inherited arrhythmogenic syndromes. EBioMedicine 2020, 54, 102732. [Google Scholar] [CrossRef]
- Towbin, J.A.; McKenna, W.J.; Abrams, D.J.; Ackerman, M.J.; Calkins, H.; Darrieux, F.C.C.; Daubert, J.P.; de Chillou, C.; DePasquale, E.C.; Desai, M.Y.; et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm 2019, 16, e301–e372. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Givertz, M.M.; Ho, C.Y.; Judge, D.P.; Kantor, P.F.; McBride, K.L.; Morales, A.; Taylor, M.R.G.; Vatta, M.; Ware, S.M.; et al. Genetic evaluation of cardiomyopathy: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2018, 20, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Musunuru, K.; Hershberger, R.E.; Day, S.M.; Klinedinst, N.J.; Landstrom, A.P.; Parikh, V.N.; Prakash, S.; Semsarian, C.; Sturm, A.C.; on behalf of the American Heart Association Council on Genomic and Precision Medicine; et al. Genetic Testing for Inherited Cardiovascular Diseases: A Scientific Statement from the American Heart Association. Circ. Genom. Precis. Med. 2020, 13, e000067. [Google Scholar] [CrossRef] [PubMed]
- Stiles, M.K.; Wilde, A.A.; Abrams, D.J.; Ackerman, M.J.; Albert, C.M.; Behr, E.R.; Chugh, S.S.; Cornel, M.C.; Gardner, K.; Ingles, J.; et al. 2020 APHRS/HRS expert consensus statement on the investigation of decedents with sudden unexplained death and patients with sudden cardiac arrest, and of their families. Heart Rhythm 2021, 18, e1–e50. [Google Scholar] [CrossRef] [PubMed]
- Sarquella-Brugada, G.; Fernandez-Falgueras, A.; Cesar, S.; Arbelo, E.; Coll, M.; Perez-Serra, A.; Puigmulé, M.; Iglesias, A.; Alcalde, M.; Vallverdú-Prats, M.; et al. Clinical impact of rare variants associated with inherited channelopathies: A 5-year update. Hum. Genet. 2022, 141, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.S.; Bernhardt, M.; McBride, K.L.; Reshmi, S.C.; Zmuda, E.; Kertesz, N.J.; Garg, V.; Fitzgerald-Butt, S.; Kamp, A.N. Reclassification of Variants of Uncertain Significance in Children with Inherited Arrhythmia Syndromes is Predicted by Clinical Factors. Pediatr. Cardiol. 2019, 40, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Vallverdú-Prats, M.; Alcalde, M.; Sarquella-Brugada, G.; Cesar, S.; Arbelo, E.; Fernandez-Falgueras, A.; Coll, M.; Pérez-Serra, A.; Puigmulé, M.; Iglesias, A.; et al. Rare Variants Associated with Arrhythmogenic Cardiomyopathy: Reclassification Five Years Later. J. Pers. Med. 2021, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Quiat, D.; Witkowski, L.; Zouk, H.; Daly, K.P.; Roberts, A.E. Retrospective Analysis of Clinical Genetic Testing in Pediatric Primary Dilated Cardiomyopathy: Testing Outcomes and the Effects of Variant Reclassification. J. Am. Heart Assoc. 2020, 9, e016195. [Google Scholar] [CrossRef]
