Proteomic Analysis of Salivary Extracellular Vesicles from COVID-19 Patients Reveals a Specific Anti-COVID-19 Response Protein Signature
Abstract
1. Introduction
2. Results
2.1. Demographic and Clinical Variables
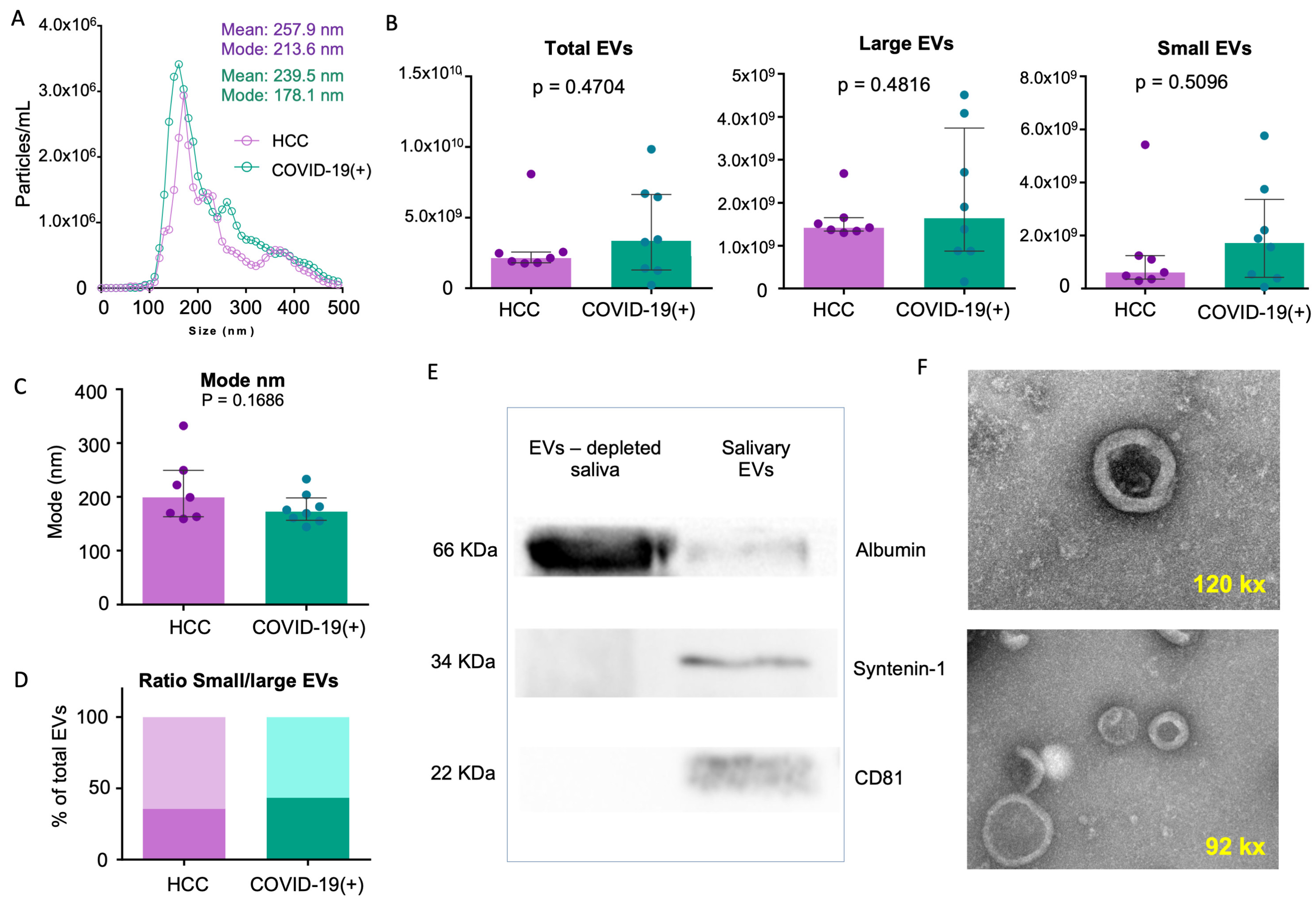
2.2. sEV Characterization
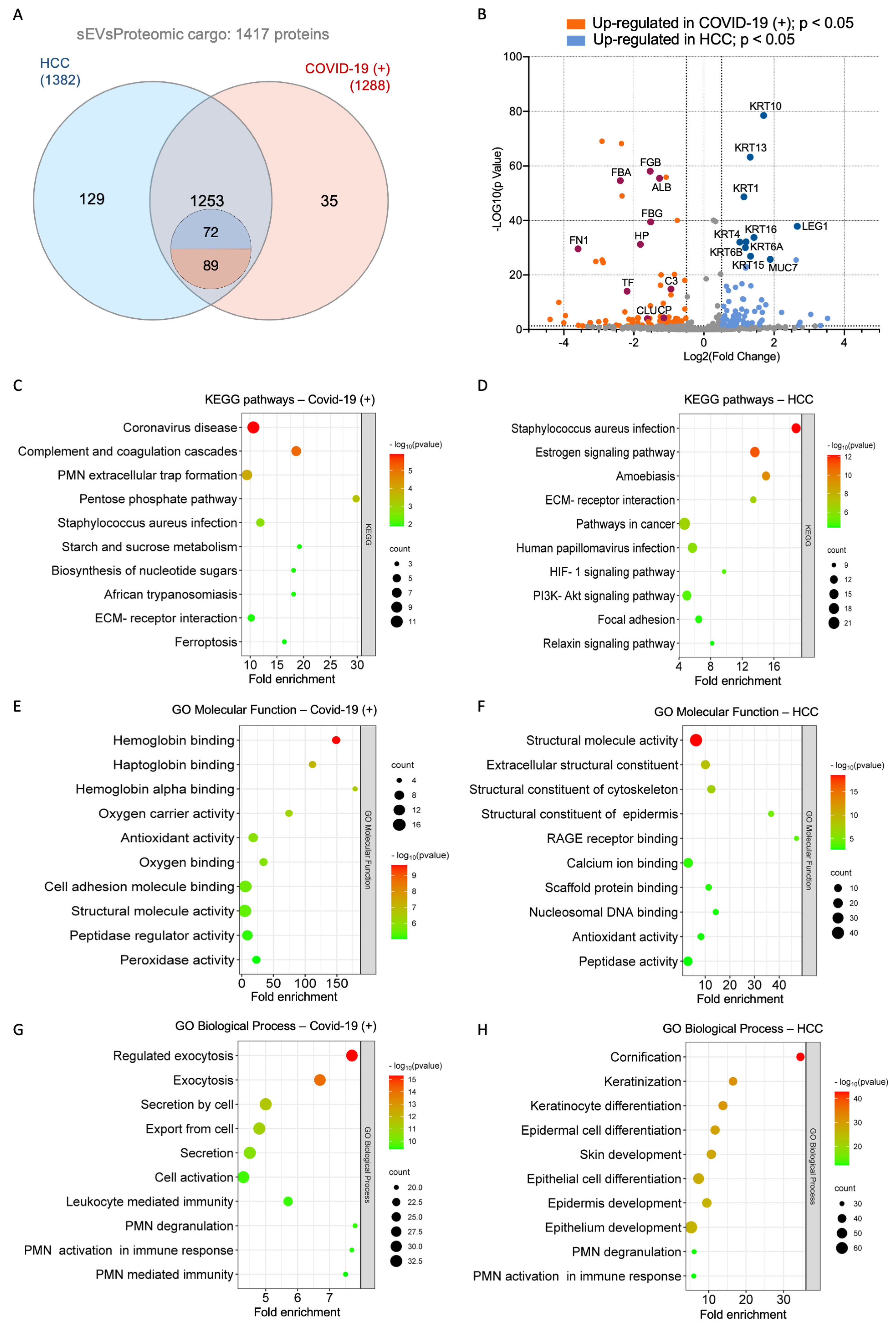
2.3. Proteome Identification
2.4. Functional Enrichment Analysis of Differentially Expressed Proteins
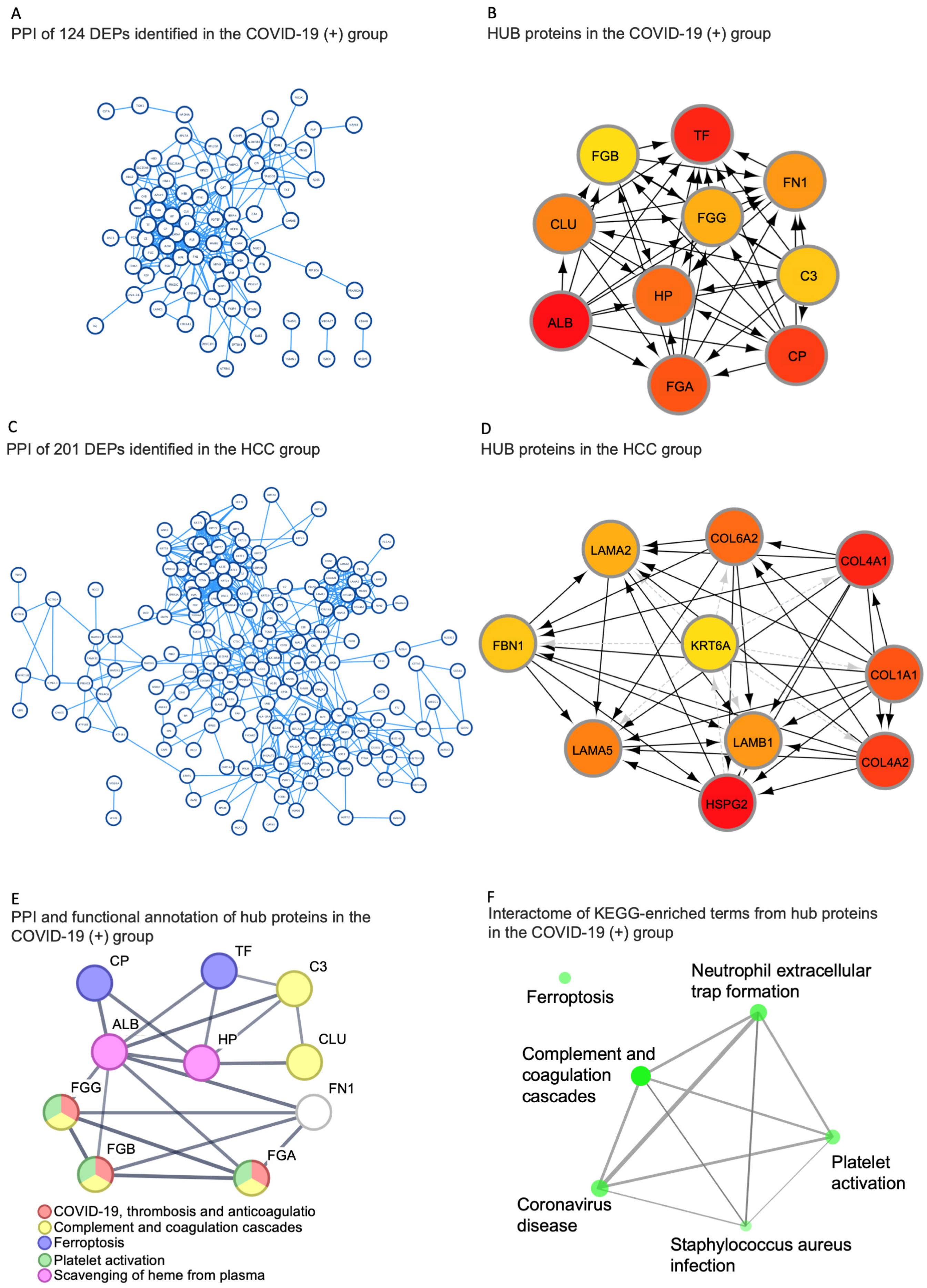
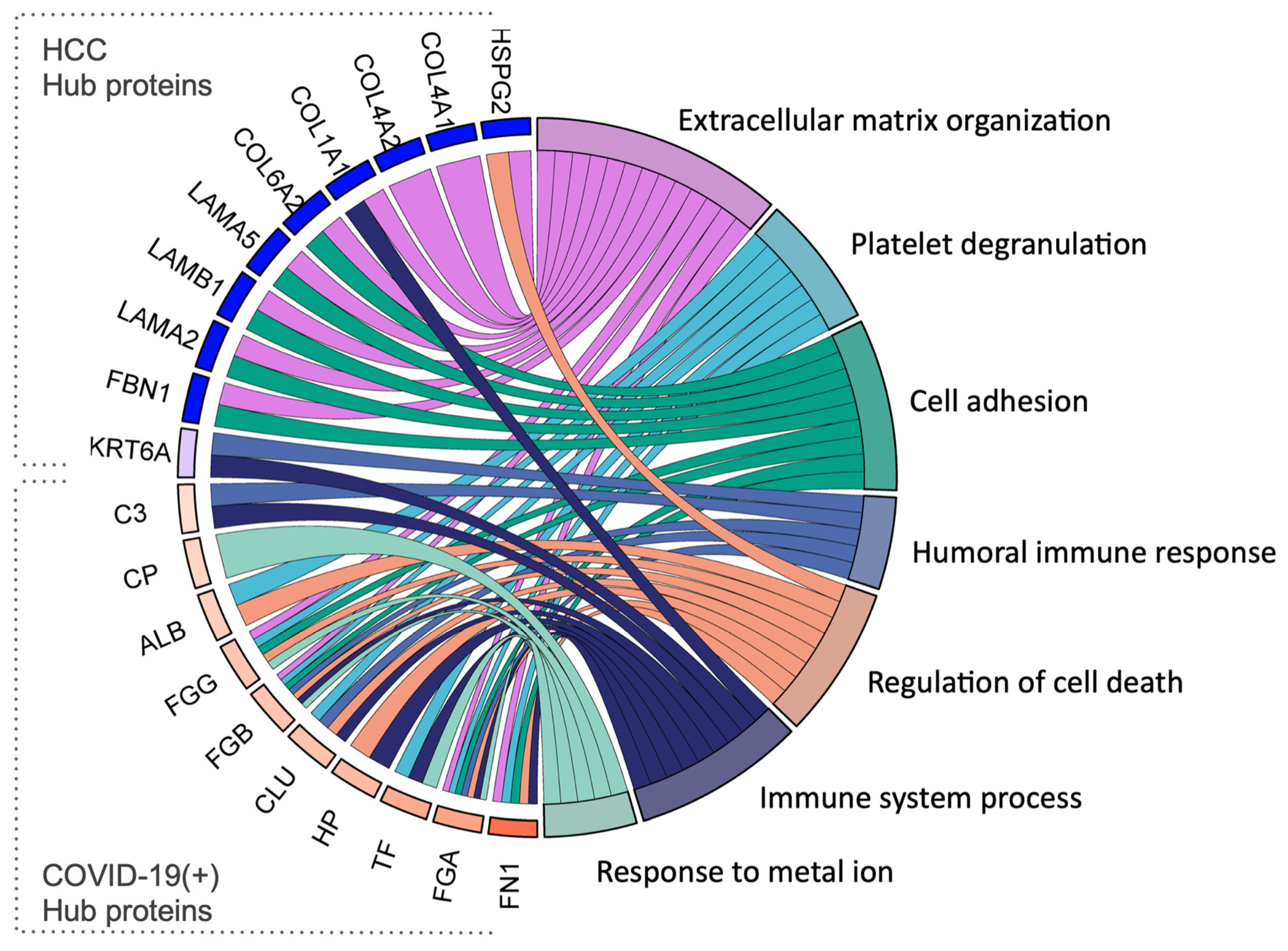
2.5. Identification of Hub Proteins in the Key Modules
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Saliva Samples Collection
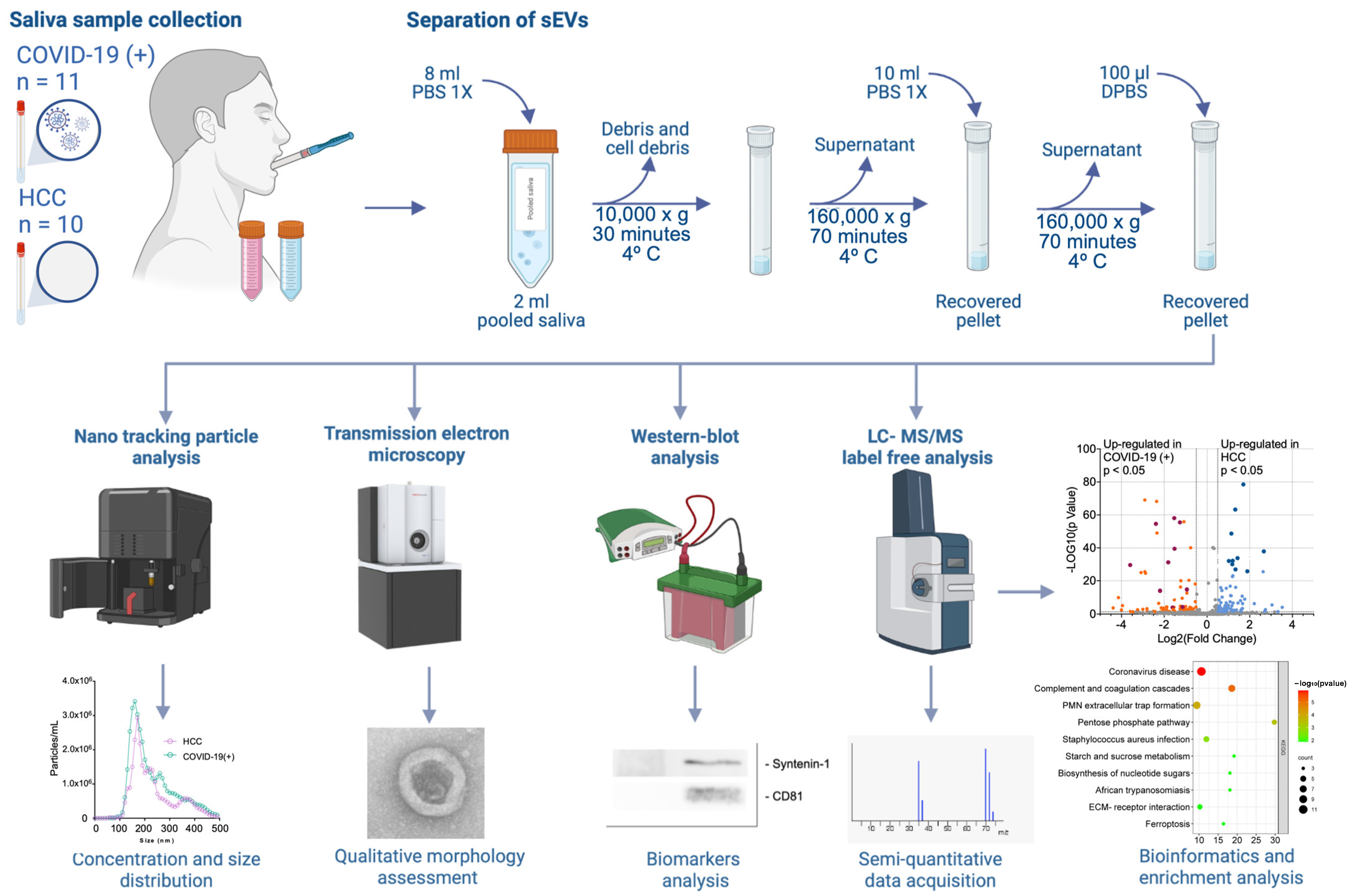
4.3. Separation of sEVs
4.4. Nano-Tracking Particle Analysis
4.5. Western Blot (WB) Analysis
4.6. Transmission Electron Microscopy (TEM)
4.7. Protein Extraction from sEVs
4.8. Proteolytic Digestion
4.9. Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS) Analysis
4.10. Protein Identification
4.11. Protein Quantification by Label-Free Quantification
4.12. Functional Enrichment Analysis of Differentially Expressed Proteins
4.13. Identification of Hub Proteins in the Key Modules
4.14. Data and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grant, M.C.; Geoghegan, L.; Arbyn, M.; Mohammed, Z.; McGuinness, L.; Clarke, E.L.; Wade, R.G. The Prevalence of Symptoms in 24,410 Adults Infected by the Novel Coronavirus (SARS-CoV-2; COVID-19): A Systematic Review and Meta-Analysis of 148 Studies from 9 Countries. PLoS ONE 2020, 15, e0234765. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.B.; June, C.H. Cytokine Release Syndrome in Severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A Novel Coronavirus Outbreak of Global Health Concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Matson, M.J.; Yinda, C.K.; Seifert, S.N.; Bushmaker, T.; Fischer, R.J.; van Doremalen, N.; Lloyd-Smith, J.O.; Munster, V.J. Effect of Environmental Conditions on SARS-CoV-2 Stability in Human Nasal Mucus and Sputum. Emerg. Infect. Dis. 2020, 26, 2276–2278. [Google Scholar] [CrossRef] [PubMed]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva Is a Reliable Tool to Detect SARS-CoV-2. J. Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Fujisawa, S.; Nakakubo, S.; Kamada, K.; Yamashita, Y.; Fukumoto, T.; Sato, K.; Oguri, S.; Taki, K.; Senjo, H.; et al. Comparison of SARS-CoV-2 Detection in Nasopharyngeal Swab and Saliva. J. Infect. 2020, 81, e145–e147. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High Expression of ACE2 Receptor of 2019-NCoV on the Epithelial Cells of Oral Mucosa. Int. J. Oral Sci. 2020, 12, 8. [Google Scholar] [CrossRef]
