EBS in Children with De Novo Pathogenic Variants Disturbing Krt14
Abstract
1. Introduction
2. Case Description
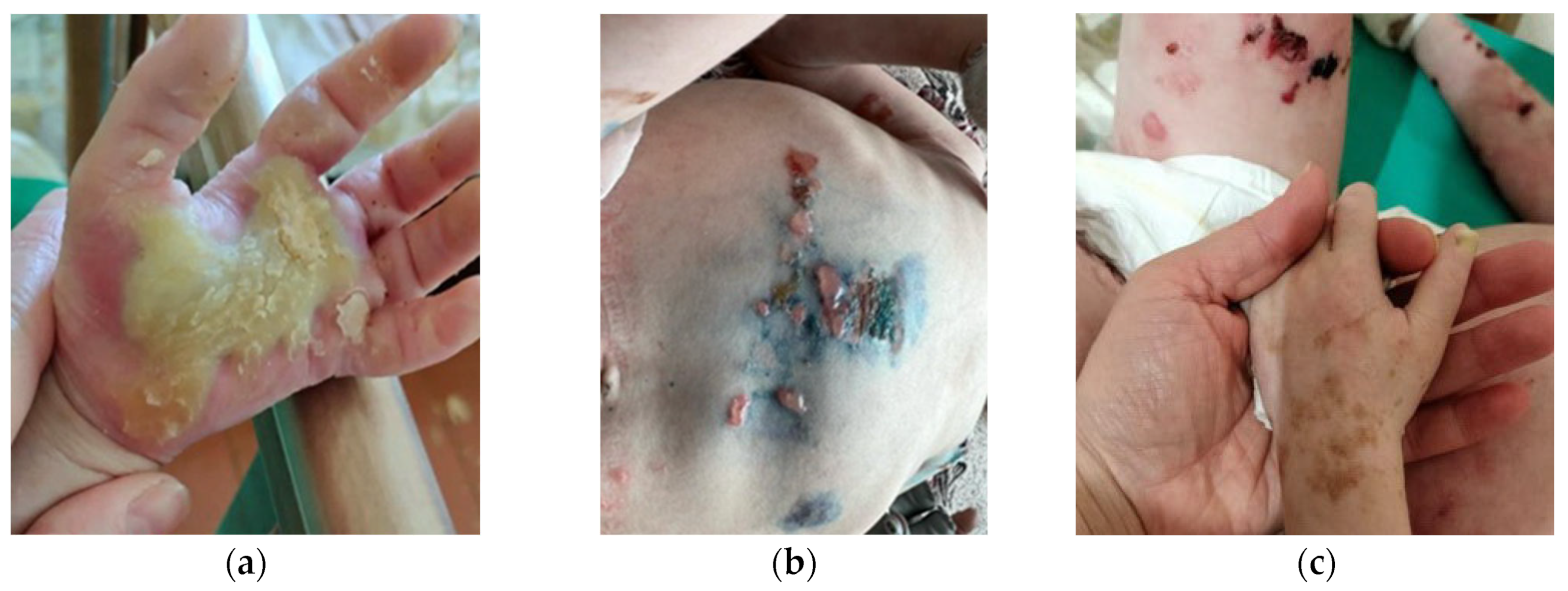
2.1. EBS Severe with Mottled Pigmentation (EBS-MP)
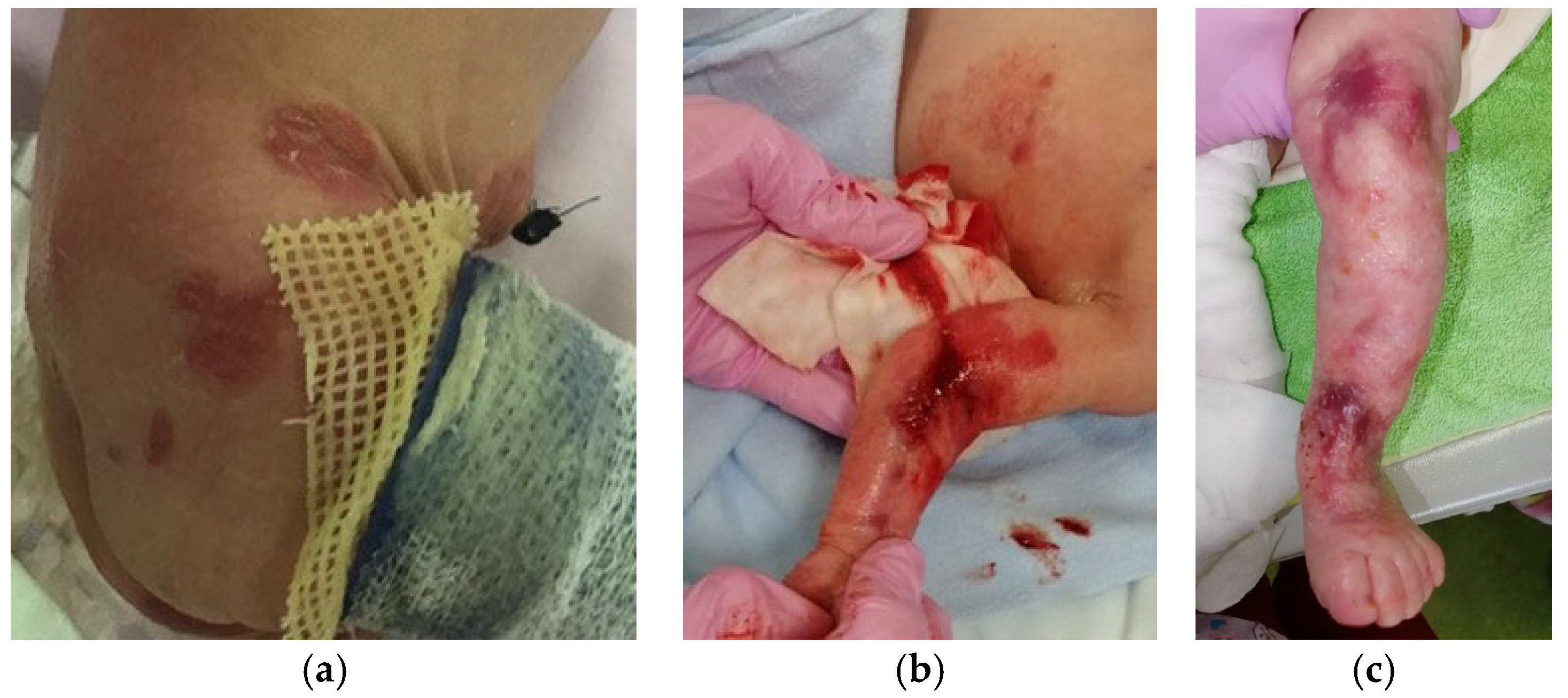
2.2. Epidermolysis Bullosa Simplex, Generalized Intermediate, with or without Cardiomyopathy
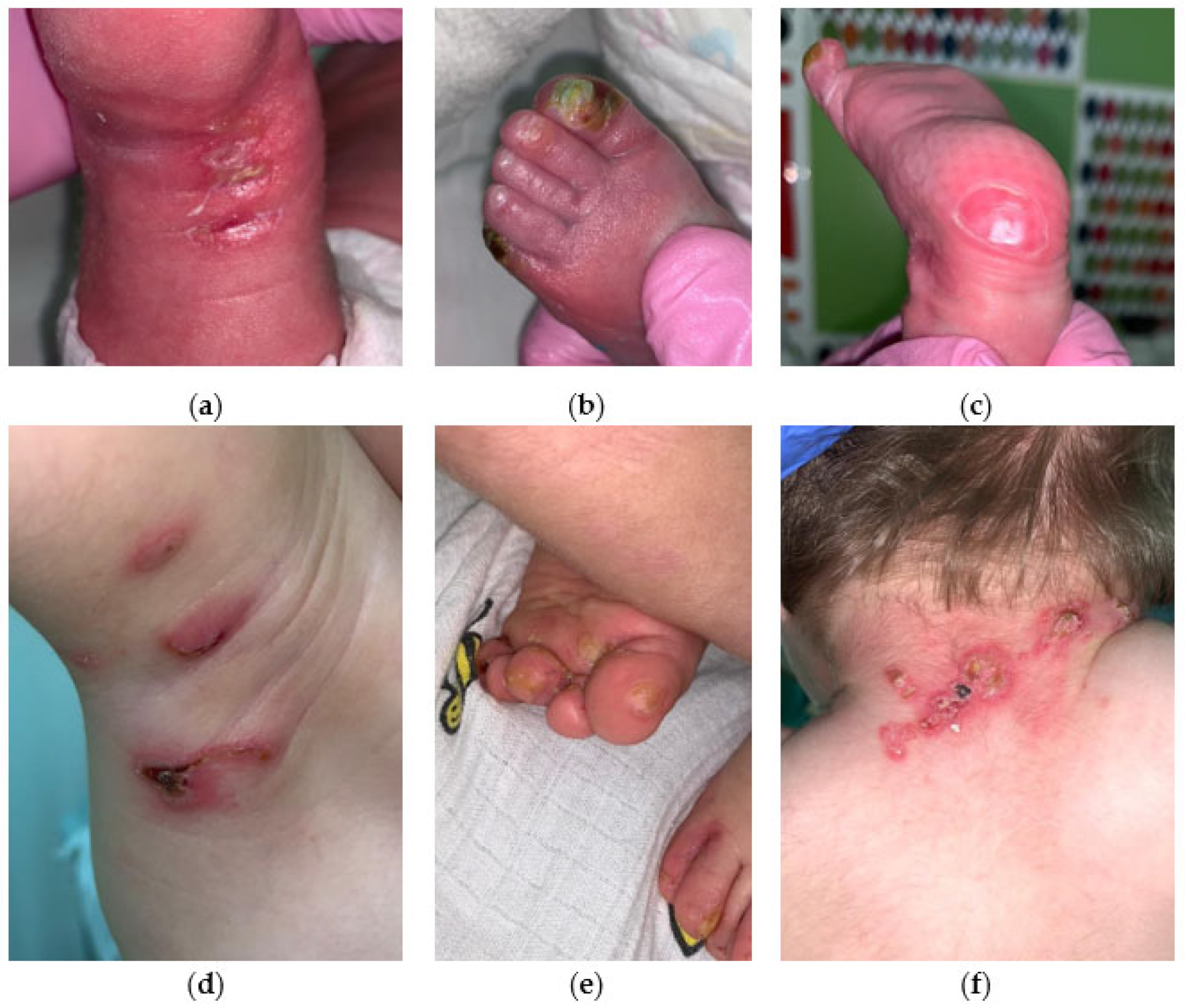
2.3. EBS Severe with Recurrent Arg125His Krt14 Variant
3. Discussion
3.1. The Pathogenic Variants Induced Severe and Intermediate Type of EBS
3.2. Severe EBS-MP
3.3. Intermediate EBS with KLHL24 Mutation
3.4. EBS Severe with Recurrent Arg125His
4. Material and Methods
4.1. Keratinocytes Culture Obtaining
4.2. Obtaining DNA from Human Peripheral Blood Mononuclear Cells (hPBMC)
4.3. Exome Sequencing and Data Analysis
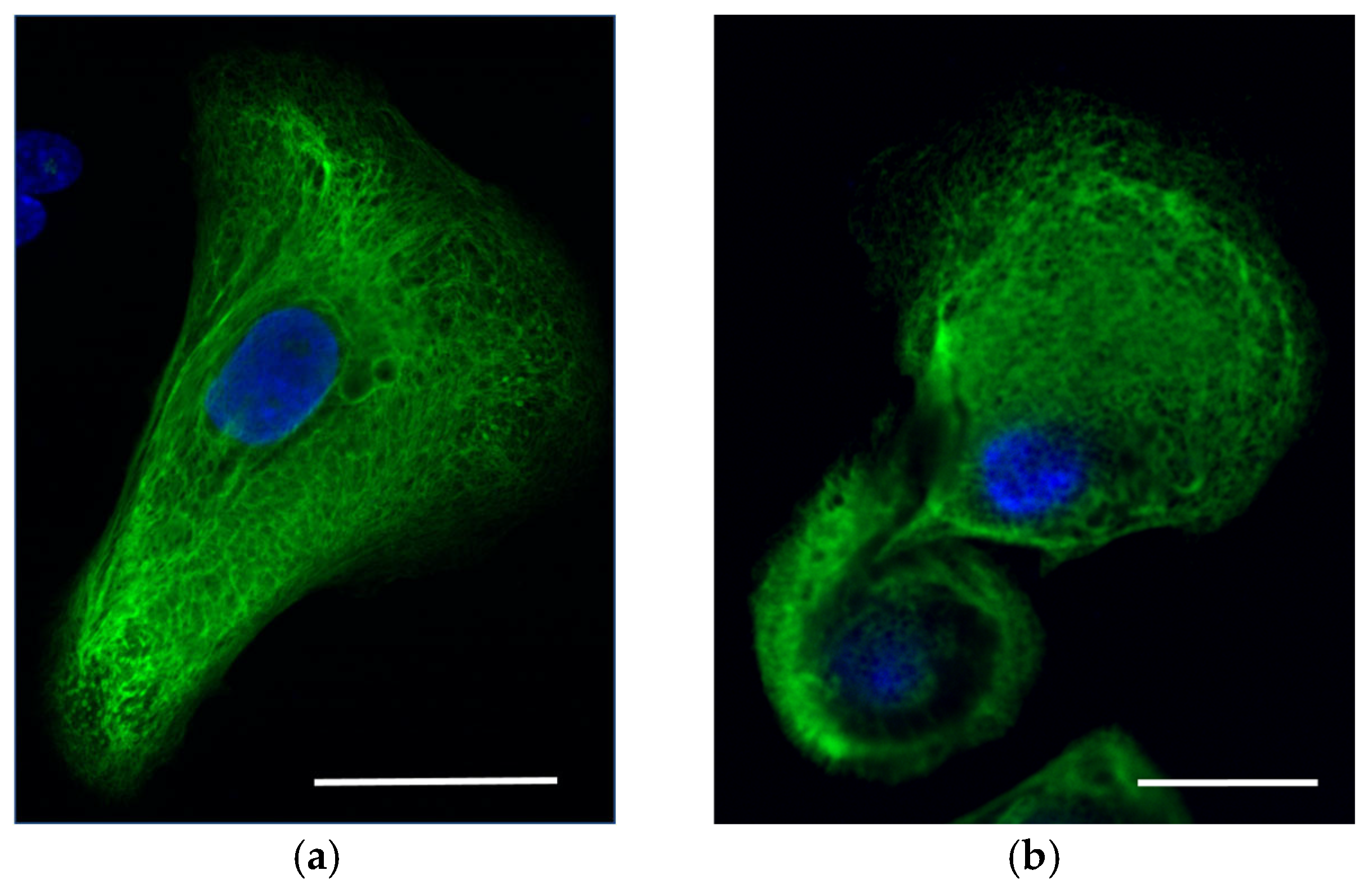
4.4. Immunohistochemistry of KRT14 in Skin Sections
4.5. Confocal Imaging
4.6. Software
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Titeux, M.; Mazereeuw-Hautier, J.; Hadj-Rabia, S.; Prost, C.; Tonasso, L.; Fraitag, S.; Prost, Y.d.; Hovnanian, A.; Bodemer, C. Three Severe Cases of EBS Dowling-Meara Caused by Missense and Frameshift Mutations in the Keratin 14 Gene. J. Investig. Dermatol. 2006, 126, 773–776. [Google Scholar] [CrossRef]
- Harel, A.; Bergman, R.; Indelman, M.; Sprecher, E. Epidermolysis Bullosa Simplex with Mottled Pigmentation Resulting from a Recurrent Mutation in KRT14. J. Investig. Dermatol. 2006, 126, 1654–1657. [Google Scholar] [CrossRef]