- Stroeks, S.; Hellebrekers, D.; Claes, G.R.F.; Tayal, U.; Krapels, I.P.C.; Vanhoutte, E.K.; van den Wijngaard, A.; Henkens, M.T.H.M.; Ware, J.S.; Heymans, S.R.B.; et al. Clinical impact of re-evaluating genes and variants implicated in dilated cardiomyopathy. Genet. Med. 2021, 23, 2186–2193. [Google Scholar] [CrossRef]
- Martínez-Barrios, E.; Sarquella-Brugada, G.; Pérez-Serra, A.; Fernández-Falgueras, A.; Cesar, S.; Coll, M.; Puigmulé, M.; Iglesias, A.; Alcalde, M.; Vallverdú-Prats, M.; et al. Discerning the Ambiguous Role of Missense TTN Variants in Inherited Arrhythmogenic Syndromes. J. Pers. Med. 2022, 12, 241. [Google Scholar] [CrossRef]
- Martinez-Barrios, E.; Sarquella-Brugada, G.; Perez-Serra, A.; Fernandez-Falgueras, A.; Cesar, S.; Alcalde, M.; Coll, M.; Puigmulé, M.; Iglesias, A.; Ferrer-Costa, C.; et al. Reevaluation of ambiguous genetic variants in sudden unexplained deaths of a young cohort. Int. J. Leg. Med. 2023, 137, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Ware, S.M.; Bhatnagar, S.; Dexheimer, P.J.; Wilkinson, J.D.; Sridhar, A.; Fan, X.; Shen, Y.; Tariq, M.; Schubert, J.A.; Colan, S.D.; et al. The genetic architecture of pediatric cardiomyopathy. Am. J. Hum. Genet. 2022, 109, 282–298. [Google Scholar] [CrossRef]
- Stroeks, S.L.V.M.; Hellebrekers, D.; Claes, G.R.F.; Krapels, I.P.C.; Henkens, M.H.T.M.; Sikking, M.; Vanhoutte, E.K.; Enden, A.H.-V.D.; Brunner, H.G.; Wijngaard, A.v.D.; et al. Diagnostic and prognostic relevance of using large gene panels in the genetic testing of patients with dilated cardiomyopathy. Eur. J. Hum. Genet. 2023, 31, 776–783. [Google Scholar] [CrossRef] [PubMed]
- McAfee, Q.; Chen, C.Y.; Yang, Y.; Caporizzo, M.A.; Morley, M.; Babu, A.; Jeong, S.; Brandimarto, J.; BediJr, K.C.; Flam, E.; et al. Truncated titin proteins in dilated cardiomyopathy. Sci. Transl. Med. 2021, 13, eabd7287. [Google Scholar] [CrossRef]
- Begay, R.L.; Graw, S.; Sinagra, G.; Merlo, M.; Slavov, D.; Gowan, K.; Jones, K.L.; Barbati, G.; Spezzacatene, A.; Brun, F.; et al. Role of Titin Missense Variants in Dilated Cardiomyopathy. J. Am. Heart Assoc. 2015, 4, e002645. [Google Scholar] [CrossRef] [PubMed]
- Rich, K.A.; Moscarello, T.; Siskind, C.; Brock, G.; Tan, C.A.; Vatta, M.; Winder, T.L.; Elsheikh, B.; Vicini, L.; Tucker, B.; et al. Novel heterozygous truncating titin variants affecting the A-band are associated with cardiomyopathy and myopathy/muscular dystrophy. Mol. Genet. Genom. Med. 2020, 8, e1460. [Google Scholar] [CrossRef] [PubMed]
- Akinrinade, O.; Heliö, T.; Deprez, R.H.L.; Jongbloed, J.D.H.; Boven, L.G.; Berg, M.P.v.D.; Pinto, Y.M.; Alastalo, T.-P.; Myllykangas, S.; van Spaendonck-Zwarts, K.; et al. Relevance of Titin Missense and Non-Frameshifting Insertions/Deletions Variants in Dilated Cardiomyopathy. Sci. Rep. 2019, 9, 4093. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, F.; Lalaguna, L.; Martínez-Martín, I.; Piqueras-Flores, J.; Rasmussen, T.B.; Zorio, E.; Giovinazzo, G.; Prados, B.; Ochoa, J.P.; Bornstein, B.; et al. Titin Missense Variants as a Cause of Familial Dilated Cardiomyopathy. Circulation 2023, 147, 1711–1713. [Google Scholar] [CrossRef] [PubMed]
