MicroRNA Associations with Preterm Labor—A Systematic Review
Abstract
1. Introduction
2. Methods
2.1. Inclusion Criteria
2.2. Exclusion Criteria
2.3. Search Strategy
2.4. Study Selection
2.5. Quality Assessment
2.6. Data Extraction
2.7. Study Selection
3. Results
4. Discussion
Clinical Implication
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Cook, J.; Bennett, P.R.; Kim, S.H.; Teoh, T.G.; Sykes, L.; Kindinger, L.M.; Garrett, A.; Binkhamis, R.; MacIntyre, D.A.; Terzidou, V. First Trimester Circulating MicroRNA Biomarkers Predictive of Subsequent Preterm Delivery and Cervical Shortening. Sci. Rep. 2019, 9, 5861. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.N.; Fàbregues, S.; Bartlett, G.; Boardman, F.; Cargo, M.; Dagenais, P.; Gagnon, M.-P.; Griffiths, F.; Nicolau, B.; O’cathain, A.; et al. The Mixed Methods Appraisal Tool (MMAT) version 2018 for information professionals and researchers. Educ. Inf. 2018, 34, 285–291. [Google Scholar] [CrossRef]
- Mavreli, D.; Papantoniou, N.; Kolialexi, A. miRNAs in pregnancy-related complications: An update. Expert Rev. Mol. Diagn. 2018, 18, 587–589. [Google Scholar] [CrossRef]
- Addo, K.A.; Palakodety, N.; Hartwell, H.J.; Tingare, A.; Fry, R.C. Placental microRNAs: Responders to environmental chemicals and mediators of the pathophysiology of the human placenta. Toxicol. Rep. 2020, 7, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Hadlich, F.; Abbas, M.W.; Iqbal, M.A.; Tesfaye, D.; Bouma, G.J.; Winger, Q.A.; Ponsuksili, S. MicroRNA–mRNA Networks in Pregnancy Complications: A Comprehensive Downstream Analysis of Potential Biomarkers. Int. J. Mol. Sci. 2021, 22, 2313. [Google Scholar] [CrossRef]
- Aryan, L.; Medzikovic, L.; Umar, S.; Eghbali, M. Pregnancy-associated cardiac dysfunction and the regulatory role of microRNAs. Biol. Sex Differ. 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Dinesen, S.; El-Faitarouni, A.; Frisk, N.L.S.; Sørensen, A.E.; Dalgaard, L.T. Circulating microRNA as Biomarkers for Gestational Diabetes Mellitus—A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 6186. [Google Scholar] [CrossRef] [PubMed]
- Elhag, D.A.; Al Khodor, S. Exploring the potential of microRNA as a diagnostic tool for gestational diabetes. J. Transl. Med. 2023, 21, 392. [Google Scholar] [CrossRef]
- Fu, X.; Li, Y.; Zhang, Z.; Wang, B.; Wei, R.; Chu, C.; Xu, K.; Li, L.; Liu, Y.; Li, X. Emerging role of miRNAs, lncRNAs, and circRNAs in pregnancy-associated diseases. Chin. Med. J. 2023, 136, 1300–1310. [Google Scholar] [CrossRef]
- Hu, X.-Q.; Zhang, L. MicroRNAs in Uteroplacental Vascular Dysfunction. Cells 2019, 8, 1344. [Google Scholar] [CrossRef]
- Jin, M.; Xu, Q.; Li, J.; Xu, S.; Tang, C. Micro-RNAs in Human Placenta: Tiny Molecules, Immense Power. Molecules 2022, 27, 5943. [Google Scholar] [CrossRef]
- Juchnicka, I.; Kuzmicki, M. Influence of MiRNAs in gestational diabetes mellitus development. Ginekol. Polska 2021, 92, 579–582. [Google Scholar] [CrossRef]
- Li, L.; Feng, T.; Zhou, W.; Liu, Y.; Li, H. miRNAs in decidual NK cells: Regulators worthy of attention during pregnancy. Reprod. Biol. Endocrinol. 2021, 19, 150. [Google Scholar] [CrossRef]
- Liang, L.; Chen, Y.; Wu, C.; Cao, Z.; Xia, L.; Meng, J.; He, L.; Yang, C.; Wang, Z. MicroRNAs: Key regulators of the trophoblast function in pregnancy disorders. J. Assist. Reprod. Genet. 2023, 40, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-N.; Jiang, Y.; Liu, X.-Q.; Yang, M.-M.; Chen, C.; Zhao, B.-H.; Huang, H.-F.; Luo, Q. MiRNAs in Gestational Diabetes Mellitus: Potential Mechanisms and Clinical Applications. J. Diabetes Res. 2021, 2021, 4632745. [Google Scholar] [CrossRef] [PubMed]
- Masete, M.; Dias, S.; Malaza, N.; Adam, S.; Pheiffer, C. A Big Role for microRNAs in Gestational Diabetes Mellitus. Front. Endocrinol. 2022, 13, 892587. [Google Scholar] [CrossRef]
