Biomolecular Actions by Intestinal Endotoxemia in Metabolic Syndrome
Abstract
1. Introduction
1.1. The Metabolic Syndrome (MetS)
1.2. Metabolomics Concepts
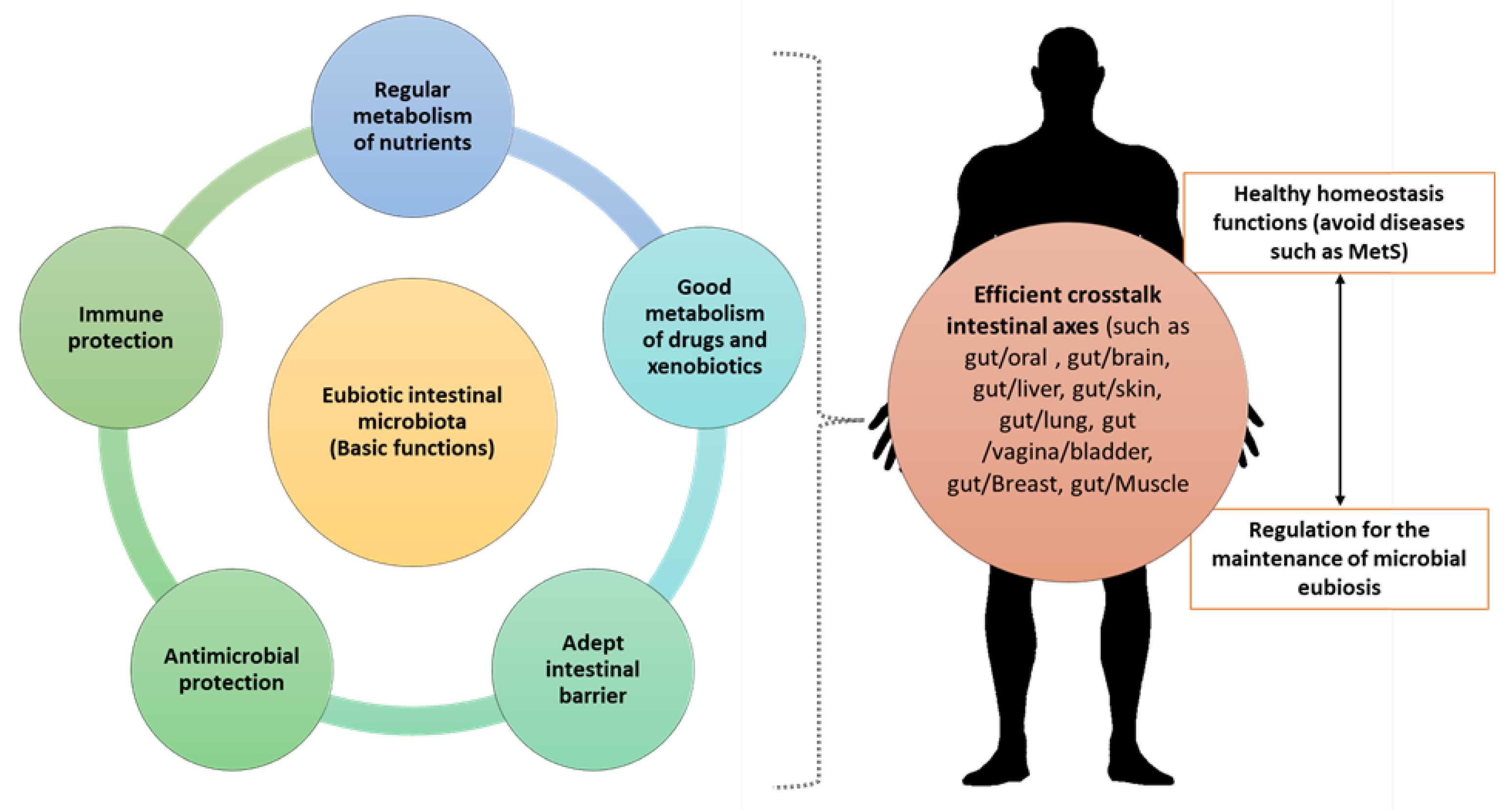
2. The Importance of IM Composition over Time
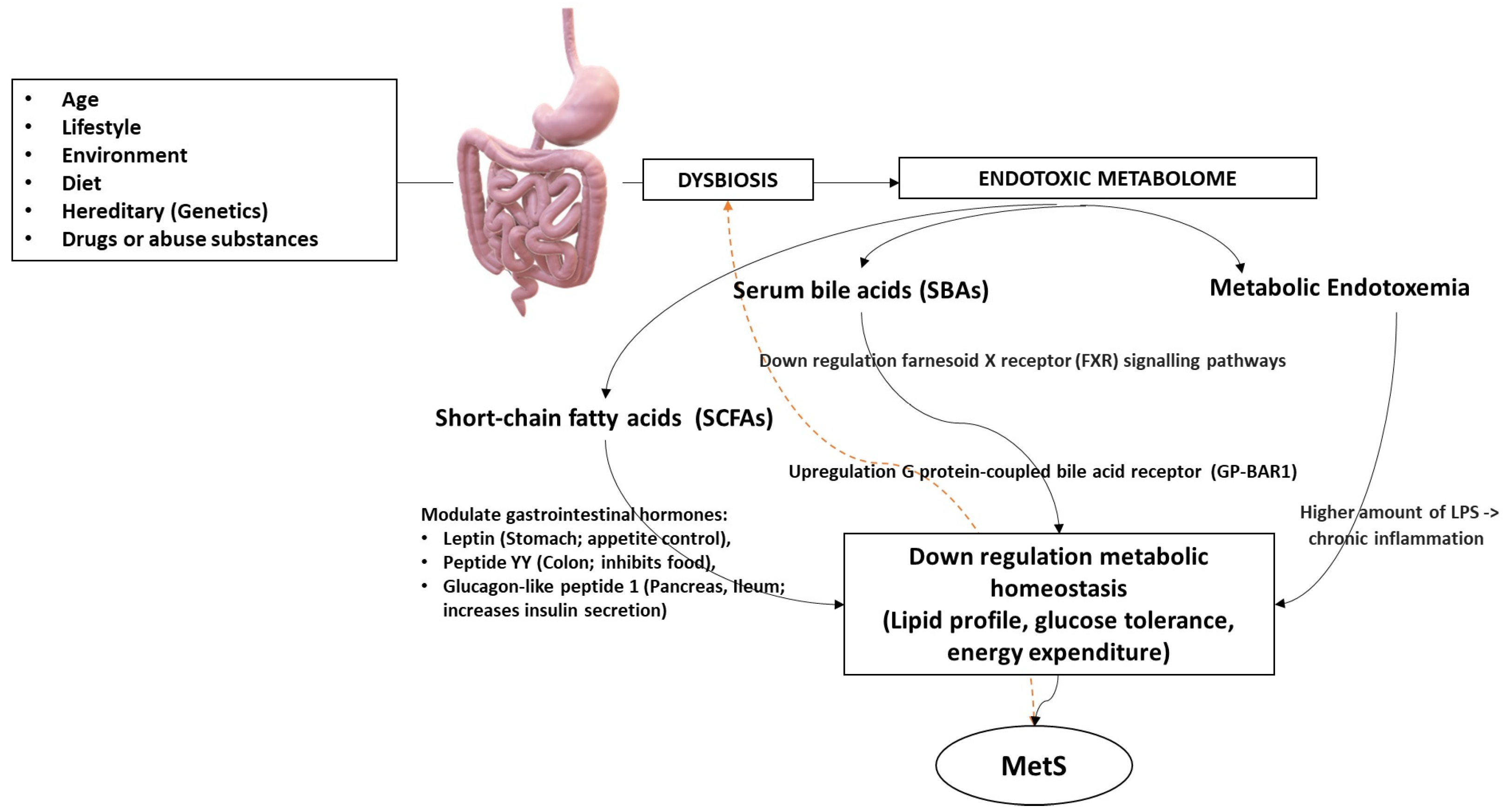
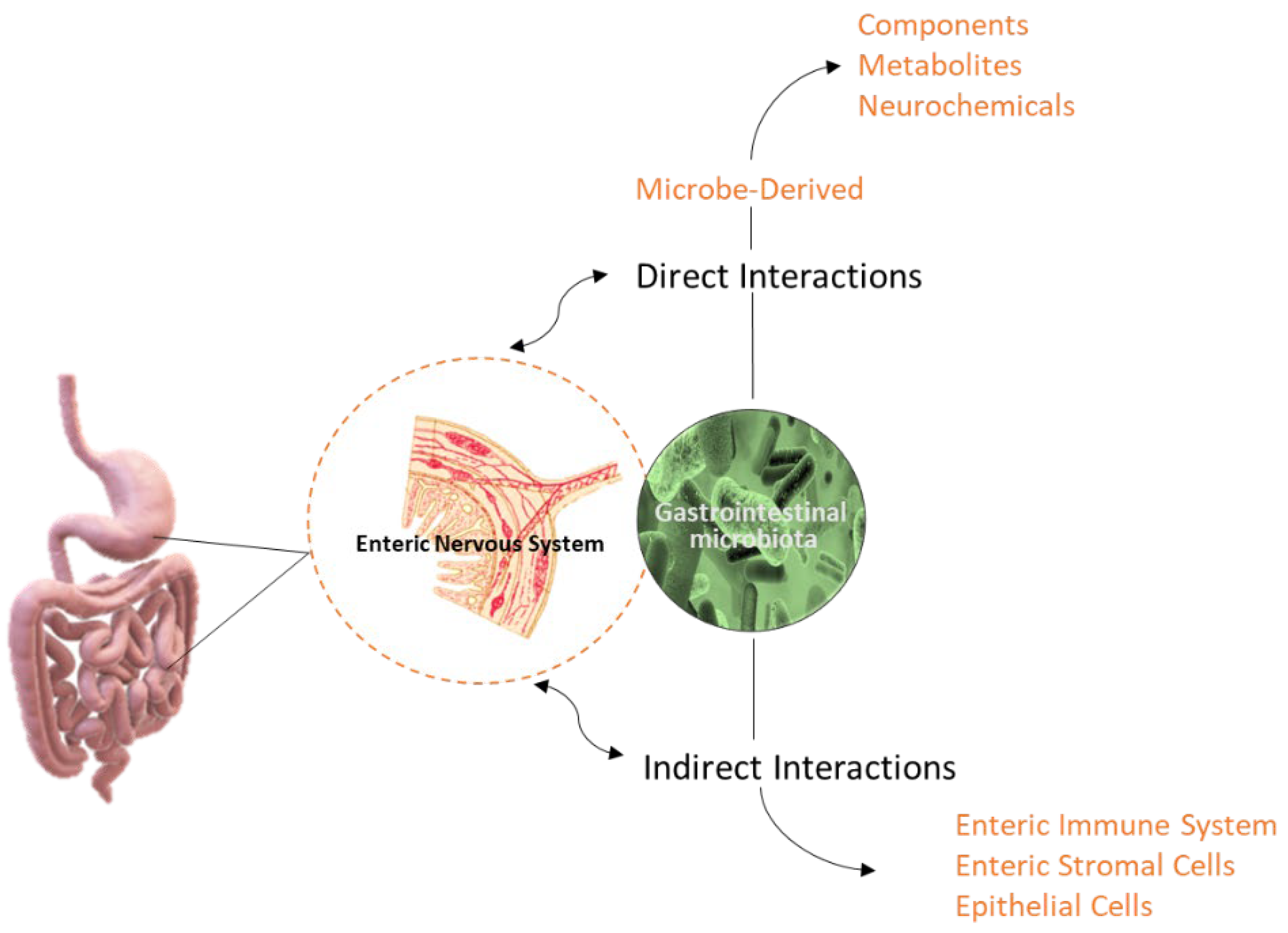
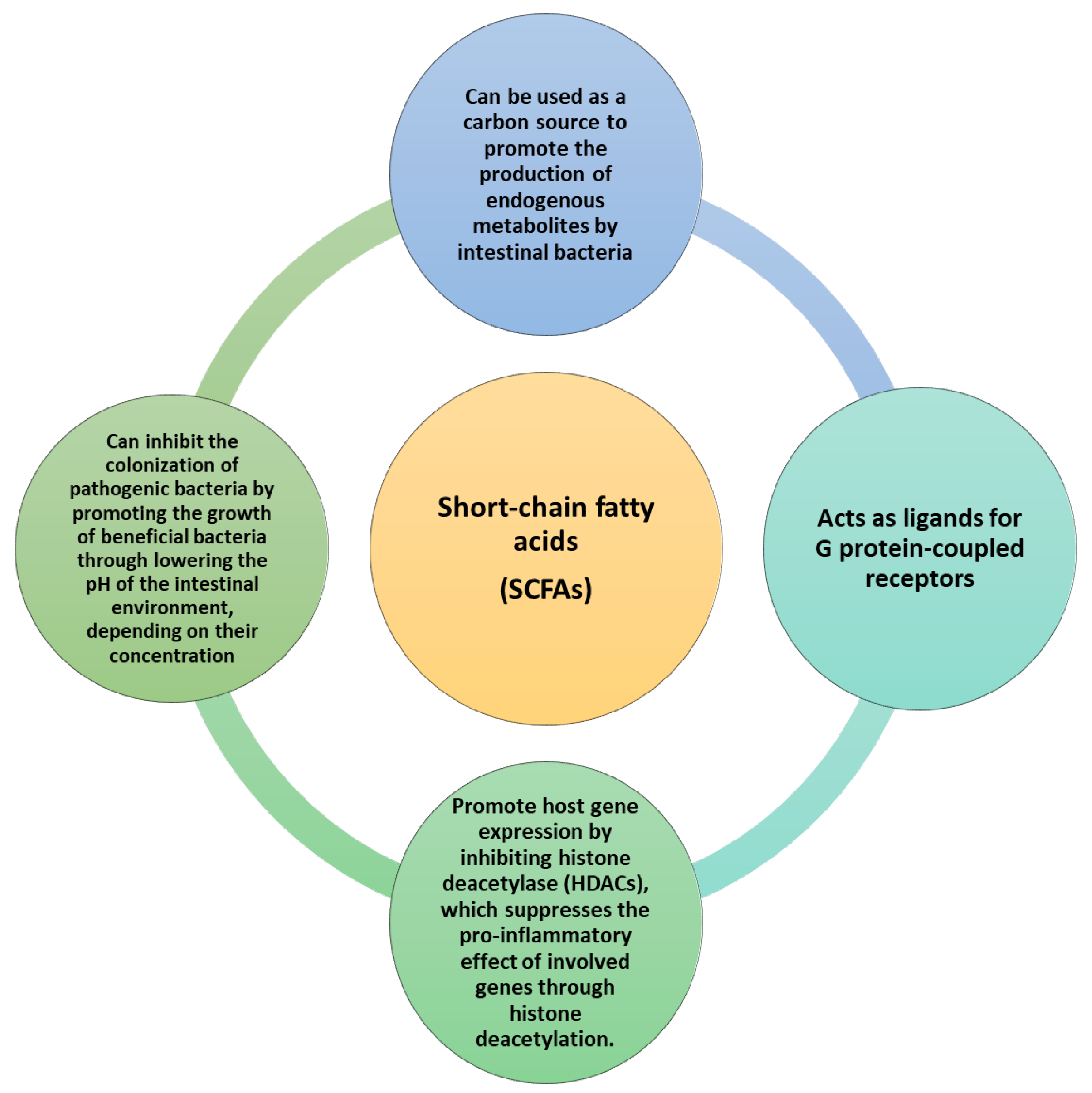
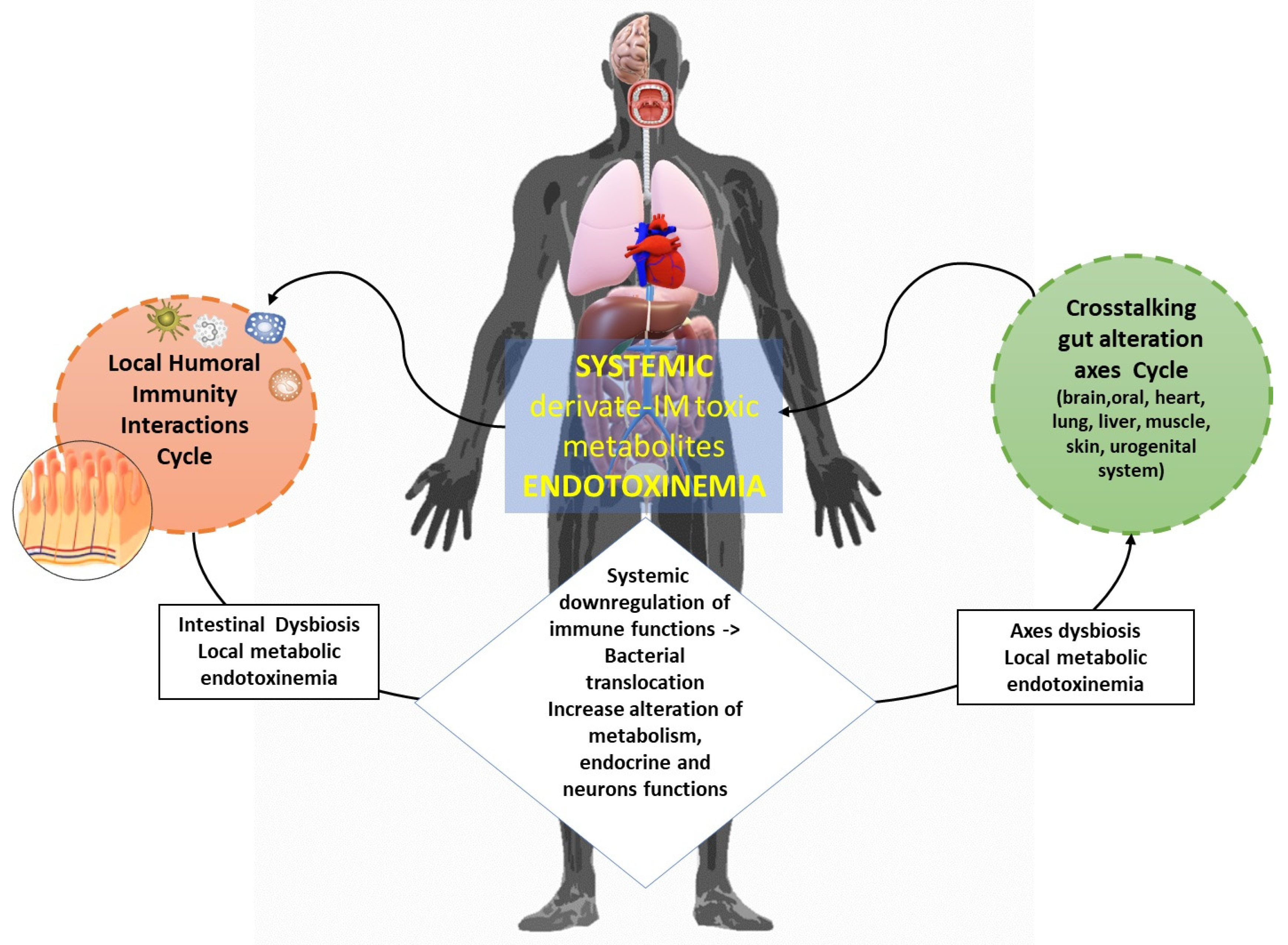
3. The IM-Derived Metabolites
4. Metabolome’s Pathogenic Biomolecular Mechanics in MetS
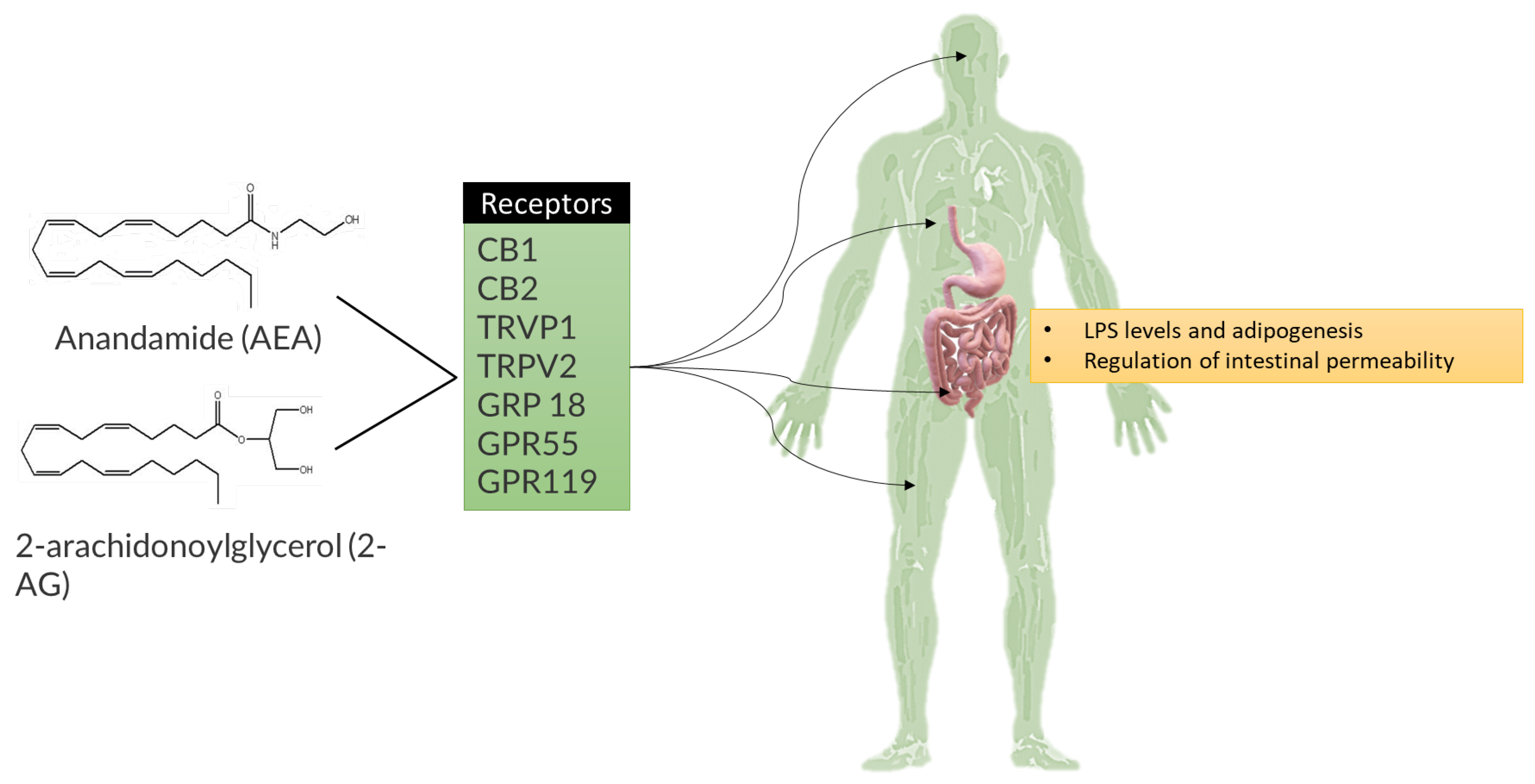
4.1. Obesity
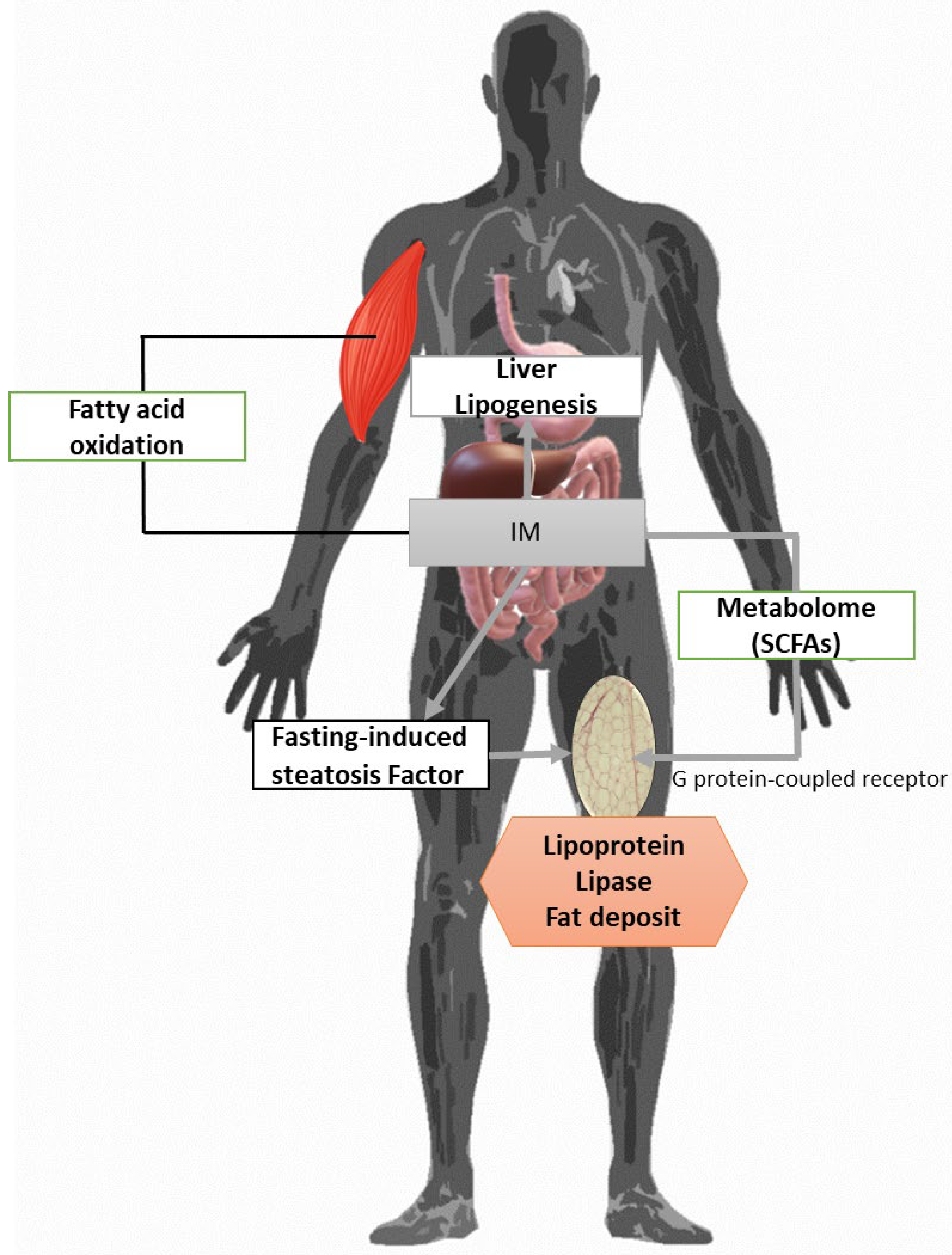
4.2. Non-Alcoholic Fatty Liver Disease (NAFLD)
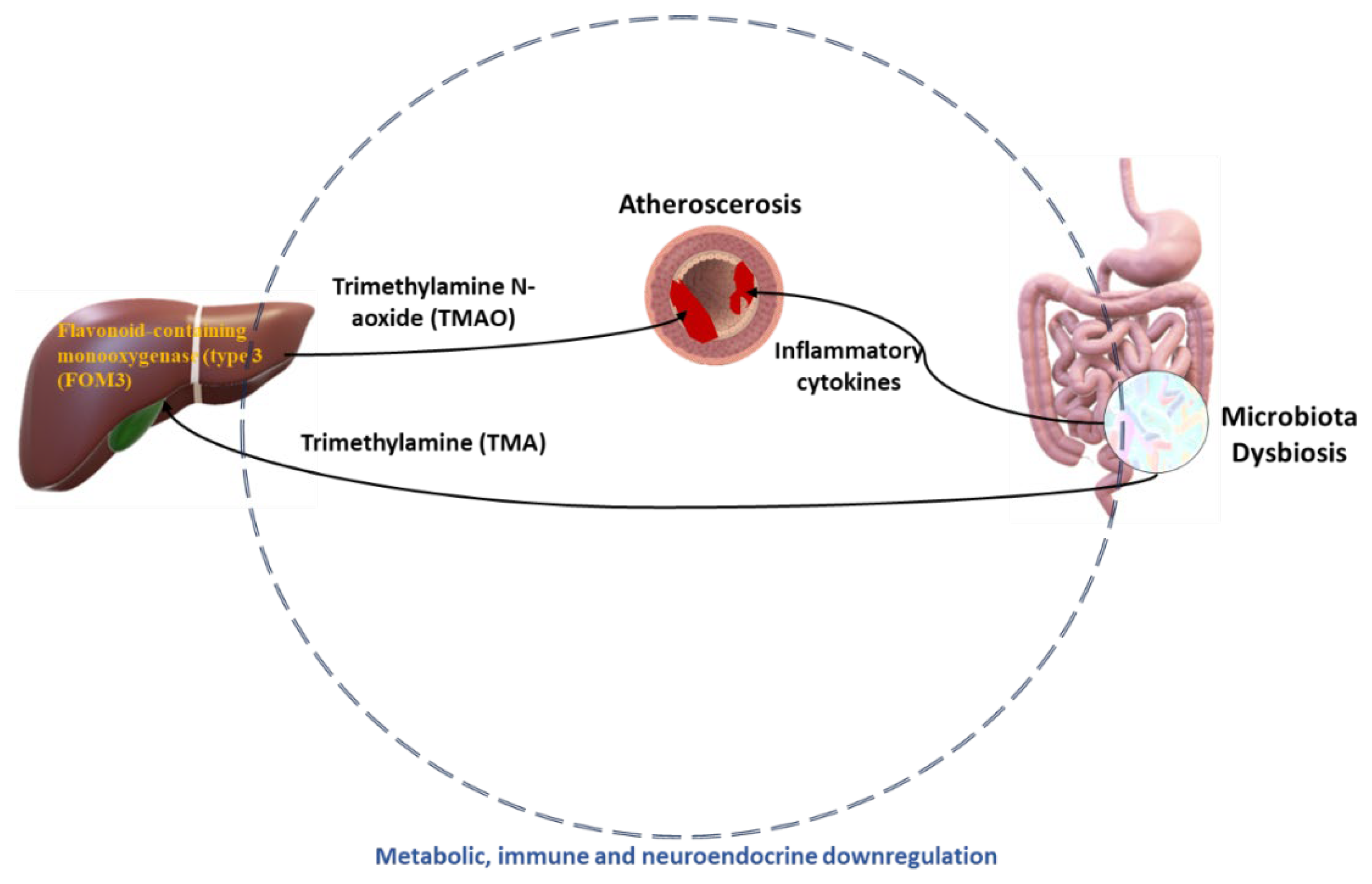
4.3. Atherosclerosis
4.4. Type 1 and 2 Diabetes Mellitus
5. The Perspectives from the Individual Research Metabolome to Personalized Therapy
5.1. Nutritional Interventions
5.2. Faecal Microbiota Transplantation (FMT)
5.3. Probiotic and Prebiotic Formulation in Metabolic Diseases
5.4. Antibiotics
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Papanastasiou, E. The prevalence and mechanisms of metabolic syndrome in schizophrenia: A review. Ther. Adv. Psychopharmacol. 2012, 3, 33–51. [Google Scholar] [CrossRef]
- Noubiap, J.J.; Nansseu, J.R.; Lontchi-Yimagou, E.; Nkeck, J.R.; Nyaga, U.F.; Ngouo, A.T.; Tounouga, D.N.; Tianyi, F.L.; Foka, A.J.; Ndoadoumgue, A.L.; et al. Geographic distribution of metabolic syndrome and its components in the general adult population: A meta-analysis of global data from 28 million individuals. Diabetes Res. Clin. Pract. 2022, 188, 109924. [Google Scholar] [CrossRef]