- Rosamilia, M.B.; Markunas, A.M.; Kishnani, P.S.; Landstrom, A.P. Underrepresentation of Diverse Ancestries Drives Uncertainty in Genetic Variants Found in Cardiomyopathy-Associated Genes. JACC 2024, 3, 100767. [Google Scholar] [CrossRef]
- Priori, S.G.; Blomstrom-Lundqvist, C. 2015 European Society of Cardiology Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death summarized by co-chairs. Eur. Heart J. 2015, 36, 2757–2759. [Google Scholar] [CrossRef]
- Al-Khatib, S.M.; Stevenson, W.G.; Ackerman, M.J.; Bryant, W.J.; Callans, D.J.; Curtis, A.B.; Deal, B.J.; Dickfeld, T.; Field, M.E.; Fonarow, G.C.; et al. 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: Executive summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Heart Rhythm 2018, 15, e190–e252. [Google Scholar] [PubMed]
- Kobayashi, Y.; Yang, S.; Nykamp, K.; Garcia, J.; Lincoln, S.E.; Topper, S.E. Pathogenic variant burden in the ExAC database: An empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Jordan, D.M.; Kiezun, A.; Baxter, S.M.; Agarwala, V.; Green, R.C.; Murray, M.F.; Pugh, T.; Lebo, M.S.; Rehm, H.L.; Funke, B.H.; et al. Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2011, 88, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.A.; Caleshu, C.; Morales, A.; Buchan, J.; Wolf, Z.; Harrison, S.M.; Cook, S.; Dillon, M.W.; Garcia, J.; Haverfield, E.; et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: Recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet. Med. 2018, 20, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Bains, S.; Dotzler, S.M.; Krijger, C.; Giudicessi, J.R.; Ye, D.; Bikker, H.; Rohatgi, R.K.; Tester, D.J.; Bos, J.M.; Wilde, A.A.; et al. A phenotype-enhanced variant classification framework to decrease the burden of missense variants of uncertain significance in type 1 long QT syndrome. Heart Rhythm 2022, 19, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Giudicessi, J.R.; Lieve, K.V.V.; Rohatgi, R.K.; Koca, F.; Tester, D.J.; van der Werf, C.; Martijn Bos, J.; Wilde, A.A.M.; Ackerman, M.J. Assessment and Validation of a Phenotype-Enhanced Variant Classification Framework to Promote or Demote RYR2 Missense Variants of Uncertain Significance. Circ. Genom. Precis. Med. 2019, 12, e002510. [Google Scholar] [CrossRef] [PubMed]
- Arbustini, E.; Behr, E.R.; Carrier, L.; van Duijn, C.; Evans, P.; Favalli, V.; van der Harst, P.; Haugaa, K.H.; Jondeau, G.; Kääb, S.; et al. Interpretation and actionability of genetic variants in cardiomyopathies: A position statement from the European Society of Cardiology Council on cardiovascular genomics. Eur. Heart J. 2022, 43, 1901–1916. [Google Scholar] [CrossRef]
