Stabilization of G-Quadruplex Structures of the SARS-CoV-2 Genome by TMPyP4, BRACO19, and PhenDC3
Abstract
1. Introduction
2. Results
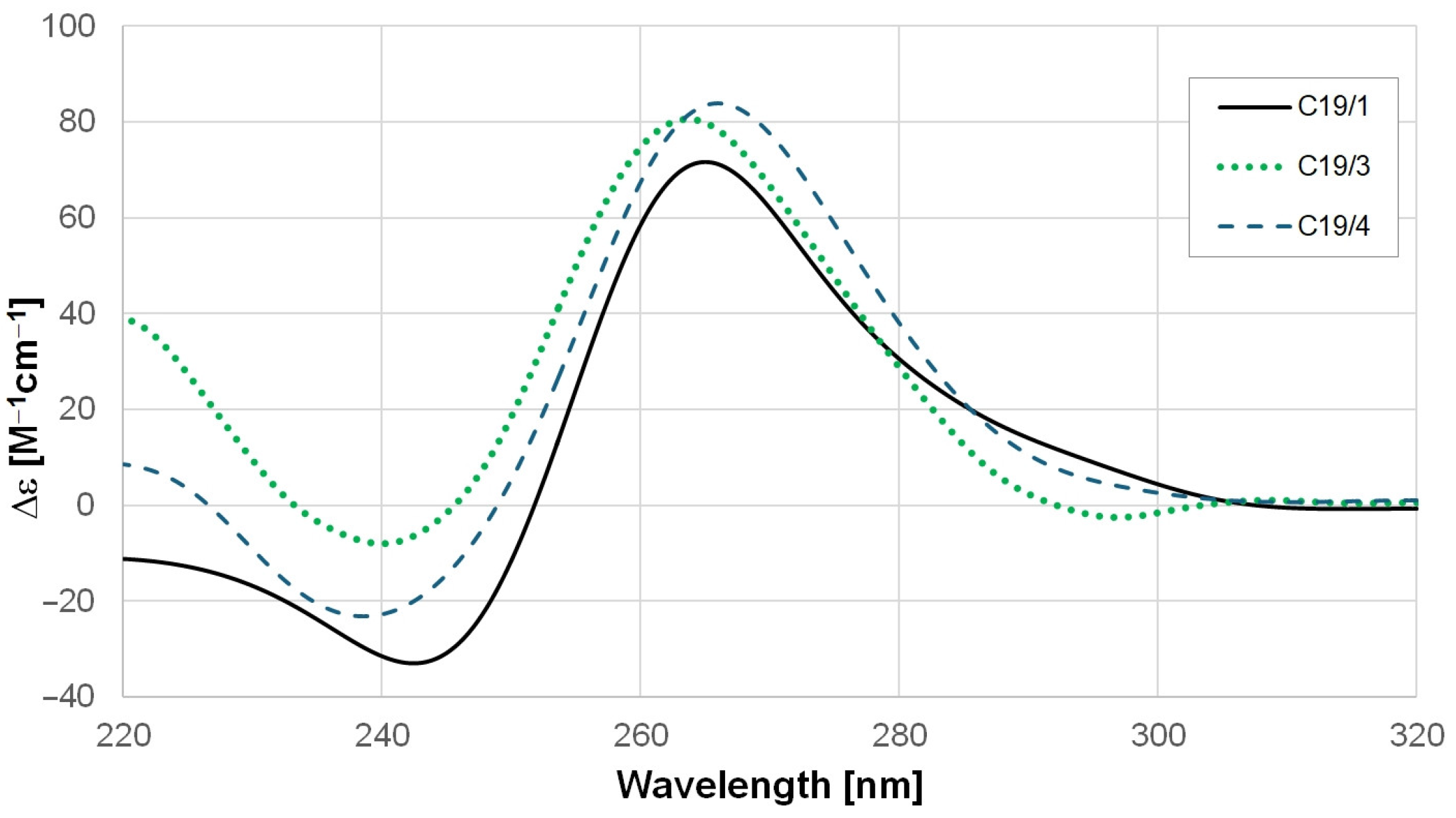
2.1. GQ-Forming Ability and Stability of the SARS-CoV-2 RNA Oligonucleotides
2.2. Effect of pH on the Stability of SARS-CoV-2 GQs
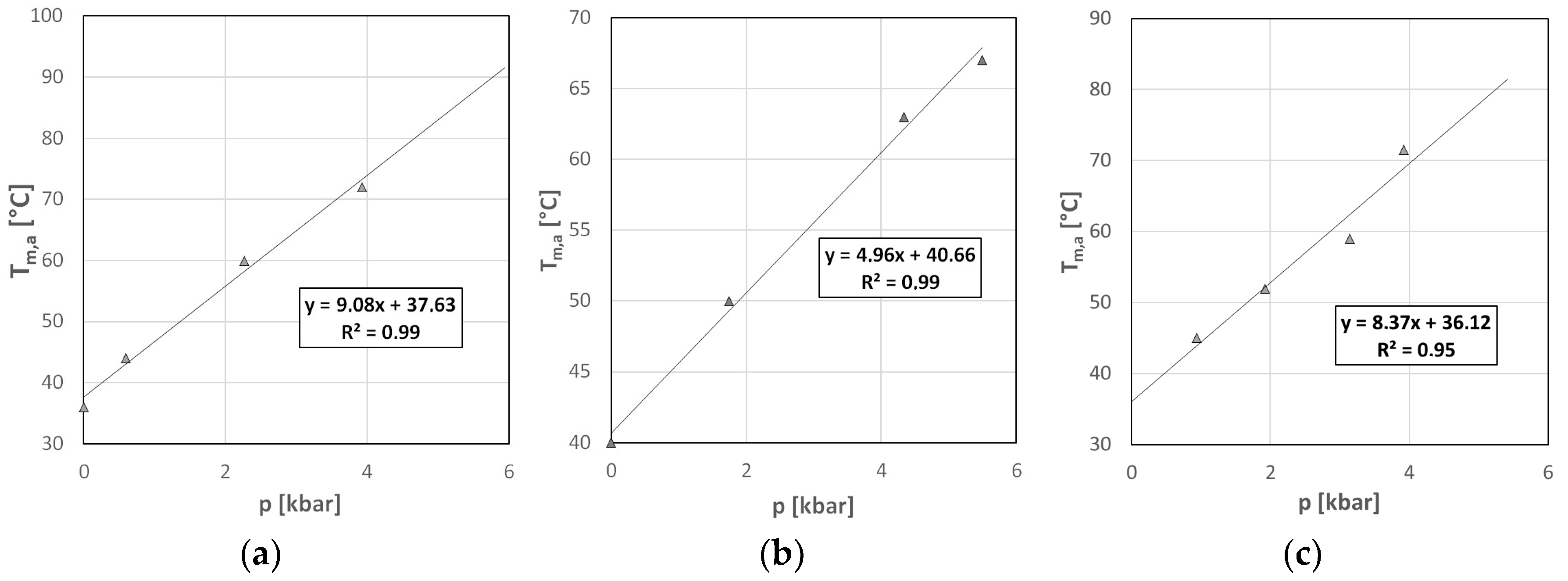
2.3. Volumetric Characterization of SARS-CoV-2 GQs
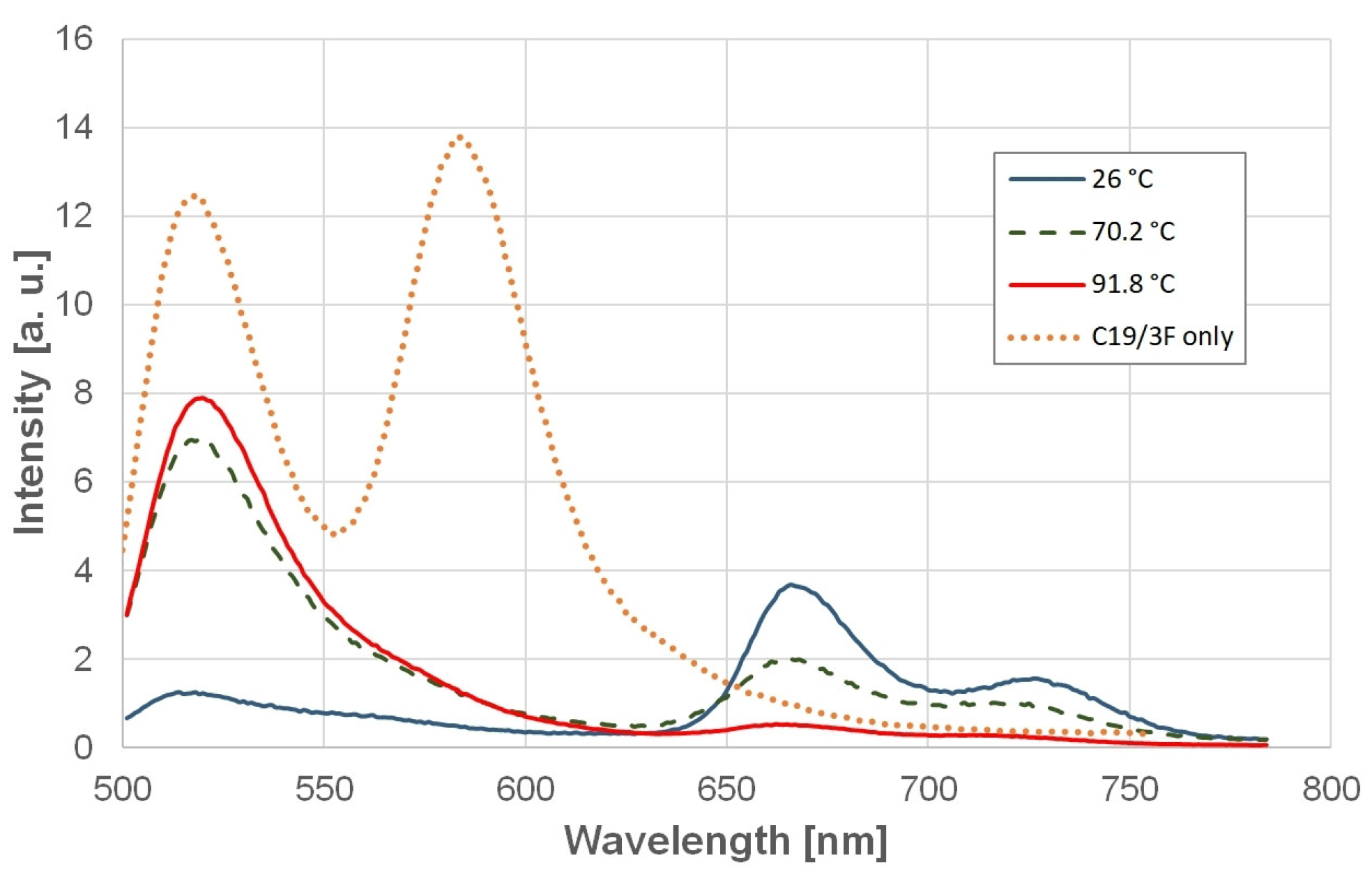
2.4. Stabilization of SARS-CoV-2 GQs by Ligands
3. Discussion
3.1. Stability of GQs Formed by the SARS-CoV-2 RNA Oligonucleotides
3.2. Effect of pH on the Stability of the GQs
3.3. Volumetric Characterization of the GQs
3.4. Stabilization of SARS-CoV-2 GQs by Ligands
4. Materials and Methods
4.1. Materials
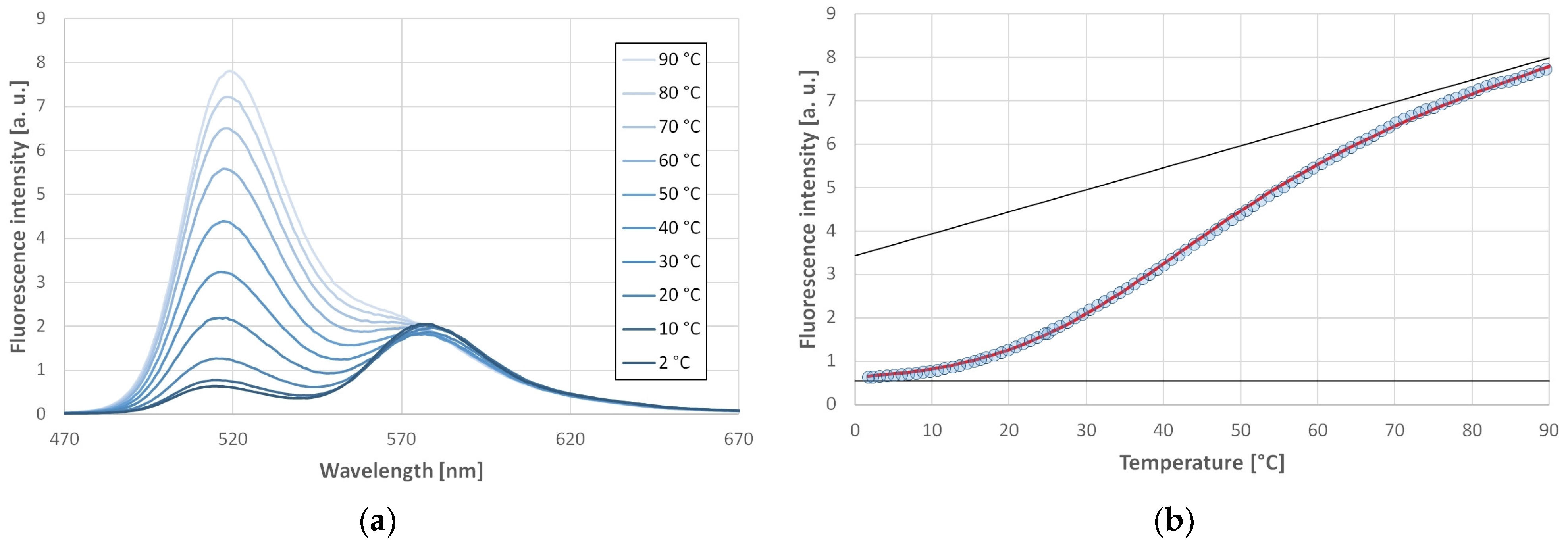
4.2. Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef] [PubMed]
- Weisz, K. A world beyond double-helical nucleic acids: The structural diversity of tetra-stranded G-quadruplexes. Chemtexts 2021, 7, 1–9. [Google Scholar] [CrossRef]
- Sugimoto, N. Noncanonical structures and their thermodynamics of DNA and RNA under molecular crowding: Beyond the Watson-Crick double helix. Int. Rev. Cell Mol. Biol. 2014, 307, 205–273. [Google Scholar] [CrossRef] [PubMed]
- Hennecker, C.; Yamout, L.; Zhang, C.; Zhao, C.; Hiraki, D.; Moitessier, N.; Mittermaier, A. Structural Polymorphism of Guanine Quadruplex-Containing Regions in Human Promoters. Int. J. Mol. Sci. 2022, 23, 16020. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef]
- Hardin, C.C.; Watson, T.; Corregan, M.; Bailey, C. Cation-Dependent Transition between the Quadruplex and Watson-Crick Hairpin Forms of D(Cgcg3gcg). Biochemistry 1992, 31, 833–841. [Google Scholar] [CrossRef]
- Venczel, E.A.; Sen, D. Parallel and Antiparallel G-DNA Structures from a Complex Telomeric Sequence. Biochemistry 1993, 32, 6220–6228. [Google Scholar] [CrossRef]
- Saintome, C.; Amrane, S.; Mergny, J.L.; Alberti, P. The exception that confirms the rule: A higher-order telomeric G-quadruplex structure more stable in sodium than in potassium. Nucleic Acids Res. 2016, 44, 2926–2935. [Google Scholar] [CrossRef]