- Kovatcheva-Datchary, P.; Arora, T. Nutrition, the gut microbiome and the metabolic syndrome. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Palmnäs-Bédard, M.S.; Costabile, G.; Vetrani, C.; Åberg, S.; Hjalmarsson, Y.; Dicksved, J.; Riccardi, G.; Landberg, R. The human gut microbiota and glucose metabolism: A scoping review of key bacteria and the potential role of SCFAs. Am. J. Clin. Nutr. 2022, 116, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed]
- Pohl, K.; Moodley, P.; Dhanda, A. The effect of increasing intestinal short-chain fatty acid concentration on gut permeability and liver injury in the context of liver disease: A systematic review. J. Gastroenterol. Hepatol. 2022, 37, 1498–1506. [Google Scholar] [CrossRef] [PubMed]
- Ziętek, M.; Celewicz, Z.; Kikut, J.; Szczuko, M. Implications of SCFAs on the Parameters of the Lipid and Hepatic Profile in Pregnant Women. Nutrients 2021, 13, 1749. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, L.; Man, A.; Charitos, I.A.; Haxhirexha, K.; Topi, S. Current knowledge about the connection between health status and gut microbiota from birth to elderly. Front. Biosci. 2021, 26, 135–148. [Google Scholar] [CrossRef]
- Nemes, P.; Rubakhin, S.S.; Aerts, J.T.; Sweedler, J.V. Qualitative and quantitative metabolomic investigation of single neurons by capillary electrophoresis electrospray ionization mass spectrometry. Nat. Protoc. 2013, 8, 783–799. [Google Scholar] [CrossRef]
- Mohammad, S.; Thiemermann, C. Role of Metabolic Endotoxemia in Systemic Inflammation and Potential Interventions. Front. Immunol. 2021, 11, 594150. [Google Scholar] [CrossRef]
- Eberl, G.; Colonna, M.; Di Santo, J.P.; McKenzie, A.N.J. Innate lymphoid cells: A new paradigm in immunology. Science 2015, 348, aaa6566. [Google Scholar] [CrossRef] [PubMed]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef] [PubMed]