- Arbustini, E.; Urtis, M.; Elliott, P. Interpretation of genetic variants depends on a clinically guided integration of phenotype and molecular data. Eur. Heart J. 2022, 43, 2638–2639. [Google Scholar] [CrossRef]

| Patient | Gene | Nucleotide | Protein | dbSNP | ClinVar | GnomAD (MAF%) | Classification (Year) | Classification 2023 |
|---|---|---|---|---|---|---|---|---|
| 1 | TNNT2 | c.860G>A | p.Trp287Ter | rs727504247 | LP | NA | VUS (2016) | LP |
| 1 | TTN | c.47951G>A | p.Arg15984His | rs201774108 | VUSc | 0.0001 | VUS (2016) | VUS |
| 2 | TTN | c.73195G>A | p.Val24399Ile | rs1257567608 | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 2 | TTN | c.57388C>T | p.Arg19130Cys | rs72646861 | LB | 0.827 | VUS (2016) | LB |
| 2 | TTN | c.53117C>T | p.Pro17706Leu | rs72646845 | LB | 0.369 | VUS (2016) | LB |
| 3 | ACTN2 | c.2051A>T | p.Asn684Ile | rs576783493 | VUSc | 0.0001 | VUS (2016) | VUS |
| 3 | TTN | c.93125G>A | p.Gly31042Asp | rs373754986 | VUS | 0.007 | VUS (2016) | VUS (VUS-LP) |
| 3 | TTN | c.76456G>C | p.Asp25486His | rs780958039 | VUS | 0.0008 | VUS (2016) | VUS (VUS-LP) |
| 4 | ACTN2 | c.1426G>A | p.Ala476Thr | rs142943120 | VUSc | 0.027 | VUS (2016) | VUS (VUS-LB) |
| 4 | TTN | c.57978del | p.Val19327PhefsTer10 | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 4 | TTN | c.76559G>A | p.Ser25520Asn | rs200450022 | VUSc | 0.061 | VUS (2016) | VUS (VUS-LB) |
| 4 | TTN | c.72764G>C | p.Gly24225Ala | rs114071241 | VUS | 0.001 | VUS (2016) | VUS (VUS-LP) |
| 4 | TTN | c.17066G>C | p.Gly5689Ala | rs200118743 | VUSc | 0.063 | VUS (2016) | VUS (VUS-LB) |
| 4 | TTN | c.4675G>A | p.Val1559Ile | rs538451328 | NA | 0.0003 | VUS (2016) | VUS (VUS-LP) |
| 5 | TTN | c.98971G>C | p.Glu32991Gln | rs199632397 | VUSc | 0.042 | VUS (2016) | VUS (VUS-LB) |
| 5 | TTN | c.27659G>A | p.Arg9220Gln | rs727504757 | VUS | 0.003 | VUS (2016) | VUS (VUS-LP) |
| 6 | TTN | c.45392G>A | p.Arg15131His | rs72646808 | VUSc | 0.185 | VUS (2016) | LB |
| 7 | TNNT2 | c.629_631del | p.Lys210del | rs45578238 | VUS | NA | VUS (2016) | VUS (VUS-LP) |
| 8 | MYH7 | c.1106G>A | p.Arg369Gln | rs397516089 | LP | 0.00006 | VUS (2016) | LP |
| 9 | TTN | c.73967A>G | p.Asn24656Ser | rs368443217 | VUSc | 0.008 | VUS (2016) | VUS (VUS-LB) |
| 9 | TTN | c.65012T>A | p.Met21671Lys | rs750298083 | VUSc | 0.004 | VUS (2016) | VUS |
| 9 | TTN | c.51830G>A | p.Arg17277His | rs201457934 | VUSc | 0.01 | VUS (2016) | VUS (VUS-LB) |
| 9 | TTN | c.9247G>A | p.Glu3083Lys | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 9 | MYH7 | c.2711G>A | p.Arg904His | rs397516165 | LP | 0.0001 | VUS (2016) | LP |
| 10 | DSP | c.1266+6G>T | NA | rs73375345 | LB | 0.037 | VUS (2016) | VUS (VUS-LB) |
