Cloning and Characterization of Chitin Deacetylase from Euphausia superba
Abstract
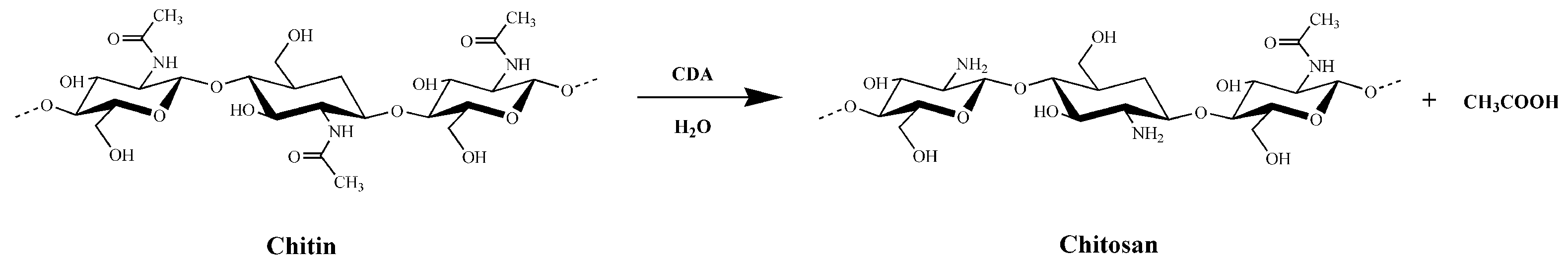
1. Introduction
2. Results
2.1. Cloning and Bioinformatics Analysis
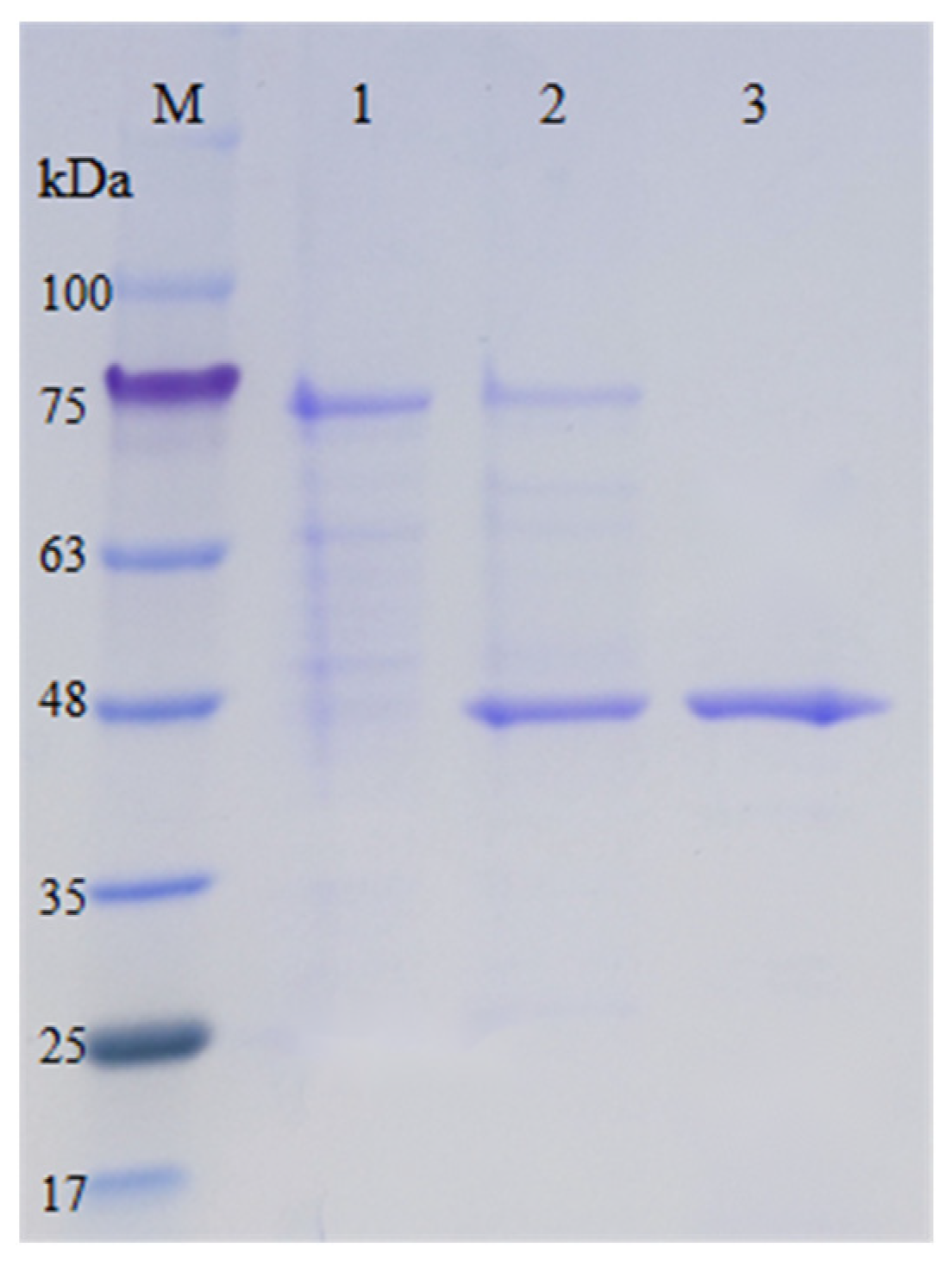