- Kermali, M.; Khalsa, R.K.; Pillai, K.; Ismail, Z.; Harky, A. The Role of Biomarkers in Diagnosis of COVID-19—A Systematic Review. Life Sci. 2020, 254, 117788. [Google Scholar] [CrossRef]
- Chiappin, S.; Antonelli, G.; Gatti, R.; De Palo, E.F. Saliva Specimen: A New Laboratory Tool for Diagnostic and Basic Investigation. Clin. Chim. Acta 2007, 383, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Lamy, E.; Mau, M. Saliva Proteomics as an Emerging, Non-Invasive Tool to Study Livestock Physiology, Nutrition and Diseases. J. Proteom. 2012, 75, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, G.H. The Secretion, Components, and Properties of Saliva. Annu. Rev. Food Sci. Technol. 2013, 4, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Apweiler, R.; Balgley, B.M.; Boontheung, P.; Bundy, J.L.; Cargile, B.J.; Cole, S.; Fang, X.; Gonzalez-Begne, M.; Griffin, T.J.; et al. Systematic Comparison of the Human Saliva and Plasma Proteomes. Proteom. Clin. Appl. 2009, 3, 116–134. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.B.; Botchway, C.A. Circadian Variations in the Flow Rate and Composition of Whole Saliva Stimulated by Mastication. Arch. Oral Biol. 1979, 24, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Mackie, D.A.; Pangborn, R.M. Mastication and Its Influence on Human Salivary Flow and Alpha-Amylase Secretion. Physiol. Behav. 1990, 47, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Bandhakavi, S.; Stone, M.D.; Onsongo, G.; Van Riper, S.K.; Griffin, T.J. A Dynamic Range Compression and Three-Dimensional Peptide Fractionation Analysis Platform Expands Proteome Coverage and the Diagnostic Potential of Whole Saliva. J. Proteome Res. 2009, 8, 5590–5600. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell. Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes—Vesicular Carriers for Intercellular Communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Palanisamy, V.; Sharma, S.; Deshpande, A.; Zhou, H.; Gimzewski, J.; Wong, D.T. Nanostructural and Transcriptomic Analyses of Human Saliva Derived Exosomes. PLoS ONE 2010, 5, e8577. [Google Scholar] [CrossRef]
- Ogawa, Y.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yanoshita, R. Exosome-like Vesicles with Dipeptidyl Peptidase IV in Human Saliva. Biol. Pharm. Bull. 2008, 31, 1059–1062. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Miura, Y.; Harazono, A.