| 10 | TTN | c.20920A>G | p.Ser6974Gly | rs72648980 | VUSc | 0.062 | VUS (2016) | VUS (VUS-LB) |
| 10 | VCL | c.2862_2864del | p.Leu955del | rs397517237 | VUSc | 0.021 | VUS (2016) | VUS (VUS-LB) |
| 11 | SCN5A | c.6010T>C | p.Phe2004Leu | rs41311117 | VUSc | 0.198 | VUS (2016) | LB |
| 12 | TTN | c.72970C>T | p.Gln24324Ter | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 13 | TTN | c.49249G>A | p.Asp16417Asn | rs1244503464 | NA | 0.0004 | VUS (2016) | VUS (VUS-LP) |
| 14 | TTN | c.12814G>T | p.Asp4272Tyr | rs72648940 | VUSc | 0.0005 | VUS (2016) | VUS |
| 15 | TTN | c.77076A>C | p.Glu25692Asp | rs370547473 | NA | 0.004 | VUS (2016) | VUS (VUS-LP) |
| 16 | TTN | c.32932C>A | p.Pro10978Thr | rs1393076582 | VUSc | NA | VUS (2016) | VUS |
| 17 | DES | c.179C>T | p.Ser60Leu | rs868853251 | NA | 0.0001 | VUS (2016) | VUS (VUS-LP) |
| 17 | TTN | c.46877G>A | p.Gly15626Asp | rs201802447 | VUS | 0.007 | VUS (2016) | VUS |
| 17 | TTN | c.1066G>C | p.Glu356Gln | rs144531477 | VUSc | 0.015 | VUS (2016) | VUS (VUS-LB) |
| 18 | TTN | c.95137A>G | p.Ile31713Val | rs758945559 | NA | 0.0008 | VUS (2016) | VUS (VUS-LP) |
| 18 | TTN | c.30484C>A | p.Pro10162Thr | rs532102837 | VUSc | 0.058 | VUS (2016) | VUS (VUS-LB) |
| 18 | DSP | c.6799A>T | p.Thr2267Ser | rs181378432 | LB | 0.009 | VUS (2016) | VUS |
| 18 | LMNA | c.1621C>T | p.Arg541Cys | rs56984562 | LP | 0.0001 | VUS (2016) | LP |
| 19 | RBM20 | c.1900C>T | p.Arg634Trp | rs796734066 | LP | NA | VUS (2016) | LP |
| 20 | DSP | del ex. 21_24 | NA | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 21 | MYH7 | c.1371A>G | p.Ile457Met | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 22 | TTN | c.94724T>C | p.Met31575Thr | rs397517786 | VUSc | 0.007 | VUS (2016) | VUS (VUS-LB) |
| 22 | TTN | c.62687G>T | p.Gly20896Val | rs549938348 | VUS | 0.0004 | VUS (2016) | VUS (VUS-LP) |
| 23 | RBM20 | c.1906C>T | p.Arg636Cys | rs267607002 | VUSc | 0.0001 | VUS (2016) | VUS |
| 24 | JPH2 | c.1736C>T | p.Pro579Leu | rs953353202 | VUS | 0.001 | VUS (2016) | VUS (VUS-LP) |
| 24 | RBM20 | c.1980C>A | p.Ser660Arg | NA | NA | 0.00007 | VUS (2016) | VUS (VUS-LP) |
| 25 | RBM20 | c.2200C>G | p.Arg734Gly | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 26 | TTN | c.57212G>A | p.Arg19071Gln | rs373282633 | VUS | 0.003 | VUS (2016) | VUS |
| 26 | TTN | c.48221T>A | p.Leu16074Gln | rs140714512 | VUSc | 0.051 | VUS (2016) | VUS (VUS-LB) |
| 26 | TTN | c.29645A>C | p.Lys9882Thr | rs760742068 | VUS | 0.001 | VUS (2016) | VUS (VUS-LP) |
| 27 | LMNA | c.1930C>T | p.Arg644Cys | rs1420000963 | VUSc | 0.201 | VUS (2016) | LB |
| 27 | TTN | c.77188C>T | p.Arg25730Trp | rs779581886 | VUS | 0.002 | VUS (2016) | VUS (VUS-LP) |
| 27 | TTN | c.60581T>C | p.Leu20194Pro | rs1359881893 | VUS | 0.0008 | VUS (2016) | VUS (VUS-LP) |
| 28 | RBM20 | c.1913C>T | p.Pro638Leu | rs267607003 | LP | 0.0003 | VUS (2016) | LP |