- Rosato, V.; Masarone, M.; Dallio, M.; Federico, A.; Aglitti, A.; Persico, M. NAFLD and Extra-Hepatic Comorbidities: Current Evidence on a Multi-Organ Metabolic Syndrome. Int. J. Environ. Res. Public Health 2019, 16, 3415. [Google Scholar] [CrossRef] [PubMed]
- Torres-Sánchez, A.; Ruiz-Rodríguez, A.; Ortiz, P.; Aguilera, M. Key Stratification of Microbiota Taxa and Metabolites in the Host Metabolic Health–Disease Balance. Int. J. Mol. Sci. 2023, 24, 4519. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Bai, Y.; Zha, L.; Ullah, N.; Ullah, H.; Shah, S.R.H.; Sun, H.; Zhang, C. Mechanism of the Gut Microbiota Colonization Resistance and Enteric Pathogen Infection. Front. Cell. Infect. Microbiol. 2021, 11, 716299. [Google Scholar] [CrossRef] [PubMed]
- Charitos, I.A.; Topi, S.; Castellaneta, F.; D’agostino, D. Current Issues and Perspectives in Patients with Possible Sepsis at Emergency Departments. Antibiotics 2019, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.-H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Leyrolle, Q.; Koistinen, V.; Kärkkäinen, O.; Layé, S.; Delzenne, N.; Hanhineva, K. Microbiota-derived metabolites as drivers of gut–brain communication. Gut Microbes 2022, 14, 2102878. [Google Scholar] [CrossRef]
- Wang, K.; Baldassano, R.; Zhang, H.; Qu, H.-Q.; Imielinski, M.; Kugathasan, S.; Annese, V.; Dubinsky, M.; Rotter, J.I.; Russell, R.K.; et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum. Mol. Genet. 2010, 19, 2059–2067. [Google Scholar] [CrossRef]
- Henneman, P.; Beer, F.V.D.S.-D.; Moghaddam, P.H.; Huijts, P.; Stalenhoef, A.F.; Kastelein, J.J.; Van Duijn, C.M.; Havekes, L.M.; Frants, R.R.; Van Dijk, K.W.; et al. The expression of type III hyperlipoproteinemia: Involvement of lipolysis genes. Eur. J. Hum. Genet. 2009, 17, 620–628. [Google Scholar] [CrossRef]