- Molnar, O.R.; Vegh, A.; Somkuti, J.; Smeller, L. Characterization of a G-quadruplex from hepatitis B virus and its stabilization by binding TMPyP4, BRACO19 and PhenDC3. Sci. Rep. 2021, 11, 23243. [Google Scholar] [CrossRef]
- Lightfoot, H.L.; Hagen, T.; Tatum, N.J.; Hall, J. The diverse structural landscape of quadruplexes. Febs Lett. 2019, 593, 2083–2102. [Google Scholar] [CrossRef]
- Bertini, L.; Libera, V.; Ripanti, F.; Natali, F.; Paolantoni, M.; Orecchini, A.; Nucara, A.; Petrillo, C.; Comez, L.; Paciaroni, A. Polymorphism and Ligand Binding Modulate Fast Dynamics of Human Telomeric G-Quadruplexes. Int. J. Mol. Sci. 2023, 24, 4280. [Google Scholar] [CrossRef]
- Neupane, A.; Chariker, J.H.; Rouchka, E.C. Structural and Functional Classification of G-Quadruplex Families within the Human Genome. Genes 2023, 14, 645. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.X.; Bialis, T.; Jones, R.A.; Yang, D.Z. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef]
- Dai, J.X.; Carver, M.; Yang, D.Z. Polymorphism of human telomeric quadruplex structures. Biochimie 2008, 90, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Palacky, J.; Vorlickova, M.; Kejnovska, I.; Mojzes, P. Polymorphism of human telomeric quadruplex structure controlled by DNA concentration: A Raman study. Nucleic Acids Res. 2013, 41, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Jana, J.; Vianney, Y.M.; Schroder, N.; Weisz, K. Guiding the folding of G-quadruplexes through loop residue interactions. Nucleic Acids Res. 2022, 50, 7161–7175. [Google Scholar] [CrossRef]
- Guedin, A.; Gros, J.; Alberti, P.; Mergny, J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef]
- Takahashi, S.; Sugimoto, N. Volumetric contributions of loop regions of G-quadruplex DNA to the formation of the tertiary structure. Biophys. Chem. 2017, 231, 146–154. [Google Scholar] [CrossRef]
- Neidle, S.; Parkinson, G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003, 13, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1, 0041. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Neidle, S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [Google Scholar] [CrossRef]
- Mathad, R.I.; Yang, D. G-quadruplex structures and G-quadruplex-interactive compounds. Methods Mol. Biol. 2011, 735, 77–96. [Google Scholar] [CrossRef]