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yamaguchi, T.; Toda, T.; Endo, T.; Tsubuki, M.; et al. Proteomic Analysis of Two Types of Exosomes in Human Whole Saliva. Biol. Pharm. Bull. 2011, 34, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane Vesicles as Conveyors of Immune Responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I.; György, B.; Nagy, G.; Falus, A.; Gay, S. Emerging Role of Extracellular Vesicles in Inflammatory Diseases. Nat. Rev. Rheumatol. 2014, 10, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Reiner, A.T.; Witwer, K.W.; van Balkom, B.W.M.; de Beer, J.; Brodie, C.; Corteling, R.L.; Gabrielsson, S.; Gimona, M.; Ibrahim, A.G.; de Kleijn, D.; et al. Concise Review: Developing Best-Practice Models for the Therapeutic Use of Extracellular Vesicles. Stem. Cells Transl. Med. 2017, 6, 1730–1739. [Google Scholar] [CrossRef] [PubMed]
- Demichev, V.; Tober-Lau, P.; Lemke, O.; Nazarenko, T.; Thibeault, C.; Whitwell, H.; Röhl, A.; Freiwald, A.; Szyrwiel, L.; Ludwig, D.; et al. A Time-Resolved Proteomic and Prognostic Map of COVID-19. Cell Syst. 2021, 12, 780–794.e7. [Google Scholar] [CrossRef] [PubMed]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.-S.; Kreidl, M.; et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020, 11, 11–24.e4. [Google Scholar] [CrossRef]
- Haas, P.; Muralidharan, M.; Krogan, N.J.; Kaake, R.M.; Hüttenhain, R. Proteomic Approaches to Study SARS-CoV-2 Biology and COVID-19 Pathology. J. Proteome Res. 2021, 20, 1133–1152. [Google Scholar] [CrossRef]
- McArdle, A.; Washington, K.E.; Chazarin Orgel, B.; Binek, A.; Manalo, D.-M.; Rivas, A.; Ayres, M.; Pandey, R.; Phebus, C.; Raedschelders, K.; et al. Discovery Proteomics for COVID-19: Where We Are Now. J. Proteome Res. 2021, 20, 4627–4639. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, C.M.; Motta, L.C.; Folli, G.S.; Marcarini, W.D.; Costa, C.A.; Vilela, A.C.S.; Barauna, V.G.; Martin, F.L.; Singh, M.N.; Campos, L.C.G.; et al. MALDI(+) FT-ICR Mass Spectrometry (MS) Combined with Machine Learning toward Saliva-Based Diagnostic Screening for COVID-19. J. Proteome Res. 2022, 21, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Esteves, E.; Mendes, V.M.; Manadas, B.; Lopes, R.; Bernardino, L.; Correia, M.J.; Barros, M.; Esteves, A.C.; Rosa, N. COVID-19 Salivary Protein Profile: Unravelling Molecular Aspects of SARS-CoV-2 Infection. J. Clin. Med. 2022, 11, 5571. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Prieto, A.; Rubić, I.; Gonzalez-Sanchez, J.C.; Kuleš, J.; Martínez-Subiela, S.; Cerón, J.J.; Bernal, E.; Torres-Cantero, A.; Vicente-Romero, M.R.; Mrljak, V.; et al. Saliva Changes in Composition Associated to COVID-19: A Preliminary Study. Sci. Rep. 2022, 12, 10879. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Byappanahalli, A.M.; Vannoy, M.; Omoniyi, V.; Evans, M.K. Influences of Age, Race, and Sex on Extracellular Vesicle Characteristics. Theranostics 2022, 12, 4459–4476. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rasool, H.I.; Palanisamy, V.; Mathisen, C.; Schmidt, M.; Wong, D.T.; Gimzewski, J.K. Structural-Mechanical Characterization of Nanoparticle Exosomes in Human Saliva, Using Correlative AFM, FESEM, and Force Spectroscopy. ACS Nano 2010, 4, 1921–1926. [Google Scholar] [CrossRef] [PubMed]
- Zlotogorski-Hurvitz, A.; Dayan, D.; Chaushu, G.; Korvala, J.; Salo, T.; Sormunen, R.; Vered, M. Human Saliva-Derived Exosomes. J. Histochem. Cytochem. 2015, 63, 181–189. [Google Scholar] [CrossRef]
- Chaparro Padilla, A.; Weber Aracena, L.; Realini Fuentes, O.; Albers Busquetts, D.; Hernández Ríos, M.; Ramírez Lobos, V.; Pascual La Rocca, A.; Nart Molina, J.; Beltrán Varas, V.; Acuña-Gallardo, S.; et al. Molecular Signatures of Extracellular Vesicles in Oral Fluids of Periodontitis Patients. Oral Dis. 2020, 26, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Giannobile, W.V.; McDevitt, J.T.; Niedbala, R.S.; Malamud, D. Translational and Clinical Applications of Salivary Diagnostics. Adv. Dent. Res. 2011, 23, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, L.J.; Varas-Godoy, M.; Monckeberg, M.; Realini, O.; Hernández, M.; Rice, G.; Romero, R.; Saavedra, J.F.; Illanes, S.E.; Chaparro, A. Oral Extracellular Vesicles in Early Pregnancy Can Identify Patients at Risk of Developing Gestational Diabetes Mellitus. PLoS ONE 2019, 14, e0218616. [Google Scholar] [CrossRef]
- Xylaki, M.; Chopra, A.; Weber, S.; Bartl, M.; Outeiro, T.F.; Mollenhauer, B. Extracellular Vesicles for the Diagnosis of Parkinson’s Disease: Systematic Review and Meta-Analysis. Mov. Disord. 2023, 38, 1585–1597. [Google Scholar] [CrossRef]
- Rolla, R.; Puricelli, C.; Bertoni, A.; Boggio, E.; Gigliotti, C.L.; Chiocchetti, A.; Cappellano, G.; Dianzani, U. Platelets: “Multiple Choice” Effectors in the Immune Response and Their Implication in COVID-19 Thromboinflammatory Process. Int. J. Lab. Hematol. 2021, 43, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Asakura, H.; Ogawa, H. COVID-19-Associated Coagulopathy and Disseminated Intravascular Coagulation. Int. J. Hematol. 