- Brauner, H.; Elemans, M.; Lemos, S.; Broberger, C.; Holmberg, D.; Flodström-Tullberg, M.; Kärre, K.; Höglund, P. Distinct Phenotype and Function of NK Cells in the Pancreas of Nonobese Diabetic Mice. J. Immunol. 2010, 184, 2272–2280. [Google Scholar] [CrossRef]
- Kornete, M.; Mason, E.S.; Piccirillo, C.A. Immune Regulation in T1D and T2D: Prospective Role of Foxp3+ Treg Cells in Disease Pathogenesis and Treatment. Front. Endocrinol. 2013, 4, 76. [Google Scholar] [CrossRef]
- Huang, J.; Ellinghaus, D.; Franke, A.; Howie, B.; Li, Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur. J. Hum. Genet. 2012, 20, 801–805. [Google Scholar] [CrossRef]
- Gjymishka, A.; Coman, R.M.; Brusko, T.M.; Glover, S.C.; Schloot, N.C.; Kumar, N.; Kaur, G.; Mehra, N.; Woittiez, N.J.; O Roep, B.; et al. Influence of host immunoregulatory genes, ER stress and gut microbiota on the shared pathogenesis of inflammatory bowel disease and Type 1 diabetes. Immunotherapy 2013, 5, 1357–1366. [Google Scholar] [CrossRef]
- Shaffer, M.; Armstrong, A.J.; Phelan, V.V.; Reisdorph, N.; Lozupone, C.A. Microbiome and metabolome data integration provides insight into health and disease. Transl. Res. 2017, 189, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.-U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Orešič, M.; Bäckhed, F. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-beta-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials; Board on Health Care Services; Board on Health Sciences Policy; Institute of Medicine. 2, Omics-Based Clinical Discovery: Science, Technology, and Applications. In Evolution of Translational Omics: Lessons Learned and the Path Forward; Michael, C.M., Nass, S.J., Omenn, G.S., Eds.; National Academies Press: Washington, DC, USA, 2012. Available online: https://www.ncbi.nlm.nih.gov/books/NBK202165/ (accessed on 12 November 2023).