- Omeljaniuk, W.J.; Laudański, P.; Miltyk, W. The role of miRNA molecules in the miscarriage process. Biol. Reprod. 2023, 109, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.B.M.; Sadek, S.T.; Mahesan, A.M. The role of microRNAs in human embryo implantation: A review. J. Assist. Reprod. Genet. 2019, 36, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Salma, U.; Alshaikh, A.B.A.; Alanazi, M.F.; Alomair, B.M.; Alruwaili, M.; Alruwaili, R. Role of MicroRNAs in Cardiac Disease with Stroke in Pregnancy. Oxidative Med. Cell. Longev. 2022, 2022, 5260085. [Google Scholar] [CrossRef]
- Shekibi, M.; Heng, S.; Nie, G. MicroRNAs in the Regulation of Endometrial Receptivity for Embryo Implantation. Int. J. Mol. Sci. 2022, 23, 6210. [Google Scholar] [CrossRef]
- Subramanian, A.; Weiss, D.; Nyhan, K.; Dewan, A.; Jukic, A.M.Z. Circulating miRNAs in the first trimester and pregnancy complications: A systematic review. Epigenetics 2023, 18, 2152615. [Google Scholar] [CrossRef]
- Tong, S.; Kaitu’U-Lino, T. Measuring circulating miRNAs in early pregnancy could identify fetuses’ destined to undergrow and be at increased risk of stillbirth. EBioMedicine 2021, 63, 103172. [Google Scholar] [CrossRef]
- Yang, H.; Ma, Q.; Wang, Y.; Tang, Z. Clinical application of exosomes and circulating microRNAs in the diagnosis of pregnancy complications and foetal abnormalities. J. Transl. Med. 2020, 18, 32. [Google Scholar] [CrossRef]
- Chavira-Suárez, E.; Hernández-Olvera, A.L.; Flores-Torres, M.; Celaya-Cruz, K.R.; Gitler, S.; De la Cerda-Ángeles, J.C.; Espinosa-Maldonado, N.C.; Flores-Jasso, C.F.; Gutiérrez, H.; Vadillo-Ortega, F. Longitudinal large-scale changes in maternal circulating microRNAs associated with gestation-related compartments, fetal sex, and growth during and post-pregnancy. Genomics 2023, 115, 110628. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Li, Z.; Yu, J.; Long, Y.; Zhao, Q.; Ding, X.; Wu, L.; Shao, S.; Zhang, L.; Xiang, W. MicroRNAs secreted by human embryos could be potential biomarkers for clinical outcomes of assisted reproductive technology. J. Adv. Res. 2021, 31, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Foley, H.B.; Howe, C.G.; Eckel, S.P.; Chavez, T.; Gevorkian, L.; Reyes, E.G.; Kapanke, B.; Martinez, D.; Xue, S.; Suglia, S.F.; et al. Extracellular vesicle-enriched miRNA profiles across pregnancy in the MADRES cohort. PLoS ONE 2021, 16, e0251259. [Google Scholar] [CrossRef]
- He, X.; Ding, D.-N. Expression and clinical significance of miR-204 in patients with hypertensive disorder complicating pregnancy. BMC Pregnancy Childbirth 2022, 22, 182. [Google Scholar] [CrossRef]
- Hosseini, M.K.; Gunel, T.; Gumusoglu, E.; Benian, A.; Aydinli, K. MicroRNA expression profiling in placenta and maternal plasma in early pregnancy loss. Mol. Med. Rep. 2018, 17, 4941–4952. [Google Scholar] [CrossRef] [PubMed]
- Illarionov, R.A.; Pachuliia, O.V.; Vashukova, E.S.; Tkachenko, A.A.; Maltseva, A.R.; Postnikova, T.B.; Nasykhova, Y.A.; Bespalova, O.N.; Glotov, A.S. Plasma miRNA Profile in High Risk of Preterm Birth during Early and Mid-Pregnancy. Genes 2022, 13, 2018. [Google Scholar] [CrossRef]
- Kim, S.H.; MacIntyre, D.A.; Binkhamis, R.; Cook, J.; Sykes, L.; Bennett, P.R.; Terzidou, V. Maternal plasma miRNAs as potential biomarkers for detecting risk of small-for-gestational-age births. EBioMedicine 2020, 62, 103145. [Google Scholar] [CrossRef]
- Légaré, C.; Clément, A.-A.; Desgagné, V.; Thibeault, K.; White, F.; Guay, S.-P.; Arsenault, B.J.; Scott, M.S.; Jacques, P.; Perron, P.; et al. Human plasma pregnancy-associated miRNAs and their temporal variation within the first trimester of pregnancy. Reprod. Biol. Endocrinol. 2022, 20, 14. [Google Scholar] [CrossRef] [PubMed]
- Manuck, T.A.; Eaves, L.A.; Rager, J.E.; Fry, R.C. Mid-pregnancy maternal blood nitric oxide-related gene and miRNA expression are associated with preterm birth. Epigenomics 2021, 13, 667–682. [Google Scholar] [CrossRef]
- Mavreli, D.; Theodora, M.; Avgeris, M.; Papantoniou, N.; Antsaklis, P.; Daskalakis, G.; Kolialexi, A. First Trimester Maternal Plasma Aberrant miRNA Expression Associated with Spontaneous Preterm Birth. Int. J. Mol. Sci. 2022, 23, 14972. [Google Scholar] [CrossRef]