- Lin, Z.; Li, S.; Feng, C.; Yang, S.; Wang, H.; Ma, D.; Zhang, J.; Gou, M.; Bu, D.; Zhang, T.; et al. Stabilizing Mutations of KLHL24 Ubiquitin Ligase Cause Loss of Keratin 14 and Human Skin Fragility. Nat. Genet. 2016, 48, 1508–1516. [Google Scholar] [CrossRef]
- Wilson, N.J.; Cole, C.; Kroboth, K.; Hunter, W.N.; Mann, J.A.; McLean, W.H.I.; Kernland Lang, K.; Beltraminelli, H.; Sabroe, R.A.; Tiffin, N.; et al. Mutations in POGLUT1 in Galli–Galli/Dowling–Degos Disease. Br. J. Dermatol. 2017, 176, 270–274. [Google Scholar] [CrossRef]
- Ralser, D.J.; Takeuchi, H.; Fritz, G.; Basmanav, F.B.; Effern, M.; Sivalingam, S.; El-Shabrawi-Caelen, L.; Degirmentepe, E.N.; Kocatürk, E.; Singh, M.; et al. Altered Notch Signaling in Dowling-Degos Disease: Additional Mutations in POGLUT1 and Further Insights into Disease Pathogenesis. J. Investig. Dermatol. 2019, 139, 960–964. [Google Scholar] [CrossRef]
- Shemanko, C.S.; Mellerio, J.E.; Tidman, M.J.; Lane, E.B.; Eady, R.A. Severe palmo-plantar hyperkeratosis in Dowling-Meara epidermolysis bullosa simplex caused by a mutation in the keratin 14 gene (KRT14). J. Investig. Dermatol. 1998, 111, 893–895. [Google Scholar] [CrossRef][Green Version]
- Bolling, M.C.; Lemmink, H.H.; Jansen, G.H.L.; Jonkman, M.F. Mutations in KRT5 and KRT14 Cause Epidermolysis Bullosa Simplex in 75% of the Patients. Br. J. Dermatol. 2011, 164, 637–644. [Google Scholar] [CrossRef]