| 28 | TTN | c.74942C>T | p.Ala24981Val | rs749950083 | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 28 | TTN | c.73501C>G | p.Pro24501Ala | rs770542451 | NA | 0.0004 | VUS (2016) | VUS (VUS-LP) |
| 28 | TTN | c.1333G>A | p.Ala445Thr | rs142414432 | VUSc | 0.021 | VUS (2016) | VUS (VUS-LB) |
| 29 | LMNA | c.1949A>G | p.Asn650Ser | rs775728847 | VUS | 0.0003 | VUS (2016) | VUS (VUS-LP) |
| 29 | TTN | c.38807A>G | p.Asn12936Ser | rs1184631064 | VUS | 0.0008 | VUS (2016) | VUS (VUS-LP) |
| 30 | ACTN2 | c.1040C>T | p.Thr347Met | rs727504590 | VUSc | 0.009 | VUS (2016) | VUS (VUS-LB) |
| 31 | RBM20 | c.3684A>G | p.Ter1228TrpextTer33 | rs1845123103 | NA | 0.0001 | VUS (2016) | VUS (VUS-LP) |
| 31 | TTN | c.40001G>A | p.Gly13334Glu | rs561284948 | VUS | 0.001 | VUS (2016) | VUS (VUS-LP) |
| 31 | TTN | c.20863C>T | p.Pro6955Ser | rs1438804317 | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 32 | MYH7 | c.602T>C | p.Ile201Thr | rs397516258 | LP | 0.0001 | VUS (2016) | LP |
| 32 | TTN | c.71188G>A | p.Gly23730Arg | rs72648205 | VUSc | 0.034 | VUS (2016) | VUS (VUS-LB) |
| 33 | TTN | c.82400G>A | p.Arg27467His | rs199895320 | VUS | 0.002 | VUS (2016) | VUS (VUS-LP) |
| 33 | TTN | c.75575A>T | p.Asn25192IIe | rs200714263 | VUS | 0.002 | VUS (2016) | VUS (VUS-LP) |
| 34 | TTN | c.9139_9150del | p.Ser3047_Thr3050del | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 35 | ACTC1 | c.455-7C>T | NA | rs768363857 | LB | 0.003 | VUS (2017) | VUS |
| 35 | TTN | c.53791C>T | p.Arg17931Ter | rs869312112 | LP | NA | VUS (2017) | LP |
| 35 | TTN | c.33568T>G | p.Cys11190Gly | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 36 | TTN | c.58846G>A | p.Gly19616Ser | rs1262240030 | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 36 | TTN | c.58590G>C | p.Gln19530His | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 36 | TTN | c.8320G>A | p.Glu2774Lys | rs763666119 | LB | 0.0003 | VUS (2017) | VUS (VUS-LP) |
| 37 | TNNI3 | c.-8G>A | NA | rs773513015 | VUSc | 0.001 | VUS (2017) | VUS |
| 37 | TTN | c.40796G>A | p.Arg13599Gln | rs778774812 | VUS | 0.001 | VUS (2017) | VUS (VUS-LP) |
| 38 | TTN | c.57709G>T | p.Val19237Leu | rs1397460981 | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 39 | TTN | c.95967dup | p.Arg31990ThrfsTer10 | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 40 | TTN | c.12389G>A | p.Cys4130Tyr | rs375577529 | VUSc | 0.008 | VUS (2017) | VUS (VUS-LB) |
| 40 | VCL | c.2905G>A | p.Ala969Thr | rs199751261 | VUS | 0.002 | VUS (2017) | VUS (VUS-LP) |
| 41 | LMNA | c.253C>G | p.Leu85Val | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 41 | TTN | c.78602G>A | p.Gly26201Asp | rs756222422 | NA | 0.0004 | VUS (2017) | VUS (VUS-LP) |
| 41 | TTN | c.64137G>C | p.Lys21379Asn | rs56019808 | VUSc | 0.014 | VUS (2017) | VUS (VUS-LB) |