- Menon, R.; Debnath, C.; Lai, A.; Guanzon, D.; Bhatnagar, S.; Kshetrapal, P.K.; Sheller-Miller, S.; Salomon, C.; The Garbhini Study Team. Circulating Exosomal miRNA Profile During Term and Preterm Birth Pregnancies: A Longitudinal Study. Endocrinology 2019, 160, 249–275. [Google Scholar] [CrossRef] [PubMed]
- Ramos, B.R.A.; Tronco, J.A.; Carvalho, M.; Felix, T.F.; Reis, P.P.; Silveira, J.C.; Silva, M.G. Circulating Extracellular Vesicles microRNAs Are Altered in Women Undergoing Preterm Birth. Int. J. Mol. Sci. 2023, 24, 5527. [Google Scholar] [CrossRef]
- Soobryan, N.; Kumar, A.; Moodley, J.; Mackraj, I. The Role and Expression of Pro/Antiangiogenic Factors and MicroRNAs in Gestational Hypertension and Pre-eclampsia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2023, 290, 38–42. [Google Scholar] [CrossRef]
- Sun, D.-G.; Tian, S.; Zhang, L.; Hu, Y.; Guan, C.-Y.; Ma, X.; Xia, H.-F. The miRNA-29b Is Downregulated in Placenta During Gestational Diabetes Mellitus and May Alter Placenta Development by Regulating Trophoblast Migration and Invasion Through a HIF3A-Dependent Mechanism. Front. Endocrinol. 2020, 11, 169. [Google Scholar] [CrossRef]
- Tian, Q.; Xia, S.; Wu, Y.; Zhang, J.; Wang, L.; Zhu, W. Comprehensive analysis of the differential expression profile of microRNAs in missed abortion. Kaohsiung J. Med. Sci. 2020, 36, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Vonkova, B.; Blahakova, I.; Hruban, L.; Janku, P.; Pospisilova, S. MicroRNA-210 expression during childbirth and postpartum as a potential biomarker of acute fetal hypoxia. Biomed. Pap. 2019, 163, 259–264. [Google Scholar] [CrossRef]
- Winger, E.E.; Reed, J.L.; Ji, X.; Gomez-Lopez, N.; Pacora, P.; Romero, R. MicroRNAs isolated from peripheral blood in the first trimester predict spontaneous preterm birth. PLoS ONE 2020, 15, e0236805. [Google Scholar] [CrossRef]
- Wommack, J.C.; Trzeciakowski, J.P.; Miranda, R.C.; Stowe, R.P.; Ruiz, R.J. Micro RNA clusters in maternal plasma are associated with preterm birth and infant outcomes. PLoS ONE 2018, 13, e0199029. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, L.; Wang, Y. Circulating microRNAs as candidate biomarkers for the ovarian response during in vitro fertilization. Medicine 2021, 100, e24612. [Google Scholar] [CrossRef]
- Burris, H.H.; Gerson, K.D.; Woodward, A.; Redhunt, A.M.; Ledyard, R.; Brennan, K.; Baccarelli, A.A.; Hecht, J.L.; Collier, A.-R.Y.; Hacker, M.R. Cervical microRNA expression and spontaneous preterm birth. Am. J. Obstet. Gynecol. MFM 2023, 5, 100783. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Lee, D.S.; Choong, O.K.; Chang, S.-K.; Hsu, T.; Nicholson, M.W.; Liu, L.-W.; Lin, P.-J.; Ruan, S.-C.; Lin, S.-W.; et al. Cardiac-specific microRNA-125b deficiency induces perinatal death and cardiac hypertrophy. Sci. Rep. 2021, 11, 2377. [Google Scholar] [CrossRef]
- Chen, S.; Pang, D.; Li, Y.; Zhou, J.; Liu, Y.; Yang, S.; Liang, K.; Yu, B. Serum miRNA biomarker discovery for placenta accreta spectrum. Placenta 2020, 101, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Su, X.; Li, X.; Shen, X.; Chen, S.; Lin, X.; Ye, M. Effect and Role of miR-196b in Ectopic Pregnancy. J. Healthc. Eng. 2022, 2022, 7797484. [Google Scholar] [CrossRef]
- Gródecka-Szwajkiewicz, D.; Ulańczyk, Z.; Zagrodnik, E.; Łuczkowska, K.; Rogińska, D.; Kawa, M.P.; Stecewicz, I.; Safranow, K.; Machaliński, B. Differential Secretion of Angiopoietic Factors and Expression of MicroRNA in Umbilical Cord Blood from Healthy Appropriate-For-Gestational-Age Preterm and Term Newborns-in Search of Biomarkers of Angiogenesis-Related Processes in Preterm Birth. Int. J. Mol. Sci. 2020, 21, 1305. [Google Scholar] [CrossRef]