- Gomase, V.S.; Changbhale, S.S.; Patil, S.A.; Kale, K.V. Metabolomics. Curr. Drug Metab. 2008, 9, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Charitos, I.A.; Topi, S.; Gagliano-Candela, R.; De Nitto, E.; Polimeno, L.; Montagnani, M.; Santacroce, L. The Toxic Effects of Endocrine Disrupting Chemicals (EDCs) on Gut Microbiota: Bisphenol A (BPA) A Review. Endocrine, Metab. Immune Disord. Drug Targets 2022, 22, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Tyler, J.K. Epigenetics and aging. Sci. Adv. 2016, 2, e1600584. [Google Scholar] [CrossRef]
- Rochfort, S. Metabolomics Reviewed: A New “Omics” Platform Technology for Systems Biology and Implications for Natural Products Research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.J.; Sureshkumar, S.; Shewade, D.G. Metabolomics: A Tool Ahead for Understanding Molecular Mechanisms of Drugs and Diseases. Indian J. Clin. Biochem. 2015, 30, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to patho-physiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef]
- Oh, S.W.; Imran, M.; Kim, E.-H.; Park, S.-Y.; Lee, S.-G.; Park, H.-M.; Jung, J.-W.; Ryu, T.-H. Approach strategies and application of metabolomics to biotechnology in plants. Front. Plant Sci. 2023, 14, 1192235. [Google Scholar] [CrossRef]
- Coen, M.; Holmes, E.; Lindon, J.C.; Nicholson, J. KNMR-based metabolic profiling and metabonomic approaches to problems in molecular toxicology. Chem. Res. Toxicol. 2008, 21, 9–27. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.I.; Atherton, H.J.; Goodacre, R.; Griffin, J.L. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem. Soc. Rev. 2011, 40, 387–426. [Google Scholar] [CrossRef]
- Ellis, D.I.; Dunn, W.B.; Griffin, J.L.; Allwood, J.W.; Goodacre, R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics 2007, 8, 1243–1266. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2012, 1, 92–107. [Google Scholar] [CrossRef]
- Di Domenico, M.; Ballini, A.; Boccellino, M.; Scacco, S.; Lovero, R.; Charitos, I.A.; Santacroce, L. The Intestinal Microbiota May Be a Potential Theranostic Tool for Personalized Medicine. J. Pers. Med. 2022, 12, 523. [Google Scholar] [CrossRef]
- Blaser, M.J.; Devkota, S.; McCoy, K.D.; Relman, D.A.; Yassour, M.; Young, V.B. Lessons learned from the prenatal microbiome controversy. Microbiome 2021, 9, 8. [Google Scholar] [CrossRef]
- de Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of Mammals and Their Gut Microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef]
- Susic, D.; Yang, J.; Upitis, A.; Henry, A. Microbiota and pregnancy. Microb. Health Dis. 2021, 3, e635. [Google Scholar] [CrossRef]
- Ziętek, M.; Celewicz, Z.; Szczuko, M. Short-Chain Fatty Acids, Maternal Microbiota and Metabolism in Pregnancy. Nutrients 2021, 13, 1244. [Google Scholar] [CrossRef] [PubMed]
- Andrade, J.C.; Almeida, D.; Domingos, M.; Seabra, C.L.; Machado, D.; Freitas, A.C.; Gomes, A.M. Commensal Obligate Anaerobic Bacteria and Health: Production, Storage, and Delivery Strategies. Front. Bioeng. Biotechnol. 2020, 8, 545071. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Urbán, E. Relevance of anaerobic bacteremia in adult patients: A never-ending story? Eur. J. Microbiol. Immunol. 2020, 10, 64–75. [Google Scholar] [CrossRef]
- Daniel, N.; Lécuyer, E.; Chassaing, B. Host/microbiota interactions in health and diseases—Time for mucosal microbiology! Mucosal Immunol. 2021, 14, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, L.; Inchingolo, F.; Topi, S.; Del Prete, R.; Di Cosola, M.; Charitos, I.A.; Montagnani, M. Potential beneficial role of probiotics on the outcome of COVID-19 patients: An evolving perspective. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Charitos, I.A.; Ballini, A.; Lovero, R.; Castellaneta, F.; Colella, M.; Scacco, S.; Cantore, S.; Arrigoni, R.; Mastrangelo, F.; Dioguardi, M. Update on COVID-19 and Effectiveness of a Vaccination Campaign in a Global Context. Int. J. Environ. Res. Public Health 2022, 19, 10712. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Covarrubias, V.; Martínez-Martínez, E.; del Bosque-Plata, L. The Potential of Metabolomics in Biomedical Applications. Metabolites 2022, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S. Gut microbial metabolites and its impact on human health. Ann. Gastroenterol. 2023, 36, 360–368. [Google Scholar] [CrossRef]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef]
- Murri, M.; Leiva, I.; Gomez-Zumaquero, J.M.; Tinahones, F.J.; Cardona, F.; Soriguer, F.; Queipo-Ortuño, M.I. Gut microbiota in children with type 1 diabetes differs from that in healthy children: A case-control study. BMC Med. 2013, 11, 46. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Manco, M.; Putignani, L.; Bottazzo, G.F. Gut Microbiota, Lipopolysaccharides, and Innate Immunity in the Pathogenesis of Obesity and Cardiovascular Risk. Endocr. Rev. 2010, 31, 817–844. [Google Scholar] [CrossRef]
- Hou, J.; Xiang, J.; Li, D.; Liu, X.; Pan, W. Gut microbial response to host metabolic phenotypes. Front. Nutr. 2022, 9, 1019430. [Google Scholar] [CrossRef]
- Xia, Y.; Ren, M.; Yang, J.; Cai, C.; Cheng, W.; Zhou, X.; Lu, D.; Ji, F. Gut microbiome and microbial metabolites in NAFLD and after bariatric surgery: Correlation and causality. Front. Microbiol. 2022, 13, 1003755. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Liu, J.; Tan, Y.; Cheng, H.; Zhang, D.; Feng, W.; Peng, C. Functions of Gut Microbiota Metabolites, Current Status and Future Perspectives. Aging Dis. 2022, 13, 1106–1126. [Google Scholar] [CrossRef]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-Chain Fatty Acids Activate GPR41 and GPR43 on Intestinal Epithelial Cells to Promote Inflammatory Responses in Mice. Gastroenterology 2013, 145, 396–406.e1-10. [Google Scholar] [CrossRef]
- Angelakis, E.; Armougom, F.; Million, M.; Raoult, D. The relationship between gut microbiota and weight gain in humans. Future Microbiol. 2012, 7, 91–109. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R.; Kaser, A. Obesity and the Microbiota. Gastroenterology 2009, 136, 1476–1483. [Google Scholar] [CrossRef]
- Chhibber-Goel, J.; Gaur, A.; Singhal, V.; Parakh, N.; Bhargava, B.; Sharma, A. The complex metabolism of trimethylamine in humans: Endogenous and exogenous sources. Expert Rev. Mol. Med. 2016, 18, e8. [Google Scholar] [CrossRef]
- Badami, E.; Sorini, C.; Coccia, M.; Usuelli, V.; Molteni, L.; Bolla, A.M.; Scavini, M.; Mariani, A.; King, C.; Bosi, E.; et al. Defective Differentiation of Regulatory FoxP3+ T Cells by Small-Intestinal Dendritic Cells in Patients with Type 1 Diabetes. Diabetes 2011, 60, 2120–2124. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Arslan, N. Obesity, fatty liver disease and intestinal microbiota. World J. Gastroenterol. 2014, 20, 16452–16463. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277, Erratum in Front. Immunol. 2019, 10, 1486. [Google Scholar] [CrossRef]
- Montagnani, M.; Bottalico, L.; Potenza, M.A.; Charitos, I.A.; Topi, S.; Colella, M.; Santacroce, L. The Crosstalk between Gut Microbiota and Nervous System: A Bidirectional Interaction between Microorganisms and Metabolome. Int. J. Mol. Sci. 2023, 24, 10322. [Google Scholar] [CrossRef]
- Cakmak, A.; Nemutlu, E.; Yabanoglu-Ciftci, S.; Baysal, I.; Kocaaga, E.; Coplu, L.; Inal-Ince, D. Metabolomic, oxidative, and inflammatory responses to acute exercise in chronic obstructive pulmonary disease. Heart Lung 2023, 59, 52–60. [Google Scholar] [CrossRef]
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate mediates a microbiome–brain–β-cell axis to promote metabolic syndrome. Nature 2016, 534, 213–217. [Google Scholar] [CrossRef]
- Cunningham, A.L.; Stephens, J.W.; Harris, D.A. Intestinal microbiota and their metabolic contribution to type 2 diabetes and obesity. J. Diabetes Metab. Disord. 2021, 20, 1855–1870. [Google Scholar] [CrossRef]
- Gonçalves, P.; Araújo, J.R.; Di Santo, J.P. A Cross-Talk between Microbiota-Derived Short-Chain Fatty Acids and the Host Mucosal Immune System Regulates Intestinal Homeostasis and Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2018, 24, 558–572. [Google Scholar] [CrossRef] [PubMed]
- Gewirtz, A.T.; Navas, T.A.; Lyons, S.; Godowski, P.J.; Madara, J.L. Cutting Edge: Bacterial Flagellin Activates Basolaterally Expressed TLR5 to Induce Epithelial Proinflammatory Gene Expression. J. Immunol. 2001, 167, 1882–1885. [Google Scholar] [CrossRef] [PubMed]
- di Gregorio, M.C.; Cautela, J.; Galantini, L. Physiology and Physical Chemistry of Bile Acids. Int. J. Mol. Sci. 2021, 22, 1780. [Google Scholar] [CrossRef]
- Larabi, A.B.; Masson, H.L.P.; Bäumler, A.J. Bile acids as modulators of gut microbiota composition and function. Gut Microbes 2023, 15, 2172671. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am. J. Physiol. Liver Physiol. 2020, 318, G554–G573. [Google Scholar] [CrossRef]
- Pathak, P.; Liu, H.; Boehme, S. Farnesoid X receptor induces Takeda G-protein receptor 5 cross-talk to regulate bile acid synthesis and hepatic metabolism. J. Biol. Chem. 2017, 292, 11055–11069. [Google Scholar] [CrossRef]
- Nyambuya, T.M.; Dludla, P.V.; Mxinwa, V.; Nkambule, B.B. Obesity-induced inflammation and insulin resistance: A mini-review on T-cells. Metab. Open 2019, 3, 100015. [Google Scholar] [CrossRef]
- Zarei, I.; Koistinen, V.M.; Kokla, M.; Klåvus, A.; Babu, A.F.; Lehtonen, M.; Auriola, S.; Hanhineva, K. Tissue-wide metabolomics reveals wide impact of gut microbiota on mice metabolite composition. Sci. Rep. 2022, 12, 15018. [Google Scholar] [CrossRef]
- King, C.; Sarvetnick, N. The Incidence of Type-1 Diabetes in NOD Mice Is Modulated by Restricted Flora Not Germ-Free Conditions. PLoS ONE 2011, 6, e17049. [Google Scholar] [CrossRef] [PubMed]
- Roesch, L.F.W.; Lorca, G.L.; Casella, G.; Giongo, A.; Naranjo, A.; Pionzio, A.M.; Li, N.; Mai, V.; Wasserfall, C.H.; Schatz, D.; et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. 2009, 3, 536–548. [Google Scholar] [CrossRef]