- Jerábková, B.; Marek, J.; Bucková, H.; Kopecková, L.; Veselý, K.; Valícková, J.; Fajkus, J.; Fajkusová, L. Keratin mutations in patients with epidermolysis bullosa simplex: Correlations between phenotype severity and disturbance of intermediate filament molecular structure. Br. J. Dermatol. 2010, 162, 1004–1013. [Google Scholar] [CrossRef]
- Lane, E.B.; McLean, W.H. Keratins and skin disorders. J. Pathol. 2004, 204, 355–366. [Google Scholar] [CrossRef]
- Müller, F.B.; Küster, W.; Wodecki, K.; Almeida, H.; Bruckner-Tuderman, L.; Krieg, T.; Korge, B.P.; Arin, M.J. Novel and Recurrent Mutations in Keratin KRT5 and KRT14 Genes in Epidermolysis Bullosa Simplex: Implications for Disease Phenotype and Keratin Filament Assembly. Hum. Mutat. 2006, 27, 719–720. [Google Scholar] [CrossRef]
- Shemanko, C.S.; Horn, H.M.; Keohane, S.G.; Hepburn, N.; Kerr, A.I.; Atherton, D.J.; Tidman, M.J.; Lane, E.B. Laryngeal Involvement in the Dowling-Meara Variant of Epidermolysis Bullosa Simplex with Keratin Mutations of Severely Disruptive Potential. Br. J. Dermatol. 2000, 142, 315–320. [Google Scholar] [CrossRef]
- Coulombe, P.A.; Hutton, M.E.; Letai, A.; Hebert, A.; Paller, A.S.; Fuchs, E. Point Mutations in Human Keratin 14 Genes of Epidermolysis Bullosa Simplex Patients: Genetic and Functional Analyses. Cell 1991, 66, 1301–1311. [Google Scholar] [CrossRef]
- Kim, E.N.; Harris, A.G.; Bingham, L.J.; Yan, W.; Su, J.C.; Murrell, D.F. A Review of 52 Pedigrees with Epidermolysis Bullosa Simplex Identifying Ten Novel Mutations in KRT5 and KRT14 in Australia. Acta Derm. Venereol. 2017, 97, 1114–1119. [Google Scholar] [CrossRef]