- Sawangpanyangkura, T.; Laohapand, P.; Boriboonhirunsarn, D.; Boriboonhirunsarn, C.; Bunpeng, N.; Tansriratanawong, K. Upregulation of microRNA-223 expression in gingival crevicular blood of women with gestational diabetes mellitus and periodontitis. J. Dent. Sci. 2022, 17, 863–869. [Google Scholar] [CrossRef]
- Cavoretto, P.I.; Farina, A.; Salmeri, N.; Syngelaki, A.; Tan, M.Y.; Nicolaides, K.H. First trimester risk of preeclampsia and rate of spontaneous birth in patients without preeclampsia. Am. J. Obstet. Gynecol. 2024. epub ahead of print. [Google Scholar] [CrossRef]
- Romero, R.; Jung, E.; Chaiworapongsa, T.; Erez, O.; Gudicha, D.W.; Kim, Y.M.; Kim, J.-S.; Kim, B.; Kusanovic, J.P.; Gotsch, F.; et al. Toward a new taxonomy of obstetrical disease: Improved performance of maternal blood biomarkers for the great obstetrical syndromes when classified according to placental pathology. Am. J. Obstet. Gynecol. 2022, 227, 615.e1–615.e25. [Google Scholar] [CrossRef]
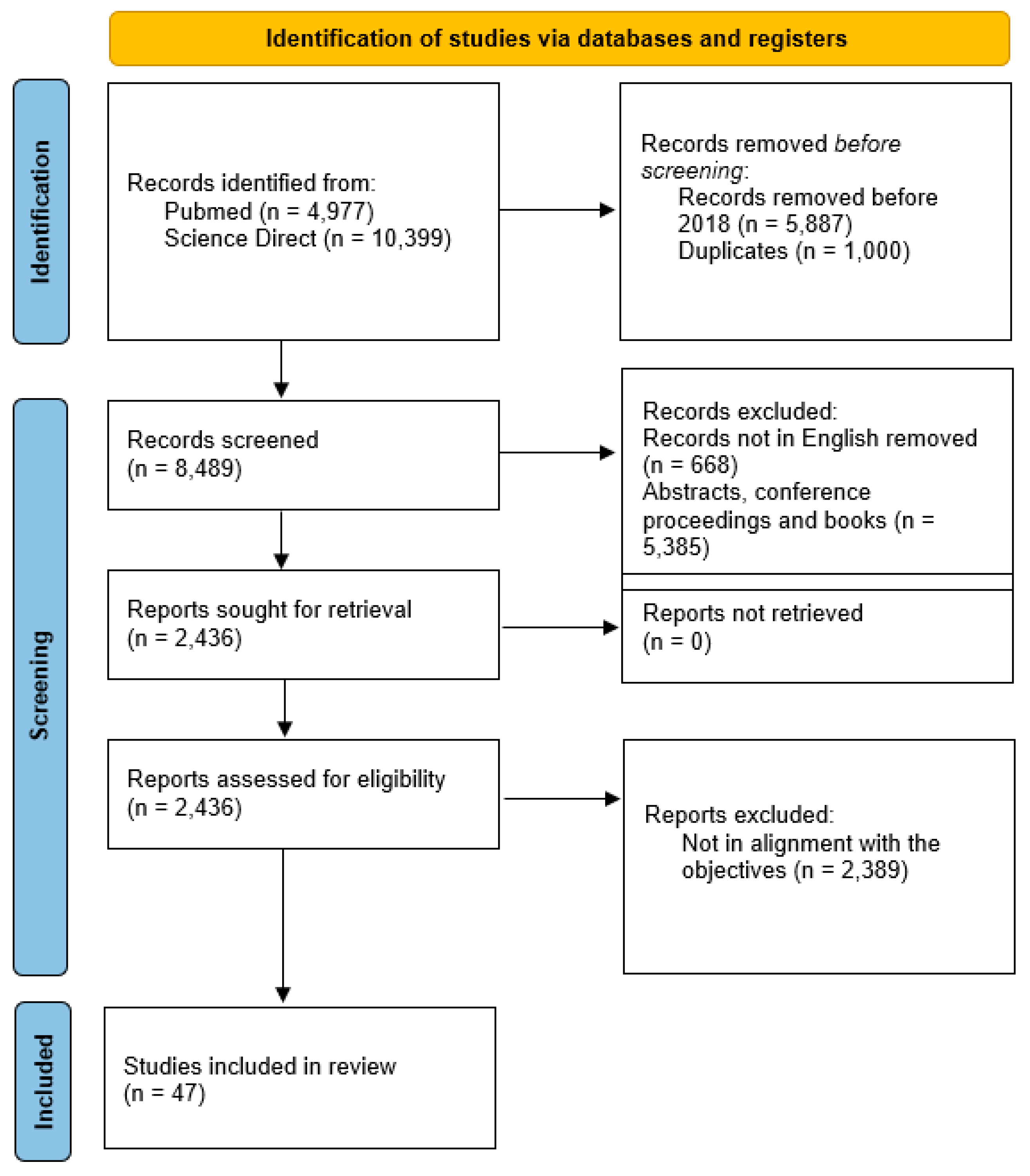
| Category of Study Designs | Responses | ||||
|---|---|---|---|---|---|
| Methodological Quality Criteria | Yes | No | Can’t Tell | ||
| Screening questions (for all types) | S1. Are there clear research questions? | ✔ | |||
| S2. Do the collected data allow me to address the research questions? | ✔ | ||||
| Further appraisal may not be feasible or appropriate when the answer is ‘No’ or ‘Can’t tell’ to one or both screening questions. | |||||
| 1. Qualitative | 1.1. Is the qualitative approach appropriate to answer the research question? | ✔ | [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23] | ||
| 1.2. Are the qualitative data collection methods adequate to address the research question? | ✔ | ||||
| 1.3. Are the findings adequately derived from the data? | ✔ | ||||
| 1.4. Is the interpretation of results sufficiently substantiated by data? | ✔ | ||||
| 1.5. Is there coherence between qualitative data sources, collection, analysis and interpretation? | ✔ | ||||
| 2. Quantitative randomized controlled trials | 2.1. Is randomization appropriately performed? | ✔ | [1,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43] | ||
| 2.2. Are the groups comparable at baseline? | ✔ | ||||
| 2.3. Are there complete outcome data? | ✔ | ||||
| 2.4. Are outcome assessors blinded to the intervention provided? | ✔ | ||||
| 2.5. Did the participants adhere to the assigned intervention? | ✔ | ||||
| 3. Quantitative non-randomized | 3.1. Are the participants representative of the target population? | ✔ | [44,45,46,47,48] | ||
| 3.2. Are measurements appropriate regarding both the outcome and intervention (or exposure)? | ✔ | ||||
| 3.3. Are there complete outcome data? | ✔ | ||||
| 3.4. Are the confounders accounted for in the design and analysis? | |||||
| 3.5. During the study period, is the intervention administered (or exposure occurred) as intended? | ✔ | ||||
| 4. Quantitative descriptive | 4.1. Is the sampling strategy relevant to address the research question? | ✔ | |||
| 4.2. Is the sample representative of the target population? | ✔ | ||||
| 4.3. Are the measurements appropriate? | ✔ | ||||
| 4.4. Is the risk of nonresponse bias low? | ✔ | ||||
| 4.5. Is statistical analysis appropriate to answer the research question? | ✔ | ||||
| 5. Mixed methods | 5.1. Is there an adequate rationale for using a mixed methods design to address the research question? | ||||
| 5.2. Are the different components of the study effectively integrated to answer the research question? | |||||
| 5.3. Are the outputs of the integration of qualitative and quantitative components adequately interpreted? | |||||
| 5.4. Are divergences and inconsistencies between quantitative and qualitative results adequately addressed? | |||||
| Authors & No of. Reference | Year of Publication | Aim | Conclusion |
|---|---|---|---|
| Mavreli et al., [3] | 2018 | Investigation of circulating miRNAs as promising noninvasive biomarkers for pregnancy-related complications. | Highlights the potential significance of circulating miRNAs as prognostic and diagnostic indicators for conditions associated with pregnancy. |