- Zorena, K.; Jachimowicz-Duda, O.; Ślęzak, D.; Robakowska, M.; Mrugacz, M. Adipokines and Obesity. Potential Link to Metabolic Disorders and Chronic Complications. Int. J. Mol. Sci. 2020, 21, 3570. [Google Scholar] [CrossRef]
- Moschen, A.R.; Kaser, S.; Tilg, H. Non-alcoholic steatohepatitis: A microbiota-driven disease. Trends Endocrinol. Metab. 2013, 24, 537–545. [Google Scholar] [CrossRef]
- Khan, M.J.; Gerasimidis, K.; Edwards, C.A.; Shaikh, M.G. Role of Gut Microbiota in the Aetiology of Obesity: Proposed Mechanisms and Review of the Literature. J. Obes. 2016, 2016, 7353642. [Google Scholar] [CrossRef]
- Ørskov, C.; Holst, J.J.; Knuhtsen, S.; Baldissera, F.G.A.; Poulsen, S.S.; Nielsen, O.V. Glucagon-Like Peptides GLP-1 and GLP-2, Predicted Products of the Glucagon Gene, Are Secreted Separately from Pig Small Intestine but Not Pancreas. Endocrinology 1986, 119, 1467–1475. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, B.; Lan, F.; Zhong, C.; Jin, J.; Li, X.; Zhou, Q.; Li, J.; Yang, N.; Wen, C.; et al. Host genetics and gut microbiota jointly regulate blood biochemical indicators in chickens. Appl. Microbiol. Biotechnol. 2023, 107, 7601–7620. [Google Scholar] [CrossRef]
- Beaumont, M.; Goodrich, J.K.; Jackson, M.A.; Yet, I.; Davenport, E.R.; Vieira-Silva, S.; Debelius, J.; Pallister, T.; Mangino, M.; Raes, J.; et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. 2016, 17, 189. [Google Scholar] [CrossRef]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in Gut Microbiota Control Metabolic Endotoxemia-Induced Inflammation in High-Fat Diet-Induced Obesity and Diabetes in Mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef]
- Ohkura, N.; Sakaguchi, S. Transcriptional and epigenetic basis of Treg cell development and function: Its genetic anomalies or variations in autoimmune diseases. Cell Res. 2020, 30, 465–474. [Google Scholar] [CrossRef]
- Lau, D.C.; Dhillon, B.; Yan, H.; Szmitko, P.E.; Verma, S. Adipokines: Molecular links between obesity and atheroslcerosis. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H2031–H2041. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Cani, P.D.; Delzenne, N.M. The gut microbiome as therapeutic target. Pharmacol. Ther. 2011, 130, 202–212. [Google Scholar] [CrossRef]
- Hersoug, L.; Møller, P.; Loft, S. Gut microbiota-derived lipopolysaccharide uptake and trafficking to adipose tissue: Implications for inflammation and obesity. Obes. Rev. 2015, 17, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Kassem, E.; Na’amnih, W.; Shapira, M.; Ornoy, A.; Muhsen, K. Comparison between School-Age Children with and without Obesity in Nutritional and Inflammation Biomarkers. J. Clin. Med. 2022, 11, 6973. [Google Scholar] [CrossRef] [PubMed]
- de La Serre, C.B.; Ellis, C.L.; Lee, J.; Hartman, A.L.; Rutledge, J.C.; Raybould, H.E. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am. J. Physiol. Liver Physiol. 2010, 299, G440–G448. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.F.; Murphy, E.F.; Nilaweera, K.; Ross, P.R.; Shanahan, F.; O’toole, P.W.; Cotter, P.D. The gut microbiota and its relationship to diet and obesity. Gut Microbes 2012, 3, 186–202. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-X.; Schwabe, R.F. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef]
- Greiner, T.; Bäckhed, F. Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol Metab. 2011, 22, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.-Q.; Chen, W.; Ma, K.; Liu, Z.-Z.; Gao, Y.; Zhang, J.-G.; Wang, T.; Yang, Y.-J. Akkermansia muciniphila Suppresses High-Fat Diet-Induced Obesity and Related Metabolic Disorders in Beagles. Molecules 2022, 27, 6074. [Google Scholar] [CrossRef]
- Geerlings, S.Y.; Kostopoulos, I.; De Vos, W.M.; Belzer, C. Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Barrea, L.; Muscogiuri, G.; Annunziata, G.; Laudisio, D.; Pugliese, G.; Salzano, C.; Colao, A.; Savastano, S. From gut microbiota dysfunction to obesity: Could short-chain fatty acids stop this dangerous course? Hormones 2019, 18, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Fei, N.; Zhao, L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2012, 7, 880–884. [Google Scholar] [CrossRef]
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojärvi, J.; Kootte, R.S.; Bartelsman, J.F.; Dallinga-Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of Intestinal Microbiota from Lean Donors Increases Insulin Sensitivity in Individuals with Metabolic Syndrome. Gastroenterology 2012, 143, 913–916.e7. [Google Scholar] [CrossRef]
- Gupta, S.; Allen-Vercoe, E.; Petrof, E.O. Fecal microbiota transplantation: In perspective. Ther. Adv. Gastroenterol. 2015, 9, 229–239. [Google Scholar] [CrossRef]
- Li, W.-Z.; Stirling, K.; Yang, J.-J.; Zhang, L. Gut microbiota and diabetes: From correlation to causality and mechanism. World J. Diabetes 2020, 11, 293–308. [Google Scholar] [CrossRef]
- Grigor’eva, I.N. Gallstone Disease, Obesity and the Firmicutes/Bacteroidetes Ratio as a Possible Biomarker of Gut Dysbiosis. J. Pers. Med. 2020, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Castaner, O.; Goday, A.; Park, Y.-M.; Lee, S.-H.; Magkos, F.; Shiow, S.-A.T.E.; Schröder, H. The Gut Microbiome Profile in Obesity: A Systematic Review. Int. J. Endocrinol. 2018, 2018, 4095789. [Google Scholar] [CrossRef]
- Zuo, H.-J. Gut bacteria alteration in obese people and its relationship with gene polymorphism. World J. Gastroenterol. 2011, 17, 1076–1081. [Google Scholar] [CrossRef] [PubMed]
- Festi, D.; Schiumerini, R.; Eusebi, L.H.; Marasco, G.; Taddia, M.; Colecchia, A. Gut microbiota and metabolic syndrome. World J. Gastroenterol. 2014, 20, 16079–16094. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.; Giudici, F.; Fiorindi, C.; Ficari, F.; Scaringi, S.; Amedei, A. Immunomodulating Activity and Therapeutic Effects of Short Chain Fatty Acids and Tryptophan Post-biotics in Inflammatory Bowel Disease. Front. Immunol. 2019, 10, 2754. [Google Scholar] [CrossRef]
- Vetrani, C.; Di Nisio, A.; Paschou, S.A.; Barrea, L.; Muscogiuri, G.; Graziadio, C.; Savastano, S.; Colao, A.; on behalf of the Obesity Programs of Nutrition, Education, Research and Assessment (OPERA) Group. From Gut Microbiota through Low-Grade Inflammation to Obesity: Key Players and Potential Targets. Nutrients 2022, 14, 2103. [Google Scholar] [CrossRef]
- Boutagy, N.E.; McMillan, R.P.; Frisard, M.I.; Hulver, M.W. Metabolic endotoxemia with obesity: Is it real and is it relevant? Biochimie 2016, 124, 11–20. [Google Scholar] [CrossRef]
- An, L.; Wirth, U.; Koch, D.; Schirren, M.; Drefs, M.; Koliogiannis, D.; Nieß, H.; Andrassy, J.; Guba, M.; Bazhin, A.V.; et al. The Role of Gut-Derived Lipopolysaccharides and the Intestinal Barrier in Fatty Liver Diseases. J. Gastrointest. Surg. 2021, 26, 671–683. [Google Scholar] [CrossRef]
- Parikh, S.; Pollock, N.K.; Bhagatwala, J.; Guo, D.-H.; Gutin, B.; Zhu, H.; Dong, Y. Adolescent Fiber Consumption Is Associated with Visceral Fat and Inflammatory Markers. J. Clin. Endocrinol. Metab. 2012, 97, E1451–E1457. [Google Scholar] [CrossRef]
- Hersoug, L.-G.; Møller, P.; Loft, S. Role of microbiota-derived lipopolysaccharide in adipose tissue inflammation, adipocyte size and pyroptosis during obesity. Nutr. Res. Rev. 2018, 31, 153–163. [Google Scholar] [CrossRef]
- Nd, A.M. Non-Alcoholic Fatty Liver Disease, an Overview. Integr. Med. 2019, 18, 42–49. [Google Scholar]
- Llorente, C.; Schnabl, B. The Gut Microbiota and Liver Disease. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 275–284. [Google Scholar] [CrossRef]
- Kim, K.-A.; Gu, W.; Lee, I.-A.; Joh, E.-H.; Kim, D.-H. High Fat Diet-Induced Gut Microbiota Exacerbates Inflammation and Obesity in Mice via the TLR4 Signaling Pathway. PLoS ONE 2012, 7, e47713. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Xie, C.; Li, F.; Zhang, L.; Nichols, R.G.; Krausz, K.W.; Cai, J.; Qi, Y.; Fang, Z.-Z.; Takahashi, S.; et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Investig. 2014, 125, 386–402. [Google Scholar] [CrossRef] [PubMed]
- Jasirwan, C.O.M.; Lesmana, C.R.A.; Hasan, I.; Sulaiman, A.S.; Gani, R.A. The role of gut microbiota in non-alcoholic fatty liver disease: Pathways of mechanisms. Biosci. Microbiota Food Health 2019, 38, 81–88. [Google Scholar] [CrossRef]
- Chakaroun, R.M.; Massier, L.; Kovacs, P. Gut Microbiome, Intestinal Permeability, and Tissue Bacteria in Metabolic Disease: Perpetrators or Bystanders? Nutrients 2020, 12, 1082. [Google Scholar] [CrossRef]
- Odeh, M.; Sabo, E.; Srugo, I.; Oliven, A. Serum levels of tumor necrosis factor-α correlate with severity of hepatic encephalopathy due to chronic liver failure. Liver Int. 2004, 24, 110–116. [Google Scholar] [CrossRef]
- Miele, L.; Valenza, V.; La Torre, G.; Montalto, M.; Cammarota, G.; Ricci, R.; Mascianà, R.; Forgione, A.; Gabrieli, M.L.; Perotti, G.; et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 2009, 49, 1877–1887. [Google Scholar] [CrossRef]
- Verdam, F.J.; Fuentes, S.; de Jonge, C.; Zoetendal, E.G.; Erbil, R.; Greve, J.W.; Buurman, W.A.; de Vos, W.M.; Rensen, S.S. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity 2013, 21, E607–E615. [Google Scholar] [CrossRef]
- Spencer, M.D.; Hamp, T.J.; Reid, R.W.; Fischer, L.M.; Zeisel, S.H.; Fodor, A.A. Association between Composition of the Human Gastrointestinal Microbiome and Development of Fatty Liver with Choline Deficiency. Gastroenterology 2011, 140, 976–986. [Google Scholar] [CrossRef]
- Arias, N.; Arboleya, S.; Allison, J.; Kaliszewska, A.; Higarza, S.G.; Gueimonde, M.; Arias, J.L. The Relationship between Choline Bioavailability from Diet, Intestinal Microbiota Composition, and Its Modulation of Human Diseases. Nutrients 2020, 12, 2340. [Google Scholar] [CrossRef]
- Olvera Lopez, E.; Ballard, B.D.; Jan, A. Cardiovascular Disease. [Updated 2023 Aug 22]. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK535419/ (accessed on 2 October 2023).