- Takahashi, S.; Bhowmik, S.; Sato, S.; Takenaka, S.; Sugimoto, N. Replication Control of Human Telomere G-Quadruplex DNA by G-Quadruplex Ligands Dependent on Solution Environment. Life 2022, 12, 553. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Teng, F.Y.; Jiang, Z.Z.; Guo, M.; Tan, X.Z.; Chen, F.; Xi, X.G.; Xu, Y. G-quadruplex DNA: A novel target for drug design. Cell Mol. Life Sci. 2021, 78, 6557–6583. [Google Scholar] [CrossRef]
- Shu, H.; Zhang, R.; Xiao, K.; Yang, J.; Sun, X. G-Quadruplex-Binding Proteins: Promising Targets for Drug Design. Biomolecules 2022, 12, 648. [Google Scholar] [CrossRef] [PubMed]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palu, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef]
- Tomaszewska, M.; Szabat, M.; Zielinska, K.; Kierzek, R. Identification and Structural Aspects of G-Quadruplex-Forming Sequences from the Influenza A Virus Genome. Int. J. Mol. Sci. 2021, 22, 6031. [Google Scholar] [CrossRef]
- Yadav, P.; Kim, N.; Kumari, M.; Verma, S.; Sharma, T.K.; Yadav, V.; Kumar, A. G-Quadruplex Structures in Bacteria: Biological Relevance and Potential as an Antimicrobial Target. J. Bacteriol. 2021, 203, e0057720. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Ruggiero, E.; Zanin, I.; Terreri, M.; Richter, S.N. G-Quadruplex Targeting in the Fight against Viruses: An Update. Int. J. Mol. Sci. 2021, 22, 10984. [Google Scholar] [CrossRef]
- Somkuti, J.; Molnar, O.R.; Grad, A.; Smeller, L. Pressure Perturbation Studies of Noncanonical Viral Nucleic Acid Structures. Biology 2021, 10, 1173. [Google Scholar] [CrossRef] [PubMed]
- Le, D.D.; Di Antonio, M.; Chan, L.K.M.; Balasubramanian, S. G-quadruplex ligands exhibit differential G-tetrad selectivity. Chem. Commun. 2015, 51, 8048–8050. [Google Scholar] [CrossRef]
- Oliva, R.; Mukherjee, S.; Winter, R. Unraveling the binding characteristics of small ligands to telomeric DNA by pressure modulation. Sci. Rep. 2021, 11, 9714. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef]