- Cummins, R.E.; Klingberg, S.; Wesley, J.; Rogers, M.; Zhao, Y.; Murrell, D.F. Keratin 14 Point Mutations at Codon 119 of Helix 1A Resulting in Different Epidermolysis Bullosa Simplex Phenotypes. J. Investig. Dermatol. 2001, 117, 1103–1107. [Google Scholar] [CrossRef]
- Lin, J.Y.; Fisher, D.E. Melanocyte biology and skin pigmentation. Nature 2007, 445, 843–850. [Google Scholar] [CrossRef]
- Hossain, M.R.; Ansary, T.M.; Komine, M.; Ohtsuki, M. Diversified Stimuli-Induced Inflammatory Pathways Cause Skin Pigmentation. Int. J. Mol. Sci. 2021, 22, 3970. [Google Scholar] [CrossRef]
- He, Y.; Maier, K.; Leppert, J.; Hausser, I.; Schwieger-Briel, A.; Weibel, L.; Theiler, M.; Kiritsi, D.; Busch, H.; Boerries, M.; et al. Monoallelic Mutations in the Translation Initiation Codon of KLHL24 Cause Skin Fragility. Am. J. Hum. Genet. 2016, 99, 1395–1404. [Google Scholar] [CrossRef]
- Vermeer, M.C.S.C.; Bolling, M.C.; Bliley, J.M.; Gomez, K.F.A.; Pavez-Giani, M.G.; Kramer, D.; Romero-Herrera, P.H.; Westenbrink, B.D.; Diercks, G.F.H.; Berg, M.P.; et al. Gain-of-Function Mutation in Ubiquitin Ligase KLHL24 Causes Desmin Degradation and Dilatation in hiPSC-Derived Engineered Heart Tissues. J. Clin. Investig. 2021, 131, e140615. [Google Scholar] [CrossRef]
- Carney, J.A.; Gaillard, R.C.; Bertherat, J.; Stratakis, C.A. Familial Micronodular Adrenocortical Disease, Cushing Syndrome, and Mutations of the Gene Encoding Phosphodiesterase 11A4 (PDE11A). Am. J. Surg. Pathol. 2010, 34, 547–555. [Google Scholar] [CrossRef]