- Pieczynska, M.D.; Yang, Y.; Petrykowski, S.; Horbanczuk, O.K.; Atanasov, A.G.; Horbanczuk, J.O. Gut Microbiota and Its Metabolites in Atherosclerosis Development. Molecules 2020, 25, 594. [Google Scholar] [CrossRef]
- National Research Council (US) Committee on Diet and Health. 19, Atherosclerotic Cardiovascular Diseases. In Diet and Health: Implications for Reducing Chronic Disease Risk; National Academies Press: Washington, DC, USA, 1989. Available online: https://www.ncbi.nlm.nih.gov/books/NBK218744/ (accessed on 10 January 2024).
- Ziganshina, E.E.; Sharifullina, D.M.; Lozhkin, A.P.; Khayrullin, R.N.; Ignatyev, I.M. Bacterial Communities Associated with Atherosclerotic Plaques from Russian Individuals with Atherosclerosis. PLoS ONE 2016, 11, e0164836. [Google Scholar] [CrossRef]
- Velasquez, M.T.; Ramezani, A.; Manal, A.; Raj, D.S. Trimethylamine N-Oxide: The Good, the Bad and the Unknown. Toxins 2016, 8, 326. [Google Scholar] [CrossRef] [PubMed]
- Zhen, J.; Zhou, Z.; He, M.; Han, H.-X.; Lv, E.-H.; Wen, P.-B.; Liu, X.; Wang, Y.-T.; Cai, X.-C.; Tian, J.-Q.; et al. The gut microbial metabolite trimethylamine N-oxide and cardiovascular diseases. Front. Endocrinol. 2023, 14, 1085041. [Google Scholar] [CrossRef] [PubMed]
- Canas, J.A.; Damaso, L.; Altomare, A.; Killen, K.; Hossain, J.; Balagopal, P.B. Insulin Resistance and Adiposity in Relation to Serum β-Carotene Levels. J. Pediatr. 2012, 161, 58–64.e2. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dai, M. Trimethylamine N-Oxide Generated by the Gut Microbiota Is Associated with Vascular Inflammation: New Insights into Atherosclerosis. Mediat. Inflamm. 2020, 2020, 4634172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Yang, P.; Liu, X.; Lu, L.; Chen, Y.; Zhong, X.; Li, Z.; Liu, H.; Ou, C.; et al. Trimethylamine-N-Oxide Promotes Vascular Calcification Through Activation of NLRP3 (Nucleotide-Binding Domain, Leucine-Rich-Containing Family, Pyrin Domain-Containing-3) Inflammasome and NF-κB (Nuclear Factor κB) Signals. Arter. Thromb. Vasc. Biol. 2020, 40, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-S.; Li, J.; Krautkramer, K.A.; Badri, M.; Battaglia, T.; Borbet, T.C.; Koh, H.; Ng, S.; Sibley, R.A.; Li, Y.; et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. eLife 2018, 7, e37816. [Google Scholar] [CrossRef]
- Winiarska-Mieczan, A.; Tomaszewska, E.; Donaldson, J.; Jachimowicz, K. The Role of Nutritional Factors in the Modulation of the Composition of the Gut Microbiota in People with Autoimmune Diabetes. Nutrients 2022, 14, 2498. [Google Scholar] [CrossRef]
- Mokhtari, P.; Metos, J.; Babu, P.V.A. Impact of type 1 diabetes on the composition and functional potential of gut microbiome in children and adolescents: Possible mechanisms, current knowledge, and challenges. Gut Microbes 2021, 13, 1–18. [Google Scholar] [CrossRef]
- Colella, M.; Charitos, I.A.; Ballini, A.; Cafiero, C.; Topi, S.; Palmirotta, R.; Santacroce, L. Microbiota revolution: How gut microbes regulate our lives. World J. Gastroenterol. 2023, 29, 4368–4383. [Google Scholar] [CrossRef]
- Yan, H.; Fei, N.; Wu, G.; Zhang, C.; Zhao, L.; Zhang, M. Regulated Inflammation and Lipid Metabolism in Colon mRNA Expressions of Obese Germfree Mice Responding to Enterobacter cloacae B29 Combined with the High Fat Diet. Front. Microbiol. 2016, 7, 1786. [Google Scholar] [CrossRef]
- Fei, N.; Bruneau, A.; Zhang, X.; Wang, R.; Wang, J.; Rabot, S.; Gérard, P.; Zhao, L. Endotoxin Producers Overgrowing in Human Gut Microbiota as the Causative Agents for Nonalcoholic Fatty Liver Disease. mBio 2020, 11, e03263-19. [Google Scholar] [CrossRef]
- Xiong, R.-G.; Zhou, D.-D.; Wu, S.-X.; Huang, S.-Y.; Saimaiti, A.; Yang, Z.-J.; Shang, A.; Zhao, C.-N.; Gan, R.-Y.; Li, H.-B. Health Benefits and Side Effects of Short-Chain Fatty Acids. Foods 2022, 11, 2863. [Google Scholar] [CrossRef]
- van Deuren, T.; Blaak, E.E.; Canfora, E.E. Butyrate to combat obesity and obesity-associated metabolic disorders: Current status and future implications for therapeutic use. Obes. Rev. 2022, 23, e13498. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Levy, M.; Grosheva, I.; Zheng, D.; Soffer, E.; Blacher, E.; Braverman, S.; Tengeler, A.C.; Barak, O.; Elazar, M.; et al. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science 2018, 359, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Beier, U.H.; Wang, L.; Han, R.; Akimova, T.; Liu, Y.; Hancock, W.W. Histone Deacetylases 6 and 9 and Sirtuin-1 Control Foxp3 + Regulatory T Cell Function Through Shared and Isoform-Specific Mechanisms. Sci. Signal. 2012, 5, ra45. [Google Scholar] [CrossRef] [PubMed]
- Harris, H.C.; Morrison, D.J.; Edwards, C.A. Impact of the source of fermentable carbohydrate on SCFA production by human gut microbiota in vitro—A systematic scoping review and secondary analysis. Crit. Rev. Food Sci. Nutr. 2020, 61, 3892–3903. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Khalil, M.; De Angelis, M.; Calabrese, F.M.; D’amato, M.; Wang, D.Q.-H.; Di Ciaula, A. Intestinal Barrier and Permeability in Health, Obesity and NAFLD. Biomedicines 2021, 10, 83. [Google Scholar] [CrossRef]
- Szczuko, M.; Kikut, J.; Maciejewska, D.; Kulpa, D.; Celewicz, Z.; Ziętek, M. The Associations of SCFA with Anthropometric Parameters and Carbohydrate Metabolism in Pregnant Women. Int. J. Mol. Sci. 2020, 21, 9212. [Google Scholar] [CrossRef] [PubMed]
- Charitos, I.A.; D’Agostino, D.; Topi, S.; Bottalico, L. 40 Years of Helicobacter pylori: A Revolution in Biomedical Thought. Gastroenterol. Insights 2021, 12, 111–135. [Google Scholar] [CrossRef]
- Forslund, S.K.; Chakaroun, R.; Zimmermann-Kogadeeva, M.; Markó, L.; Aron-Wisnewsky, J.; Nielsen, T.; Moitinho-Silva, L.; Schmidt, T.S.B.; Falony, G.; Vieira-Silva, S.; et al. Combinatorial, additive and dose-dependent drug–microbiome associations. Nature 2021, 600, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Mayengbam, S.; Lambert, J.E.; Parnell, J.A.; Tunnicliffe, J.M.; Nicolucci, A.C.; Han, J.; Sturzenegger, T.; Shearer, J.; Mickiewicz, B.; Vogel, H.J.; et al. Impact of dietary fiber supplementation on modulating microbiota–host–metabolic axes in obesity. J. Nutr. Biochem. 2018, 64, 228–236. [Google Scholar] [CrossRef]
- Castro-Barquero, S.; Ruiz-León, A.M.; Sierra-Pérez, M.; Estruch, R.; Casas, R. Dietary Strategies for Metabolic Syndrome: A Comprehensive Review. Nutrients 2020, 12, 2983. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, K.E.B. Microbial Degradation of Whole-Grain Complex Carbohydrates and Impact on Short-Chain Fatty Acids and Health. Adv. Nutr. Int. Rev. J. 2015, 6, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Kumar, V.; Sangwan, P.; Pant, N.C.; Saxena, A.; Joshi, S.; Yadav, A.N. Personalized Nutrition and -Omics. Compr. Foodomics 2021, 3, 495–507. [Google Scholar] [CrossRef]
- Shyam, S.; Lee, K.X.; Tan, A.S.W.; Khoo, T.A.; Harikrishnan, S.; Lalani, S.A.; Ramadas, A. Effect of Personalized Nutrition on Dietary, Physical Activity, and Health Outcomes: A Systematic Review of Randomized Trials. Nutrients 2022, 14, 4104. [Google Scholar] [CrossRef]
- Malnick, S.D.H.; Fisher, D.; Somin, M.; Neuman, M.G. Treating the Metabolic Syndrome by Fecal Transplantation-Current Status. Biology 2021, 10, 447. [Google Scholar] [CrossRef]
- Woting, A.; Blaut, M. The Intestinal Microbiota in Metabolic Disease. Nutrients 2016, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Kuo, C.-H.; Kuo, F.-C.; Wang, Y.-K.; Hsu, W.-H.; Yu, F.-J.; Hu, H.-M.; Hsu, P.-I.; Wang, J.-Y.; Wu, D.-C. Fecal microbiota transplantation: Review and update. J. Formos. Med Assoc. 2019, 118 (Suppl. S1), S23–S31. [Google Scholar] [CrossRef]
- Ballini, A.; Charitos, I.A.; Cantore, S.; Topi, S.; Bottalico, L.; Santacroce, L. About Functional Foods: The Probiotics and Prebiotics State of Art. Antibiotics 2023, 12, 635. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Calcinaro, F.; Dionisi, S.; Marinaro, M.; Candeloro, P.; Bonato, V.; Marzotti, S.; Corneli, R.B.; Ferretti, E.; Gulino, A.; Grasso, F.; et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 2005, 48, 1565–1575. [Google Scholar] [CrossRef]
- Yadav, H.; Jain, S.; Sinha, P. Antidiabetic effect of probiotic dahi containing Lactobacillus acidophilus and Lactobacillus casei in high fructose fed rats. Nutrition 2007, 23, 62–68. [Google Scholar] [CrossRef]