- Zhou, G.; Liu, X.; Li, Y.; Xu, S.; Ma, C.; Wu, X.; Cheng, Y.; Yu, Z.; Zhao, G.; Chen, Y. Telomere targeting with a novel G-quadruplex-interactive ligand BRACO-19 induces T-loop disassembly and telomerase displacement in human glioblastoma cells. Oncotarget 2016, 7, 14925–14939. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Uno, T.; Ishikawa, Y. Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg. Med. Chem. 2005, 13, 2423–2430. [Google Scholar] [CrossRef]
- Qin, G.; Zhao, C.; Liu, Y.; Zhang, C.; Yang, G.; Yang, J.; Wang, Z.; Wang, C.; Tu, C.; Guo, Z.; et al. RNA G-quadruplex formed in SARS-CoV-2 used for COVID-19 treatment in animal models. Cell Discov. 2022, 8, 86. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.P.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef]
- Perrone, R.; Butovskaya, E.; Daelemans, D.; Palu, G.; Pannecouque, C.; Richter, S.N. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014, 69, 3248–3258. [Google Scholar] [CrossRef]
- Piazza, A.; Boule, J.B.; Lopes, J.; Mingo, K.; Largy, E.; Teulade-Fichou, M.P.; Nicolas, A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010, 38, 4337–4348. [Google Scholar] [CrossRef]
- Li, Y.Y.; Dubins, D.N.; Le, D.M.N.T.; Leung, K.; Macgregor, R.B. The role of loops and cation on the volume of unfolding of G-quadruplexes related to HTel. Biophys. Chem. 2017, 231, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Nagatoishi, S.; Isono, N.; Tsumoto, K.; Sugimoto, N. Loop residues of thrombin-binding DNA aptamer impact G-quadruplex stability and thrombin binding. Biochimie 2011, 93, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Ou, Z.; Wang, Y.; Gao, Y.; Wang, X.; Qian, Y.; Li, Y.; Wang, X. Targeting human telomeric and c-myc G-quadruplexes with alkynylplatinum(II) terpyridine complexes under molecular crowding conditions. J. Inorg. Biochem. 2017, 166, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Fay, M.M.; Lyons, S.M.; Ivanov, P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017, 429, 2127–2147. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Mukherjee, S.; Manisegaran, M.; Campanile, M.; Del Vecchio, P.; Petraccone, L.; Winter, R. Binding Properties of RNA Quadruplex of SARS-CoV-2 to Berberine Compared to Telomeric DNA Quadruplex. Int. J. Mol. Sci. 2022, 23, 5690. [Google Scholar] [CrossRef]