| Hu and Zhang, [10] | 2019 | An exploration of the impact of abnormal miRNA expression on genes related to trophoblast invasion and uteroplacental vascular adaptation in the context of preeclampsia and intrauterine growth restriction (IUGR). | Dysregulated miRNAs play crucial roles in compromising uteroplacental vascular function by targeting numerous genes involved in multiple signal transduction pathways. |
| Paul et al., [18] | 2019 | Recent progress in the exploration of miRNAs in human embryo implantation. | Additional investigation is necessary to explore novel investigative technologies for extracting minute quantities of miRNA and precisely identifying unidentified molecules. |
| Addo et al., [4] | 2020 | Summary of miRNAs investigated in the context of human pregnancy-related disorders and their association with exposure to environmental toxins in the placenta. | Altered expression of miRNAs in both the placenta and maternal plasma has been detected in connection with exposure to environmental chemicals. Their disruption is presumed to play a role in the onset of complications related to pregnancy. |
| Aryan et al., [6] | 2020 | Review recent literature on cardiovascular complications associated with pregnancy, particularly in initially healthy women. Explore the development of cardiac dysfunction and heart failure during and after pregnancy, emphasizing the involvement of microRNAs (miRNAs) in the underlying pathophysiological mechanisms. | Although the available data from various models of heart disease show promise, additional research is required to establish a direct and causal connection between miRNAs and cardiac pathophysiology in the context of cardiovascular complications during pregnancy. This deeper understanding will contribute to enhanced diagnostic methods and the creation of innovative therapeutic approaches. |
| Yang et al., [23] | 2020 | This study focuses on recent research regarding the roles of peripheral blood exosomes and circulating miRNAs in pregnancy complications and abnormal fetal developmental disorders. It particularly highlights the potential application of these exosomes and miRNAs as disease-specific biomarkers. | Advancements in medical science and technology enable the thorough collection of gestational data, aiding in the development of predictive models for pregnancy complications and fetal developmental disorders. |
| Ali et al., [5] | 2021 | A survey of uniquely expressed miRNAs in both the placenta and maternal circulation in cases of preeclampsia (PE) and intrauterine growth restriction (IUGR). These distinct miRNAs have the potential to function as biomarkers, offering promise in the anticipation and identification of complications during pregnancy. | By employing various bioinformatics tools, this investigation revealed potential target genes linked to both PE and IUGR through miRNAs. The study further elucidated the involvement of miRNA-mRNA networks in governing crucial signaling pathways and biological processes. |
| Jin et al., [11] | 2022 | Provide a summary of the existing knowledge regarding the expression patterns and regulatory functions of miRNAs in human placental development and associated diseases. | Identifying specific miRNAs at various gestational stages and within maternal circulation suggests the potential use of miRNAs as biomarkers for monitoring the progression of normal human pregnancy and detecting potential gestational diseases in clinical settings. |
| Juchnicka and Kuzmicki, [12] | 2021 | Examine the key miRNA molecules associated with gestational diabetes. | The alterations in miRNA expression in the blood during hyperglycemia-complicated pregnancy remain unclear, and additional investigations are necessary to establish a miRNA profile for predicting gestational diabetes mellitus (GDM). |