2021, 113, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Carsana, L.; Sonzogni, A.; Nasr, A.; Rossi, R.S.; Pellegrinelli, A.; Zerbi, P.; Rech, R.; Colombo, R.; Antinori, S.; Corbellino, M.; et al. Pulmonary Post-Mortem Findings in a Series of COVID-19 Cases from Northern Italy: A Two-Centre Descriptive Study. Lancet Infect. Dis. 2020, 20, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Savla, S.R.; Prabhavalkar, K.S.; Bhatt, L.K. Cytokine Storm Associated Coagulation Complications in COVID-19 Patients: Pathogenesis and Management. Expert Rev. Anti-Infect. Ther. 2021, 19, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Cugno, M.; Meroni, P.L.; Gualtierotti, R.; Griffini, S.; Grovetti, E.; Torri, A.; Panigada, M.; Aliberti, S.; Blasi, F.; Tedesco, F.; et al. Complement Activation in Patients with COVID-19: A Novel Therapeutic Target. J. Allergy Clin. Immunol. 2020, 146, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Zhu, L.; Liu, H.; Zhang, X.; Wang, T.; Fu, Y.; Li, H.; Dong, Q.; Hu, Y.; Zhang, Z.; et al. Highly Pathogenic Coronavirus N Protein Aggravates Inflammation by MASP-2-Mediated Lectin Complement Pathway Overactivation. Signal Transduct. Target. Ther. 2022, 7, 318. [Google Scholar] [CrossRef]
- Risitano, A.M.; Mastellos, D.C.; Huber-Lang, M.; Yancopoulou, D.; Garlanda, C.; Ciceri, F.; Lambris, J.D. Complement as a Target in COVID-19? Nat. Rev. Immunol. 2020, 20, 343–344. [Google Scholar] [CrossRef] [PubMed]
- Skendros, P.; Mitsios, A.; Chrysanthopoulou, A.; Mastellos, D.C.; Metallidis, S.; Rafailidis, P.; Ntinopoulou, M.; Sertaridou, E.; Tsironidou, V.; Tsigalou, C.; et al. Complement and Tissue Factor–Enriched Neutrophil Extracellular Traps Are Key Drivers in COVID-19 Immunothrombosis. J. Clin. Investig. 2020, 130, 6151–6157. [Google Scholar] [CrossRef]
- Stakos, D.; Skendros, P.; Konstantinides, S.; Ritis, K. Traps N’ Clots: NET-Mediated Thrombosis and Related Diseases. Thromb. Haemost. 2020, 120, 373–383. [Google Scholar] [CrossRef]
- Java, A.; Apicelli, A.J.; Liszewski, M.K.; Coler-Reilly, A.; Atkinson, J.P.; Kim, A.H.J.; Kulkarni, H.S. The Complement System in COVID-19: Friend and Foe? JCI Insight 2020, 5, e140711. [Google Scholar] [CrossRef]
- Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Al-hussaniy, H.A.; Al-Harcan, N.A.H.; Alexiou, A.; Batiha, G.E.-S. Neutrophil Extracellular Traps (NETs) and COVID-19: A New Frontiers for Therapeutic Modality. Int. Immunopharmacol. 2022, 104, 108516. [Google Scholar] [CrossRef]
- Bojkova, D.; Costa, R.; Reus, P.; Bechtel, M.; Jaboreck, M.-C.; Olmer, R.; Martin, U.; Ciesek, S.; Michaelis, M.; Cinatl, J. Targeting the Pentose Phosphate Pathway for SARS-CoV-2 Therapy. Metabolites 2021, 11, 699. [Google Scholar] [CrossRef] [PubMed]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.; Krüger, A.; Tauqeer Alam, M.; et al. The Return of Metabolism: Biochemistry and Physiology of the Pentose Phosphate Pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef] [PubMed]