- Panera, N.; Tozzi, A.E.; Alisi, A. The G-Quadruplex/Helicase World as a Potential Antiviral Approach Against COVID-19. Drugs 2020, 80, 941–946. [Google Scholar] [CrossRef]
- Belmonte-Reche, E.; Serrano-Chacon, I.; Gonzalez, C.; Gallo, J.; Banobre-Lopez, M. Potential G-quadruplexes and i-Motifs in the SARS-CoV-2. PLoS ONE 2021, 16, 18. [Google Scholar] [CrossRef]
- Zhai, L.Y.; Su, A.M.; Liu, J.F.; Zhao, J.J.; Xi, X.G.; Hou, X.M. Recent advances in applying G-quadruplex for SARS-CoV-2 targeting and diagnosis: A review. Int. J. Biol. Macromol. 2022, 221, 1476–1490. [Google Scholar] [CrossRef]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chem. Int. Ed. Engl. 2021, 60, 432–438. [Google Scholar] [CrossRef]
- Sathyaseelan, C.; Vijayakumar, V.; Rathinavelan, T. CD-NuSS: A Web Server for the Automated Secondary Structural Characterization of the Nucleic Acids from Circular Dichroism Spectra Using Extreme Gradient Boosting Decision-Tree, Neural Network and Kohonen Algorithms. J. Mol. Biol. 2021, 433, 166629. [Google Scholar] [CrossRef]
- Good, N.E.; Izawa, S. Hydrogen ion buffers. Methods Enzym. 1972, 24, 53–68. [Google Scholar] [CrossRef]
- Hancock, R. The crowded nucleus. Int. Rev. Cell Mol. Biol. 2014, 307, 15–26. [Google Scholar] [CrossRef]
- Takahashi, S.; Sugimoto, N. Effect of Pressure on the Stability of G-Quadruplex DNA: Thermodynamics under Crowding Conditions. Angew. Chem. Int. Ed. 2013, 52, 13774–13778. [Google Scholar] [CrossRef]
- Somkuti, J.; Adanyi, M.; Smeller, L. Self-crowding influences the temperature—pressure stability of the human telomere G-quadruplex. Biophys. Chem. 2019, 254, 106248. [Google Scholar] [CrossRef] [PubMed]
- Dolinnaya, N.G.; Ogloblina, A.M.; Yakubovskaya, M.G. Structure, properties, and biological relevance of the DNA and RNA G-quadruplexes: Overview 50 years after their discovery. Biochem. Mosc. 2016, 81, 1602–1649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.H.; Fujimoto, T.; Saxena, S.; Yu, H.Q.; Miyoshi, D.; Sugimoto, N. Monomorphic RNA G-Quadruplex and Polymorphic DNA G-Quadruplex Structures Responding to Cellular Environmental Factors. Biochemistry 2010, 49, 4554–4563. [Google Scholar] [CrossRef]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. Engl. 2018, 57, 7171–7175. [Google Scholar] [CrossRef]