- Uusitalo, U.; Liu, X.; Yang, J.; Aronsson, C.A.; Hummel, S.; Butterworth, M.; Lernmark, Å.; Rewers, M.; Hagopian, W.; She, J.-X.; et al. Association of Early Exposure of Probiotics and Islet Autoimmunity in the TEDDY Study. JAMA Pediatr. 2016, 170, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Park, J.-H.; Seok, S.-H.; Baek, M.-W.; Kim, D.-J.; Lee, K.-E.; Paek, K.-S.; Lee, Y.; Park, J.-H. Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim. Biophys. Acta 2006, 1761, 736–744. [Google Scholar] [CrossRef]
- Heeney, D.D.; Zhai, Z.; Bendiks, Z.; Barouei, J.; Martinic, A.; Slupsky, C.; Marco, M.L. Lactobacillus plantarum bacteriocin is associated with intestinal and systemic improvements in diet-induced obese mice and maintains epithelial barrier integrity in vitro. Gut Microbes 2018, 10, 382–397. [Google Scholar] [CrossRef]
- Aronsson, L.; Huang, Y.; Parini, P.; Korach-André, M.; Håkansson, J.; Gustafsson, J.Å.; Pettersson, S.; Arulampalam, V.; Rafter, J. Decreased Fat Storage by Lactobacillus Paracasei Is Associated with Increased Levels of Angiopoietin-Like 4 Protein (ANGPTL4). PLoS ONE 2010, 5, e13087. [Google Scholar] [CrossRef]
- Fåk, F.; Bäckhed, F. Lactobacillus reuteri Prevents Diet-Induced Obesity, but not Atherosclerosis, in a Strain Dependent Fashion in Apoe−/− Mice. PLoS ONE 2012, 7, e46837. [Google Scholar] [CrossRef]
- Barathikannan, K.; Chelliah, R.; Rubab, M.; Daliri, E.B.-M.; Elahi, F.; Kim, D.-H.; Agastian, P.; Oh, S.-Y.; Oh, D.H. Gut Microbiome Modulation Based on Probiotic Application for Anti-Obesity: A Review on Efficacy and Validation. Microorganisms 2019, 7, 456. [Google Scholar] [CrossRef]
- Cani, P.D.; Neyrinck, A.M.; Fava, F.; Knauf, C.; Burcelin, R.G.; Tuohy, K.M.; Gibson, G.R.; Delzenne, N.M. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 2007, 50, 2374–2383. [Google Scholar] [CrossRef]
- Ji, Y.; Kim, H.; Park, H.; Lee, J.; Yeo, S.; Yang, J.; Park, S.; Yoon, H.; Cho, G.; Franz, C.; et al. Modulation of the murine microbiome with a concomitant anti-obesity effect by Lactobacillus rhamnosus GG and Lactobacillus sakei NR28. Benef. Microbes 2012, 3, 13–22. [Google Scholar] [CrossRef]
- Angelakis, E.; Million, M.; Kankoe, S.; Lagier, J.-C.; Armougom, F.; Giorgi, R.; Raoult, D. Abnormal Weight Gain and Gut Microbiota Modifications Are Side Effects of Long-Term Doxycycline and Hydroxychloroquine Treatment. Antimicrob. Agents Chemother. 2014, 58, 3342–3347. [Google Scholar] [CrossRef]
- Luoto, R.; Kalliomäki, M.; Laitinen, K.; Isolauri, E. The impact of perinatal probiotic intervention on the development of overweight and obesity: Follow-up study from birth to 10 years. Int. J. Obes. 2010, 34, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Hua, J.; Li, Z. Probiotics improve high fat diet-induced hepatic steatosis and insulin resistance by increasing hepatic NKT cells. J. Hepatol. 2008, 49, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Mencarelli, A.; Cipriani, S.; Renga, B.; Bruno, A.; D’Amore, C.; Distrutti, E.; Fiorucci, S. VSL#3 Resets Insulin Signaling and Protects against NASH and Atherosclerosis in a Model of Genetic Dyslipidemia and Intestinal Inflammation. PLoS ONE 2012, 7, e45425. [Google Scholar] [CrossRef]
- Esposito, E.; Iacono, A.; Bianco, G.; Autore, G.; Cuzzocrea, S.; Vajro, P.; Canani, R.B.; Calignano, A.; Raso, G.M.; Meli, R. Probiotics Reduce the Inflammatory Response Induced by a High-Fat Diet in the Liver of Young Rats. J. Nutr. 2009, 139, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Kaser, A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Investig. 2011, 121, 2126–2132. [Google Scholar] [CrossRef]
- Cani, P.D.; Delzenne, N.M. The role of the gut microbiota in energy metabolism and metabolic disease. Curr. Pharm. Des. 2009, 15, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.K.; Lutz, B.; de Azua, I.R. The Microbiome and Gut Endocannabinoid System in the Regulation of Stress Responses and Metabolism. Front. Cell. Neurosci. 2022, 16, 867267. [Google Scholar] [CrossRef]
- Cuddihey, H.; MacNaughton, W.K.; Sharkey, K.A. Role of the Endocannabinoid System in the Regulation of Intestinal Homeostasis. Cell. Mol. Gastroenterol. Hepatol. 2022, 14, 947–963. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Ma, Y.; Yan, B.; Pei, W.; Wu, Q.; Ding, C.; Huang, C. The promotion mechanism of prebiotics for probiotics: A review. Front. Nutr. 2022, 9, 1000517. [Google Scholar] [CrossRef] [PubMed]
- Parnell, J.A.; Reimer, R.A. Prebiotic fibres dose-dependently increase satiety hormones and alter Bacteroidetes and Firmicutes in lean and obese JCR:LA-cp rats. Br. J. Nutr. 2011, 107, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J., Jr.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and Propionate Protect against Diet-Induced Obesity and Regulate Gut Hormones via Free Fatty Acid Receptor 3-Independent Mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef]
- Dehghan, P.; Gargari, B.P.; Asgharijafarabadi, M. Effects of High Performance Inulin Supplementation on Glycemic Status and Lipid Profile in Women with Type 2 Diabetes: A Randomized, Placebo-Controlled Clinical Trial. Health Promot. Perspect. 2013, 3, 55–63. [Google Scholar] [CrossRef][Green Version]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-Chain Fatty Acids Stimulate Glucagon-Like Peptide-1 Secretion via the G-Protein-Coupled Receptor FFAR2. Diabetes 2012, 61, 364–371. [Google Scholar] [CrossRef]
- Parnell, J.A.; Raman, M.; Rioux, K.P.; Reimer, R.A. The potential role of prebiotic fibre for treatment and management of non-alcoholic fatty liver disease and associated obesity and insulin resistance. Liver Int. 2011, 32, 701–711. [Google Scholar] [CrossRef]
- Neyrinck, A.M.; Possemiers, S.; Verstraete, W.; De Backer, F.; Cani, P.D.; Delzenne, N.M. Dietary modulation of clostridial cluster XIVa gut bacteria (Roseburia spp.) by chitin-glucan fiber improves host metabolic alterations induced by high-fat diet in mice. J. Nutr. Biochem. 2012, 23, 51–59. [Google Scholar] [CrossRef]
- Neyrinck, A.M.; Possemiers, S.; Druart, C.; Van de Wiele, T.; De Backer, F.; Cani, P.D.; Larondelle, Y.; Delzenne, N.M. Prebiotic Effects of Wheat Arabinoxylan Related to the Increase in Bifidobacteria, Roseburia and Bacteroides/Prevotella in Diet-Induced Obese Mice. PLoS ONE 2011, 6, e20944. [Google Scholar] [CrossRef] [PubMed]
- Bakker, G.J.; Zhao, J.; Herrema, H.; Nieuwdorp, M. Gut Microbiota and Energy Expenditure in Health and Obesity. J. Clin. Gastroenterol. 2015, 49, S13–S19. [Google Scholar] [CrossRef] [PubMed]
- Membrez, M.; Blancher, F.; Jaquet, M.; Bibiloni, R.; Cani, P.D.; Burcelin, R.G.; Corthesy, I.; Macé, K.; Chou, C.J. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008, 22, 2416–2426. [Google Scholar] [CrossRef] [PubMed]
- Burton, J.; Johnson, M.; Johnson, J.; Hsia, D.; Greenway, F.L.; Heiman, M.L. Addition of a Gastrointestinal Microbiome Modulator to Metformin Improves Metformin Tolerance and Fasting Glucose Levels. J. Diabetes Sci. Technol. 2015, 9, 808–814. [Google Scholar] [CrossRef]



| BPCO and Metabolome Changes during Work Rate Exercises | |
|---|---|
| Constant | Intermittent |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Charitos, I.A.; Aliani, M.; Tondo, P.; Venneri, M.; Castellana, G.; Scioscia, G.; Castellaneta, F.; Lacedonia, D.; Carone, M. Biomolecular Actions by Intestinal Endotoxemia in Metabolic Syndrome. Int. J. Mol. Sci. 2024, 25, 2841. https://doi.org/10.3390/ijms25052841
Charitos IA, Aliani M, Tondo P, Venneri M, Castellana G, Scioscia G, Castellaneta F, Lacedonia D, Carone M. Biomolecular Actions by Intestinal Endotoxemia in Metabolic Syndrome. International Journal of Molecular Sciences. 2024; 25(5):2841. https://doi.org/10.3390/ijms25052841
Chicago/Turabian StyleCharitos, Ioannis Alexandros, Maria Aliani, Pasquale Tondo, Maria Venneri, Giorgio Castellana, Giulia Scioscia, Francesca Castellaneta, Donato Lacedonia, and Mauro Carone. 2024. "Biomolecular Actions by Intestinal Endotoxemia in Metabolic Syndrome" International Journal of Molecular Sciences 25, no. 5: 2841. https://doi.org/10.3390/ijms25052841
APA StyleCharitos, I. A., Aliani, M., Tondo, P., Venneri, M., Castellana, G., Scioscia, G., Castellaneta, F., Lacedonia, D., & Carone, M. (2024). Biomolecular Actions by Intestinal Endotoxemia in Metabolic Syndrome. International Journal of Molecular Sciences, 25(5), 2841. https://doi.org/10.3390/ijms25052841