- Beale, D.J.; Shah, R.; Karpe, A.V.; Hillyer, K.E.; McAuley, A.J.; Au, G.G.; Marsh, G.A.; Vasan, S.S. Metabolic Profiling from an Asymptomatic Ferret Model of SARS-CoV-2 Infection. Metabolites 2021, 11, 327. [Google Scholar] [CrossRef]
- Codo, A.C.; Davanzo, G.G.; Monteiro, L.d.B.; de Souza, G.F.; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; de Biagi Junior, C.A.O.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab. 2020, 32, 437–446.e5. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING Database in 2017: Quality-Controlled Protein–Protein Association Networks, Made Broadly Accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. CytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef]
- The UniProt Consortium UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [CrossRef]
- Levin, Y. The Role of Statistical Power Analysis in Quantitative Proteomics. Proteomics 2011, 11, 2565–2567. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Perez-Riverol, Y.; Sharma, V.; Carver, J.J.; Mendoza, L.; Kundu, D.J.; Wang, S.; Bandla, C.; Kamatchinathan, S.; et al. The ProteomeXchange Consortium at 10 Years: 2023 Update. Nucleic Acids Res. 2023, 51, D1539–D1548. [Google Scholar] [CrossRef]
- van Wijk, K.J.; Leppert, T.; Sun, Q.; Boguraev, S.S.; Sun, Z.; Mendoza, L.; Deutsch, E.W. The Arabidopsis PeptideAtlas: Harnessing Worldwide Proteomics Data to Create a Comprehensive Community Proteomics Resource. Plant Cell 2021, 33, 3421–3453. [Google Scholar] [CrossRef]
- Choi, M.; Carver, J.; Chiva, C.; Tzouros, M.; Huang, T.; Tsai, T.-H.; Pullman, B.; Bernhardt, O.M.; Hüttenhain, R.; Teo, G.C.; et al. MassIVE.Quant: A Community Resource of Quantitative Mass Spectrometry–Based Proteomics Datasets. Nat. Methods 2020, 17, 981–984. [Google Scholar] [CrossRef]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. JPOSTrepo: An International Standard Data Repository for Proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Ma, J.; Liu, Y.; Chen, Z.; Xiao, N.; Lu, Y.; Fu, Y.; Yang, C.; Li, M.; Wu, S.; et al. IProX in 2021: Connecting Proteomics Data Sharing with Big Data. Nucleic Acids Res. 2022, 50, D1522–D1527. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Eckels, J.; Schilling, B.; Ludwig, C.; Jaffe, J.D.; MacCoss, M.J.; MacLean, B. Panorama Public: A Public Repository for Quantitative Data Sets Processed in Skyline. Mol. Cell. Proteom. 2018, 17, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
| COVID-19(+) (n = 11) | HCC (n = 10) | Total (n = 21) | |
|---|---|---|---|
| Gender | p = 0.0588 | ||
| Female | 6 (54.5%) 5 (45.5%) | 3 (30%) 7 (70%) | 9 (42.86%) 12 (57.14%) |
| Male | |||