- Karsisiotis, A.I.; Hessari, N.M.; Novellino, E.; Spada, G.P.; Randazzo, A.; da Silva, M.W. Topological Characterization of Nucleic Acid G-Quadruplexes by UV Absorption and Circular Dichroism. Angew. Chem. Int. Ed. 2011, 50, 10645–10648. [Google Scholar] [CrossRef]
- Smeller, L. Pressure Tuning Studies of Four-Stranded Nucleic Acid Structures. Int. J. Mol. Sci. 2023, 24, 1803. [Google Scholar] [CrossRef]
- Chalikian, T.V.; Macgregor, R.B., Jr. Volumetric Properties of Four-Stranded DNA Structures. Biology 2021, 10, 813. [Google Scholar] [CrossRef]
- Improta, R. Shedding Light on the Photophysics and Photochemistry of I-Motifs Using Quantum Mechanical Calculations. Int. J. Mol. Sci. 2023, 24, 12614. [Google Scholar] [CrossRef]
- Somkuti, J.; Molnar, O.R.; Smeller, L. Revealing unfolding steps and volume changes of human telomeric i-motif DNA. Phys. Chem. Chem. Phys. 2020, 22, 23816–23823. [Google Scholar] [CrossRef]
- Takahashi, S.; Sugimoto, N. Pressure-dependent formation of i-motif and G-quadruplex DNA structures. Phys. Chem. Chem. Phys. 2015, 17, 31004–31010. [Google Scholar] [CrossRef] [PubMed]
- Lepper, C.P.; Williams, M.A.K.; Edwards, P.J.B.; Filichev, V.V.; Jameson, G.B. Effects of Pressure and pH on the Physical Stability of an I-Motif DNA Structure. Chemphyschem 2019, 20, 1567–1571. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.Y.; Shek, Y.L.; Amiri, A.; Dubins, D.N.; Heerklotz, H.; Macgregor, R.B., Jr.; Chalikian, T.V. Volumetric characterization of sodium-induced G-quadruplex formation. J. Am. Chem. Soc. 2011, 133, 4518–4526. [Google Scholar] [CrossRef] [PubMed]
- Heremans, K.; Smeller, L. Protein structure and dynamics at high pressure. Biochim. Et Biophys. Acta-Protein Struct. Mol. Enzymol. 1998, 1386, 353–370. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, Y.; Itoh, T. Reaction Volume of Protonic Ionization for Buffering Agents-Prediction of Pressure Dependence of pH and pOH. J. Solut. Chem. 1987, 16, 715–725. [Google Scholar] [CrossRef]