| 41 | TTN | c.36844+9A>G | NA | rs372725070 | LB | 0.013 | VUS (2017) | VUS (VUS-LB) |
| 41 | TTN | c.21355G>T | p.Ala7119Ser | rs200972189 | VUSc | 0.02 | VUS (2017) | VUS (VUS-LB) |
| 42 | TTN | c.92472G>C | p.Lys30824Asn | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 42 | TTN | c.25810C>T | p.Arg8604Ter | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 42 | TTN | c.28738+7T>G | NA | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 42 | TTN | c.14656A>T | p.Thr4886Ser | rs794727816 | VUS | NA | VUS (2017) | VUS (VUS-LP) |
| 43 | TNNC1 | c.400G>A | p.Glu134Lys | rs1553651640 | VUS | NA | VUS (2017) | VUS (VUS-LP) |
| 43 | TTN | c.95173A>G | p.Lys31725Glu | rs72629783 | VUSc | 0.021 | VUS (2017) | VUS (VUS-LB) |
| 43 | TTN | c.31709T>C | p.Ile10570Thr | rs72650057 | VUSc | 0.013 | VUS (2017) | VUS (VUS-LB) |
| 44 | TTN | c.56541G>A | p.Trp18847Ter | NA | LP | NA | VUS (2017) | VUS (VUS-LP) |
| 45 | FLNC | c.3612del | p.His1205ThrfsTer65 | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 46 | TTN | c.19570G>A | p.Asp6524Asn | rs72648973 | VUSc | 0.075 | VUS (2017) | VUS (VUS-LB) |
| 47 | TNNT2 | c.476G>A | p.Arg159Gln | rs45501500 | VUSc | NA | VUS (2017) | VUS |
| 48 | TTN | c.51369_51384del | p.Asp17123GlufsTer4 | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 49 | JPH2 | c.572C>G | p.Pro191Arg | rs554853074 | LB | 0.045 | VUS (2017) | VUS (VUS-LB) |
| 49 | TNNT2 | c.230C>T | p.Pro77Leu | rs144900708 | VUS | 0.003 | VUS (2017) | VUS (VUS-LP) |
| 50 | TTN | c.86270C>A | p.Ser28757Ter | NA | LP | NA | VUS (2017) | VUS (VUS-LP) |
| 50 | TTN | c.62666A>T | p.Asp20889Val | rs535816123 | NA | 0.002 | VUS (2017) | VUS (VUS-LP) |
| 50 | TTN | c.47435T>C | p.Ile15812Thr | rs72646819 | VUSc | 0.007 | VUS (2017) | VUS (VUS-LB) |
| 50 | TTN | c.16066A>G | p.Thr5356Ala | rs530353051 | VUS | 0.002 | VUS (2017) | VUS |
| 51 | BAG3 | c.903del | p.Arg301SerfsTer6 | NA | NA | NA | VUS (2018) | VUS (VUS-LP) |
| 51 | TTN | c.29327A>G | p.Tyr9776Cys | rs72650035 | VUSc | 0.02 | VUS (2018) | VUS (VUS-LB) |
| 52 | FLNC | c.1414del | p.Cys472ValfsTer20 | NA | NA | NA | VUS (2018) | VUS (VUS-LP) |
| 53 | MYH7 | c.5452C>T | p.Arg1818Trp | rs763073072 | VUS | 0.0005 | VUS (2018) | VUS (VUS-LP) |
| 54 | TNNC1 | c.394G>A | p.Asp132Asn | rs397516846 | VUS | 0.0003 | VUS (2018) | VUS (VUS-LP) |
| 55 | TTN | c.24617A>G | p.Asn8206Ser | NA | NA | NA | VUS (2018) | VUS |
| 55 | TTN | c.12889+7A>T | NA | rs10200398 | VUSc | 0.07 | VUS (2018) | VUS (VUS-LB) |
| 56 | TTN | c.20839G>A | p.Glu6947Lys | rs201326258 | VUS | 0.003 | VUS (2018) | VUS |
| 56 | VCL | c.1620T>G | p.Asp540Glu | rs533622785 | NA | 0.0006 | VUS (2018) | VUS (VUS-LP) |
| 57 | DSP | c.5673_5674dup | p.Lys1892ArgfsTer38 | NA | NA | NA | VUS (2018) | VUS (VUS-LP) |