- Horvath, A.; Boikos, S.; Giatzakis, C.; Robinson-White, A.; Groussin, L.; Griffin, K.J.; Stein, E.; Levine, E.; Delimpasi, G.; Hsiao, H.P.; et al. A Genome-Wide Scan Identifies Mutations in the Gene Encoding Phosphodiesterase 11A4 (PDE11A) in Individuals with Adrenocortical Hyperplasia. Nat. Genet. 2006, 38, 794–800. [Google Scholar] [CrossRef]
- Basmanav, F.B.; Oprisoreanu, A.M.; Pasternack, S.M.; Thiele, H.; Fritz, G.; Wenzel, J.; Größer, L.; Wehner, M.; Wolf, S.; Fagerberg, C.; et al. Mutations in POGLUT1, encoding protein O-glucosyltransferase 1, cause autosomal-dominant Dowling-Degos disease. Am. J. Hum. Genet. 2014, 94, 135–143. [Google Scholar] [CrossRef]
- Hanneken, S.; Rütten, A.; Eigelshoven, S.; Braun-Falco, M.; Pasternack, S.M.; Ruzicka, T.; Nöthen, M.M.; Betz, R.C.; Kruse, R. Galli-Galli disease. Clinical and histopathological investigation using a case series of 18 patients. Hautarzt 2011, 62, 842–851. [Google Scholar] [CrossRef]
- Fernandez-Valdivia, R.; Takeuchi, H.; Samarghandi, A.; Lopez, M.; Leonardi, J.; Haltiwanger, R.S.; Jafar-Nejad, H. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 2011, 138, 1925–1934. [Google Scholar] [CrossRef]
- Aubin-Houzelstein, G.; Djian-Zaouche, J.; Bernex, F.; Gadin, S.; Delmas, V.; Larue, L.; Panthier, J.-J. Melanoblasts’ Proper Location and Timed Differentiation Depend on Notch/RBP-J Signaling in Postnatal Hair Follicles. J. Investig. Dermatol. 2008, 128, 2686–2695. [Google Scholar] [CrossRef]