| Age | 31 (19) | 54.5 (22) | 40 (25) |
| Smoking | p = 0.4795 | ||
| No | 8 (72.7%) 3 (27.3%) | 8 (80%) 2 (20%) | 16 (76.2%) 5 (23.8%) |
| Yes | |||
| Comorbidities | p = 0.0000 | ||
| No | 10 (90.9%) 1 (9.1%) | 6 (60%) 4 (40%) | 16 (76.2%) |
| Yes | 5 (23.8%) | ||
| BMI | 24.22 (7.25) | 25.61 (9.47) | 24.8 (6.33) |
| Hub Proteins in the HCC Group | |||
|---|---|---|---|
| Gene Name | Accession | Protein Name | Function (Uniprot) |
| HSPG2 | P98160 | Heparan sulfate proteoglycan core protein | An integral component of basement membranes. Responsible for the fixed negative electrostatic membrane charge. It serves as an attachment substrate for cells. |
| COL4A1 | P02462 | Collagen alpha-1(IV) chain | Type IV collagen is the major structural component of basement membranes, interacting with laminins and proteoglycans |
| COL4A2 | P08572 | Collagen alpha-2(IV) chain | Type IV collagen is the major structural component of basement membranes, interacting with laminins and proteoglycans |
| COL1A1 | P02452 | Collagen alpha-1(I) chain | Type I collagen is a member of the fibrillar-forming collagen. |
| COL6A2 | P12110 | Collagen alpha-2(VI) chain | Collagen VI acts as a cell-binding protein. |
| LAMA5 | O15230 | Laminin subunit alpha-5 | Involved in attachment, migration, and organization of cells by interacting with other matrix components. |
| LAMB1 | P07942 | Laminin subunit beta-1 | Involved in attachment, migration, and organization of cells by interacting with other matrix components. |
| LAMA2 | P24043 | Laminin subunit alpha-2 | Involved in attachment, migration, and organization of cells by interacting with other matrix components. |
| FBN1 | P35555 | Fibrillin-1 | Plays a key role in tissue homeostasis. Structural component of microfibrils of the extracellular matrix, which provide support to load-bearing connective tissues. |
| KRT6A | P02538 | Keratin, type II cytoskeletal 6A | Involved in wound healing. Involved in the activation of follicular keratinocytes after wounding. |
| Hub Proteins in the COVID-19(+) Group | |||
| Gene Name | Accession | Protein Name | Function (Uniprot) |
| ALB | P02768 | Albumin | Binds water, Na+, K+, fatty acids, and hormones. Major zinc, calcium, and magnesium transporter in plasma. |
| TF | P02787 | Serotransferrin | Transport of iron from sites of absorption and heme degradation to those of storage and utilization. |
| CP | P00450 | Ceruloplasmin | Copper-binding glycoprotein with ferroxidase activity. It is involved in iron transport across the cell membrane. |