| Li et al., [13] | 2021 | Emphasized the role of miRNAs in overseeing the development and maturation of NK cells, as well as modulating the activation of NK cells and their subsequent activities, including the production of pro- or anti-inflammatory factors during pregnancy. | A more thorough comprehension of the regulatory mechanisms of miRNAs in decidual natural killer (dNK) and peripheral natural killer (pNK) cells, as well as their functional specificities, has the potential to establish a new framework for achieving a balance between immune function and damage at the maternal–fetal interface. |
| Liu et al., [15] | 2021 | Focused on summarizing the pathophysiological significance of miRNAs in the development of gestational diabetes mellitus (GDM) and their potential roles in clinical diagnosis and therapeutic approaches for GDM. | Additional research is required to validate miRNA profiles for the early prediction of gestational diabetes mellitus (GDM). |
| Masete et al., [16] | 2022 | Emphasizes the absence of miRNA profiling in pregnancies affected by T1DM and T2DM, underscoring the necessity for comprehensive miRNA profiling across various forms of maternal diabetes. | Subsequent investigations should prioritize miRNA profiling across various forms of maternal diabetes, aiming to elucidate the mechanisms underlying distinct types of diabetes during pregnancy. |
| Salma et al., [19] | 2022 | Comprehensive overview of existing knowledge and practices in this field, with a focus on physiological changes occurring during pregnancy. | The miRNA biomarker serves as a noninvasive, diagnostic and prognostic indicator for early detection of pregnancy-related cardiac disease and its complications. |
| Shekibi et al., [20] | 2022 | Summarized the key events of the endometrial cycle in humans and mice, followed by a review of the miRNAs identified to date in these two species, emphasizing their probable functional significance in the establishment of receptivity. | Subsequent investigations are necessary to explore the potential of miRNAs as biomarkers and/or therapeutic targets for the detection and enhancement of endometrial receptivity in human fertility treatment. |
| Elhag and Khodor, [8] | 2023 | Summarizes the existing information regarding the dynamics of microRNA (miRNA) during pregnancy, explores their involvement in gestational diabetes mellitus (GDM) and examines their potential applications as targets for diagnosis and therapy. | Altered expression of miRNAs in females with gestational diabetes mellitus (GDM) holds promise as noninvasive biomarkers, contributing to the discovery of fundamental mechanisms associated with gestational diabetes and related pregnancy complications. |
| Fu et al., [9] | 2023 | Examined the latest findings regarding the modulation of embryonic stem cells (ESCs), trophoblast cells and immune cells by microRNA, long non-coding RNA and circular RNA. Explored the potential utility of these non-coding RNAs as diagnostic and therapeutic indicators for pregnancy complications. | Further research should concentrate on exploring the functional aspects and molecular mechanisms of miRNAs, lncRNAs and circRNAs across various phases of pregnancy. |
| Liang et al., [14] | 2023 | Explores the involvement of miRNAs in modulating the function of trophoblast cells and various shared signaling pathways associated with miRNA regulation in pregnancy disorders. | Summarized microRNAs targeting respective mRNAs and governing the biological functions of trophoblasts in pregnancy disorders. These functions encompass trophoblast cell invasion, proliferation, migration, differentiation, apoptosis, autophagy, pyroptosis, ferroptosis, cellular metabolism and angiogenesis, operating through various pathways. |
| Omeljaniuk et al., [17] | 2023 | Summarize the current understanding of the involvement of miRNA molecules in the miscarriage process. | The assessment of potential miRNA molecules’ expression as minimally invasive diagnostic biomarkers can be conducted as early as the initial weeks of pregnancy. |