- Artusi, S.; Ruggiero, E.; Nadai, M.; Tosoni, B.; Perrone, R.; Ferino, A.; Zanin, I.; Xodo, L.; Flamand, L.; Richter, S.N. Antiviral Activity of the G-Quadruplex Ligand TMPyP4 against Herpes Simplex Virus-1. Viruses 2021, 13, 196. [Google Scholar] [CrossRef]
- Martino, L.; Pagano, B.; Fotticchia, I.; Neidle, S.; Giancola, C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B 2009, 113, 14779–14786. [Google Scholar] [CrossRef]
- Oblak, D.; Hadzi, S.; Podlipnik, C.; Lah, J. Binding-Induced Diversity of a Human Telomeric G-Quadruplex Stability Phase Space. Pharmaceuticals 2022, 15, 1150. [Google Scholar] [CrossRef] [PubMed]
- Dawson, R.M.C. Data for Biochemical Research, 3rd ed.; Clarendon Press: Oxford, UK, 1986; p. xii; 580p. [Google Scholar]
- Ellis, K.J.; Morrison, J.F. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzym. 1982, 87, 405–426. [Google Scholar] [CrossRef]
- Molnar, O.R.; Somkuti, J.; Smeller, L. Negative volume changes of human G-quadruplexes at unfolding. Heliyon 2020, 6, e05702. [Google Scholar] [CrossRef] [PubMed]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Smeller, L. How precise are the positions of computer-determined peaks? Appl. Spectrosc. 1998, 52, 1623–1626. [Google Scholar] [CrossRef]
- Forman, R.A.; Piermarini, G.J.; Dean, B.J.; Stanely, B. Pressure Measurement Made by the Utilization of Ruby Sharp-Line Luminescence. Science 1972, 176, 284–285. [Google Scholar] [CrossRef]
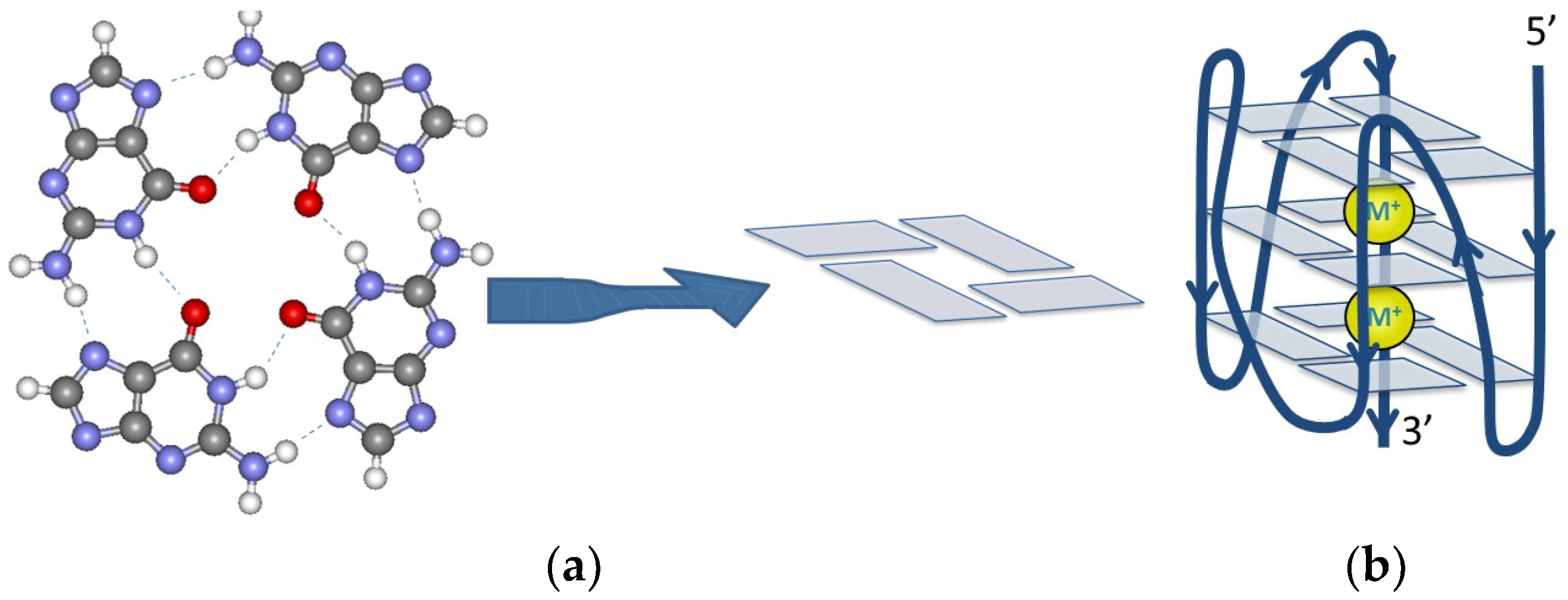
| Name | Sequence (5′–3′) | Length | Position | Gene |
|---|---|---|---|---|
| C19/1 | GG UAU GUG GAA AGG UUA UGG | 20 | 13385 | ORF1ab |
| C19/1F | Fam-CGG UAU GUG GAA AGG UUA UGG C-Tamra | 22 | 13384 | |
| C19/3 | GG UGU UGU UGG AGA AGG UUC CGA AGG | 26 | 1574 | ORF1ab |
| C19/3F | Fam-AGG UGU UGU UGG AGA AGG UUC CGA AGG U-Tamra | 28 | 1573 | |
| C19/4 | GG CUG GCA AUG GCG G | 15 | 28903 | N |
| C19/4F | Fam-UGG CUG GCA AUG GCG GU-Tamra | 17 | 28902 |
| Name | Cation | Cation Conc. (mM) | Tm (°C) | ∆H (kJ/mol) |
|---|---|---|---|---|
| C19/1F | K+ | 140 | 37.0 ± 1.0 | 54.4 ± 0.6 |
| K+ | 170 | 36.6 ± 0.4 | 95.6 ± 2.0 | |
| Na+ | 170 | 37.5 ± 0.1 | 63.9 ± 0.8 | |
| C19/3F | K+ | 140 | 47.6 ± 0.4 | 107 ± 3 |
| K+ | 170 | 49.6 ± 0.7 | 119 ± 6 | |
| Na+ | 170 | 53.8 ± 0.5 | 106 ± 4 | |
| C19/4F | K+ | 140 | 61.9 ± 0.3 | 129 ± 2 |