| HP | P00738 | Haptoglobin | Captures free hemoglobin in plasma to allow hepatic recycling of heme iron and to prevent kidney damage. Also has antioxidant and antibacterial activity and plays a role in modulating the acute phase response. |
| FGA | P02671 | Fibrinogen alpha chain | Together with FGB and FGG, polymerizes to form an insoluble fibrin matrix, acting as one of the primary components of blood clots. |
| FGG | P02679 | Fibrinogen gamma chain | Together with FGA and FGB, polymerizes to form an insoluble fibrin matrix, acting as one of the primary components of blood clots. |
| FGB | P02675 | Fibrinogen beta chain | Together with FGA and FGG, polymerizes to form an insoluble fibrin matrix, acting as one of the primary components of blood clots. |
| CLU | P10909 | Clusterin | Prevents stress-induced aggregation of blood plasma non-native proteins. |
| FN1 | P02751 | Fibronectin | Involved in cell adhesion, cell motility, opsonization, wound healing, and maintenance of cell shape. |
| C3 | P01024 | Complement C3 | C3 plays a central role in the activation of the complement system. It increases vascular permeability and causes histamine release. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weber, L.; Torres, A.; Realini, O.; Bendek, M.J.; Mizgier, M.L.; Brizuela, C.; Herrera, D.; González, F.E.; Chaparro, A. Proteomic Analysis of Salivary Extracellular Vesicles from COVID-19 Patients Reveals a Specific Anti-COVID-19 Response Protein Signature. Int. J. Mol. Sci. 2024, 25, 3704. https://doi.org/10.3390/ijms25073704
Weber L, Torres A, Realini O, Bendek MJ, Mizgier ML, Brizuela C, Herrera D, González FE, Chaparro A. Proteomic Analysis of Salivary Extracellular Vesicles from COVID-19 Patients Reveals a Specific Anti-COVID-19 Response Protein Signature. International Journal of Molecular Sciences. 2024; 25(7):3704. https://doi.org/10.3390/ijms25073704
Chicago/Turabian StyleWeber, Laura, Alfredo Torres, Ornella Realini, María José Bendek, María Luisa Mizgier, Claudia Brizuela, David Herrera, Fermín E. González, and Alejandra Chaparro. 2024. "Proteomic Analysis of Salivary Extracellular Vesicles from COVID-19 Patients Reveals a Specific Anti-COVID-19 Response Protein Signature" International Journal of Molecular Sciences 25, no. 7: 3704. https://doi.org/10.3390/ijms25073704
APA StyleWeber, L., Torres, A., Realini, O., Bendek, M. J., Mizgier, M. L., Brizuela, C., Herrera, D., González, F. E., & Chaparro, A. (2024). Proteomic Analysis of Salivary Extracellular Vesicles from COVID-19 Patients Reveals a Specific Anti-COVID-19 Response Protein Signature. International Journal of Molecular Sciences, 25(7), 3704. https://doi.org/10.3390/ijms25073704