| Reference | Study Design | Total Number of Participants | Methods | Conclusion | Results |
|---|---|---|---|---|---|
| Cook et al., 2019 [1] | Cohort study | 511 | Detect plasma miRNA biomarkers indicative of preterm birth and/or cervical shortening | The predictive efficacy of the miRNA biomarkers was validated in an independent cohort comprising 96 women with full-term deliveries, 14 with preterm deliveries, and 21 exhibiting early cervical shortening before 20 weeks of gestation. | Significantly higher mean relative expression: hsa-miR-150-5p, hsamiR-374a-5p, hsa-miR-19b-3p, hsa-miR-185-5p, hsa-miR-15b-5p, hsa-miR-191-5p, hsa-miR93-5p, hsa-let-7a-5p and hsa-miR-23a-3p at every time point except hsa-miR-191-5p, hsa-miR93-5p, hsa-let-7a-5p and hsa-miR-23a-3p at 12-14+6 weeks and hsa-miR93-5p at 15-18+6 weeks |
| Gródecka-Szwajkiewicz et al., 2021 [47] | Clinical trials | 79 | Ascertain if there are variations in the profiles of pro-angiogenic and anti-angiogenic factors in umbilical cord blood (UCB) between healthy preterm newborns appropriate for gestational age and term infants | The levels of five out of eight measured pro-angiogenic factors were notably reduced in umbilical cord blood (UCB) from preterm newborns. In contrast, two angiostatic factors showed significant upregulation in preterm UCB. | Healthy preterm newborns vs. term infants: 143 aberrantly expressed miRNAs: 33 miRNAs significantly upregulated (FC values ranged from 2.01 to 3.80) and 110 miRNAs significantly downregulated (FC values ranged from −2.01 to −6.20). Highest expression: has-miR-3135b Most downregulated: has-miR-941 |
| Illarionov et al., 2022 [29] | Prospective study | 24 | miRNA profile in plasma during the initial and subsequent trimesters among pregnant women at a high risk of preterm birth | The miRNA profile in plasma during early pregnancy has the potential to forecast a heightened risk of preterm birth. This holds significance in the proactive prevention of gestational complications. | First trimester: Upregulated: hsa-miR-122-5p, hsa-miR-34a-5p, hsa-miR-34c-5p. Downregulated: hsa-miR-487b-3p, hsa-miR-493-3p, hsa-miR-432-5p, hsa-miR-323b-3p, hsa-miR-369-3p, hsa-miR-134-5p, hsa-miR-431-5p, hsa-miR-485-5p, hsa-miR-382-5p, hsamiR-369-5p, hsa-miR-485-3p, hsa-miR-127-3p) Second trimester: no differentially expressed miRNAs |
| Mavreli et al., 2022 [33] | Case-control study | 68 | Identify differentially expressed miRNAs in maternal plasma during the first trimester, aiming to pinpoint predictive biomarkers for spontaneous preterm delivery (sPTD) and enable timely interventions for this significant pregnancy complication | Statistical analysis showed that miR-125a is an independent early predictor for sPTL, offering a basis for developing a non-invasive test to assist clinicians in estimating patient-specific risk. | Downregulation: hsa-miR-23b-5p, hsa-miR-125a-3p |
| Menon et al., 2019 [34] | Cohort study | 30 | Detail the alterations in exosomal miRNA concentrations present in maternal plasma between mothers delivering term and preterm neonates throughout gestation, employing a longitudinal study design | The miRNA composition within circulating exosomes in maternal blood could potentially serve as a distinctive biomolecular fingerprint indicative of the progression of pregnancy. | A total of 173 miRNAs were found to significantly change across gestation for normal compared with PTB pregnancies. The differences in the miRNA profile targeted signaling pathways associated with TGF-b, p53 and glucocorticoid receptor signaling. |
| Ramos et al., 2023 [35] | Cohort study | 31 | Compare the expression of miRNAs in small extracellular vesicles (sEVs) from peripheral blood between term and preterm pregnancies | miRNAs derived from circulating sEVs exhibit distinct expression patterns in both term and preterm pregnancies. These miRNAs play a regulatory role in modulating genes within pathways that are pertinent to the pathogenesis of preterm labor (PTL) and preterm premature rupture of membranes (PPROM). | Higher expression: hsa-miR-612 |