- Duchatelet, S.; Clerc, H.; Machet, L.; Gaboriaud, P.; Miskinyte, S.; Kervarrec, T.; Hovnanian, A. A New Nonsense Mutation in the POGLUT1 Gene in Two Sisters with Dowling-Degos Disease. J. Eur. Acad. Dermatol. Venereol. 2018, 32, e440–e442. [Google Scholar] [CrossRef]
- Kono, M.; Sawada, M.; Nakazawa, Y.; Ogi, T.; Muro, Y.; Akiyama, M. A Japanese Case of Galli-Galli Disease Due to a Previously Unreported POGLUT1 Mutation. Acta Derm. Venereol. 2019, 99, 458–459. [Google Scholar] [CrossRef]
- Pfendner, E.G.; Sadowski, S.G.; Uitto, J. Epidermolysis bullosa simplex: Recurrent and de novo mutations in the KRT5 and KRT14 genes, phenotype/genotype correlations, and implications for genetic counseling and prenatal diagnosis. J. Investig. Dermatol. 2005, 125, 239–243. [Google Scholar] [CrossRef]
- Grievink, H.W.; Luisman, T.; Kluft, C.; Moerland, M.; Malone, K.E. Comparison of Three Isolation Techniques for Human Peripheral Blood Mononuclear Cells: Cell Recovery and Viability, Population Composition, and Cell Functionality. Biopreservation Biobanking 2016, 14, 410–415. [Google Scholar] [CrossRef]

| Gene | Position | Variant | rs | OMIM | Phenotype MIM Number | Phenotype | Gene/ Locus MIM Number |
|---|---|---|---|---|---|---|---|
| Krt14 | Chr17:39742731 A>C | Met119Thr | rs28928893 | EBS2F (131960) | 131760 | EBS severe with mottled pigmentation | 148066 |
| KLHL24 | Chr3:119188886 A>G | Met1Val | rs886037956 | EBS6 (617294) | 131900 617294 | EBS generalized intermediate with or without cardiomyopathy | 611295 |