| 57 | SCN5A | c.2924G>A | p.Arg975Gln | rs753149586 | VUS | 0.005 | VUS (2018) | VUS (VUS-LP) |
| 58 | TNNT2 | c.835G>A | p.Gly279Arg | rs757664792 | VUS | 0.0004 | VUS (2018) | VUS (VUS-LP) |
| 59 | TTN | c.9220C>T | p.Arg3074Ter | rs780706937 | VUSc | NA | VUS (2018) | VUS |
| 60 | TTN | c.81493+1G>T | NA | NA | NA | NA | VUS (2018) | VUS (VUS-LP) |
| 61 | TNNT2 | c.391C>G | p.Arg131Gly | rs74315380 | LP | NA | VUS (2019) | VUS (VUS-LP) |
| 62 | TTN | c.78412C>T | p.Arg26138Ter | rs794729384 | LP | 0.0004 | VUS (2019) | LP |
| 63 | TTN | c.67528G>T | p.Glu22510Ter | NA | NA | NA | VUS (2019) | VUS (VUS-LP) |
| 64 | TTN | c.39790C>T | p.Arg13264Ter | rs751746401 | LP | 0.0004 | VUS (2019) | LP |
| 65 | TTN | c.59787G>A | p.Trp19929Ter | NA | NA | NA | VUS (2019) | VUS (VUS-LP) |
| 65 | VCL | c.158A>G | p.Asn53Ser | rs751938777 | VUS | 0.001 | VUS (2019) | VUS (VUS-LP) |
| 2023 | Intronic | Indels | Nonsense | Missense | Total | ||
|---|---|---|---|---|---|---|---|
| B | 0 | 0 | 0 | 0 | 0 | ||
| LB | 0 | 0 | 0 | 5 (4%) | 5 (4%) | ||
| VUS | VUS-LB | 3 (2.4%) | 1 (0.8%) | 0 | 23 (18.4%) | 27 (21.6%) | 110 (88%) |
| VUS | 2 (1.6%) | 0 | 1 (0.8%) | 13 (10.4%) | 16 (12.8%) | ||
| VUS-LP | 2 (1.6%) | 10 (8%) | 7 (5.6%) | 48 (38.4%) | 67 (53.6%) | ||
| LP | 0 | 0 | 4 (3.2%) | 6 (4.8%) | 10 (8%) | ||
| P | 0 | 0 | 0 | 0 | 0 | ||
| Total | 7 (5.6%) | 11 (8.8%) | 12 (9.6%) | 95 (76%) | 125 (100%) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Serra, A.; Toro, R.; Martinez-Barrios, E.; Iglesias, A.; Fernandez-Falgueras, A.; Alcalde, M.; Coll, M.; Puigmulé, M.; del Olmo, B.; Picó, F.; et al. Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy. Int. J. Mol. Sci. 2024, 25, 3807. https://doi.org/10.3390/ijms25073807
Pérez-Serra A, Toro R, Martinez-Barrios E, Iglesias A, Fernandez-Falgueras A, Alcalde M, Coll M, Puigmulé M, del Olmo B, Picó F, et al. Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy. International Journal of Molecular Sciences. 2024; 25(7):3807. https://doi.org/10.3390/ijms25073807
Chicago/Turabian StylePérez-Serra, Alexandra, Rocío Toro, Estefanía Martinez-Barrios, Anna Iglesias, Anna Fernandez-Falgueras, Mireia Alcalde, Mónica Coll, Marta Puigmulé, Bernat del Olmo, Ferran Picó, and et al. 2024. "Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy" International Journal of Molecular Sciences 25, no. 7: 3807. https://doi.org/10.3390/ijms25073807
APA StylePérez-Serra, A., Toro, R., Martinez-Barrios, E., Iglesias, A., Fernandez-Falgueras, A., Alcalde, M., Coll, M., Puigmulé, M., del Olmo, B., Picó, F., Lopez, L., Arbelo, E., Cesar, S., Llano, C. T. d., Mangas, A., Brugada, J., Sarquella-Brugada, G., Brugada, R., & Campuzano, O. (2024). Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy. International Journal of Molecular Sciences, 25(7), 3807. https://doi.org/10.3390/ijms25073807