| K+ | 170 | 57.9 ± 0.3 | 151 ± 5 | |
| Na+ | 170 | 53.7 ± 0.9 | 117 ± 5 |
| C19/1F | C19/3F | C19/4F | |||
|---|---|---|---|---|---|
| pH | Tm [°C] | pH | Tm [°C] | pH | Tm [°C] |
| 5.91 | 53.7 | 5.91 | 51.7 | 5.83 | 58.6 |
| 7.35 | 40.2 | 7.32 | 49.0 | 7.24 | 58.1 |
| 8.36 | 36.6 | 7.98 | 50.3 | 7.82 | 55.9 |
| 10.02 | 29.8 | 9.89 | 44.8 | 9.83 | 51.1 |
| Determined Parameter | Oligonucleotide | Unit | ||
|---|---|---|---|---|
| C19/1F | C19/3F | C19/4F | ||
| (dT/dp)exp | 9.1 ± 2.9 | 5.0 ± 1.1 | 8.4 ± 5.8 | °C/kbar |
| Ligand | Oligonucleotide | |||||
|---|---|---|---|---|---|---|
| C19/1F | C19/3F | C19/4F | ||||
| Tm [°C] | ∆Tm [°C] | Tm [°C] | ∆Tm [°C] | Tm [°C] | ∆Tm [°C] | |
| none | 37.0 | - | 47.6 | 61.9 | ||
| TMPyP4 | >80 | >40 | >90 | >40 | >90 | >40 |
| BRACO19 | 41.7 | 5.3 | 48.5 | 1 | 55.4 | −6.5 |
| PhenDC3 | 68.5 | 31.5 | 46.6 | −1 | 68.7 | 6.8 |
| Parameter | Oligonucleotide | Unit | ||
|---|---|---|---|---|
| C19/1F | C19/3F | C19/4F | ||
| measured dT/dp | 9.08 | 4.96 | 8.37 | °C/kbar |
| pH-corrected ∂T/∂p | 10.8 | 5.46 | 8.94 | °C/kbar |
| ∆Vu | 19 ± 7 | 21 ± 6 | 36 ± 26 | cm3/mol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cervenak, M.; Molnár, O.R.; Horváth, P.; Smeller, L. Stabilization of G-Quadruplex Structures of the SARS-CoV-2 Genome by TMPyP4, BRACO19, and PhenDC3. Int. J. Mol. Sci. 2024, 25, 2482. https://doi.org/10.3390/ijms25052482
Cervenak M, Molnár OR, Horváth P, Smeller L. Stabilization of G-Quadruplex Structures of the SARS-CoV-2 Genome by TMPyP4, BRACO19, and PhenDC3. International Journal of Molecular Sciences. 2024; 25(5):2482. https://doi.org/10.3390/ijms25052482
Chicago/Turabian StyleCervenak, Miklós, Orsolya Réka Molnár, Péter Horváth, and László Smeller. 2024. "Stabilization of G-Quadruplex Structures of the SARS-CoV-2 Genome by TMPyP4, BRACO19, and PhenDC3" International Journal of Molecular Sciences 25, no. 5: 2482. https://doi.org/10.3390/ijms25052482
APA StyleCervenak, M., Molnár, O. R., Horváth, P., & Smeller, L. (2024). Stabilization of G-Quadruplex Structures of the SARS-CoV-2 Genome by TMPyP4, BRACO19, and PhenDC3. International Journal of Molecular Sciences, 25(5), 2482. https://doi.org/10.3390/ijms25052482