| Subramanian et al., 2023 [21] | Systematic review | 555 | Summarize literature, compile first-trimester circulating miRNAs tied to placental pregnancy complications and pinpointed top evidence-backed miRNAs as potential early biomarkers | Early detection and interventions in pregnancy complications can be facilitated through first-trimester biomarkers. | Higher expression: hsa-miR-374a-5p and hsa-miR-191-5p -replicated by 3 studies |
| Winger et al., 2020 [40] | Retrospective nested case-control study | 486 | Forecast spontaneous preterm birth in pregnant women within an African-American population by utilizing maternal immune cell microRNAs obtained in the first trimester from peripheral blood | Quantification of microRNA levels in peripheral blood during the first trimester enables the assessment of spontaneous preterm birth risk in both early and late samples of the first trimester of pregnancy within an African-American population. | 12 microRNAs identify preterm birth risk in the first trimester: hsa-miR-181a-3p, hsa-miR-221-3p, hsa-miR-33a-5p, hsa-miR-6752-3p, hsa-miR-1244, hsa-miR-148a-3p, hsa-miR-1-3p, hsa-miR-1267, hsa-miR-223-5p, hsa-miR-199b-5p, hsa-miR-133b, hsa-miR-144-3p; hsa-miR-4485-5p, hsa-miR-340-5p, hsa-miR-132-3p, hsa-miR-219-5p also significant * no statement if up- or downregulated |
| Wommack et al., 2018 [41] | Population- based study | 515 | Explore how pregnancy-specific miRNA relates to gestation length and birth outcomes and Investigate if variations in coordinated circulating miRNA expression are linked to preterm birth (PTB) | Highlighting the necessity for additional research is crucial to elucidate the connections between circulating miRNAs and adverse pregnancy outcomes. | Positive correlation: hsa-miR-337-3p, hsa-miR-127-3p, hsa-miR-136-5p, hsa-miR-323a-3p, hsa-miR-543, hsa-miR-495-3p, hsa-miR-376c-3p, hsa-miR-376a-5p, hsa-miR-496 |
| Burris et al., 2023 [43] | Prospective, nested, case-control study | 74 | Investigate the correlations between cervical microRNA expression and spontaneous preterm birth and to pinpoint a subset of microRNAs that can serve as predictive indicators for spontaneous preterm birth. | Researchers detected a rise in global microRNA expression and the upregulation of 95 unique microRNAs linked to subsequent spontaneous preterm birth. | There were 95 miRNAs associated with PTB: * all were upregulated * there were no significantly downregulated miRNAs 20 more highly expressed: hsa-miR-145, hsa-miR-1291, hsa-miR-30a-5p, hsa-miR-30d, hsa-miR-29a, hsa-miR-7, hsa-miR-942, hsa-miR-28-3p, hsa-miR-30e-3p, hsa-miR-23b, hsa-miR-542-3p, hsa-miR-29b, hsa-miR-21, hsa-miR-130b, hsa-miR-28, hsa-miR-425-5p, hsa-miR-181c, hsa-miR-9, hsa-miR-342-3p, hsa-miR-199b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondracka, A.; Stupak, A.; Rybak-Krzyszkowska, M.; Kondracki, B.; Oniszczuk, A.; Kwaśniewska, A. MicroRNA Associations with Preterm Labor—A Systematic Review. Int. J. Mol. Sci. 2024, 25, 3755. https://doi.org/10.3390/ijms25073755
Kondracka A, Stupak A, Rybak-Krzyszkowska M, Kondracki B, Oniszczuk A, Kwaśniewska A. MicroRNA Associations with Preterm Labor—A Systematic Review. International Journal of Molecular Sciences. 2024; 25(7):3755. https://doi.org/10.3390/ijms25073755
Chicago/Turabian StyleKondracka, Adrianna, Aleksandra Stupak, Magda Rybak-Krzyszkowska, Bartosz Kondracki, Anna Oniszczuk, and Anna Kwaśniewska. 2024. "MicroRNA Associations with Preterm Labor—A Systematic Review" International Journal of Molecular Sciences 25, no. 7: 3755. https://doi.org/10.3390/ijms25073755
APA StyleKondracka, A., Stupak, A., Rybak-Krzyszkowska, M., Kondracki, B., Oniszczuk, A., & Kwaśniewska, A. (2024). MicroRNA Associations with Preterm Labor—A Systematic Review. International Journal of Molecular Sciences, 25(7), 3755. https://doi.org/10.3390/ijms25073755