| POGLUT1 | Arg279Trp | rs58330629 | 615696 611295 | 615696 | DDD4 | 615618 | |
| Krt14 | Chrl7:.39742713 C>T | Arg125His | rs886037956 | EBS1A | 131760 | 148066 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosykh, A.V.; Ryumina, I.I.; Botkina, A.S.; Evtushenko, N.A.; Zhigmitova, E.B.; Martynova, A.A.; Gurskaya, N.G.; Rebrikov, D.V. EBS in Children with De Novo Pathogenic Variants Disturbing Krt14. Int. J. Mol. Sci. 2024, 25, 2989. https://doi.org/10.3390/ijms25052989
Kosykh AV, Ryumina II, Botkina AS, Evtushenko NA, Zhigmitova EB, Martynova AA, Gurskaya NG, Rebrikov DV. EBS in Children with De Novo Pathogenic Variants Disturbing Krt14. International Journal of Molecular Sciences. 2024; 25(5):2989. https://doi.org/10.3390/ijms25052989
Chicago/Turabian StyleKosykh, Anastasiya V., Irina I. Ryumina, Alexandra S. Botkina, Nadezhda A. Evtushenko, Elena B. Zhigmitova, Aleksandra A. Martynova, Nadya G. Gurskaya, and Denis V. Rebrikov. 2024. "EBS in Children with De Novo Pathogenic Variants Disturbing Krt14" International Journal of Molecular Sciences 25, no. 5: 2989. https://doi.org/10.3390/ijms25052989
APA StyleKosykh, A. V., Ryumina, I. I., Botkina, A. S., Evtushenko, N. A., Zhigmitova, E. B., Martynova, A. A., Gurskaya, N. G., & Rebrikov, D. V. (2024). EBS in Children with De Novo Pathogenic Variants Disturbing Krt14. International Journal of Molecular Sciences, 25(5), 2989. https://doi.org/10.3390/ijms25052989