2.2. Protein Expression and Purification
2.3. Effects of Temperature and pH on the Activity and Stability of EsCDA-9k
2.4. Effects of Chemical Reagents on EsCDA-9k Enzyme Activity
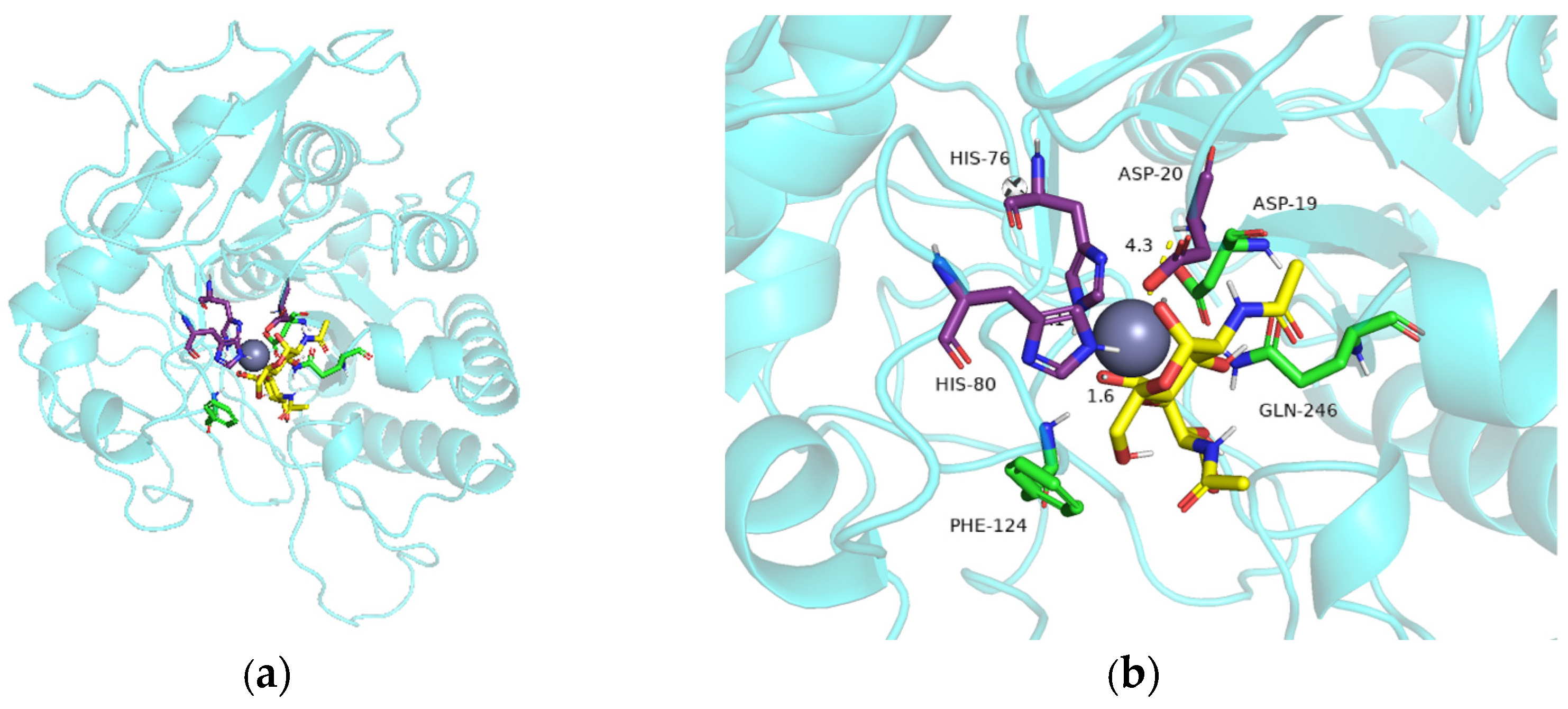
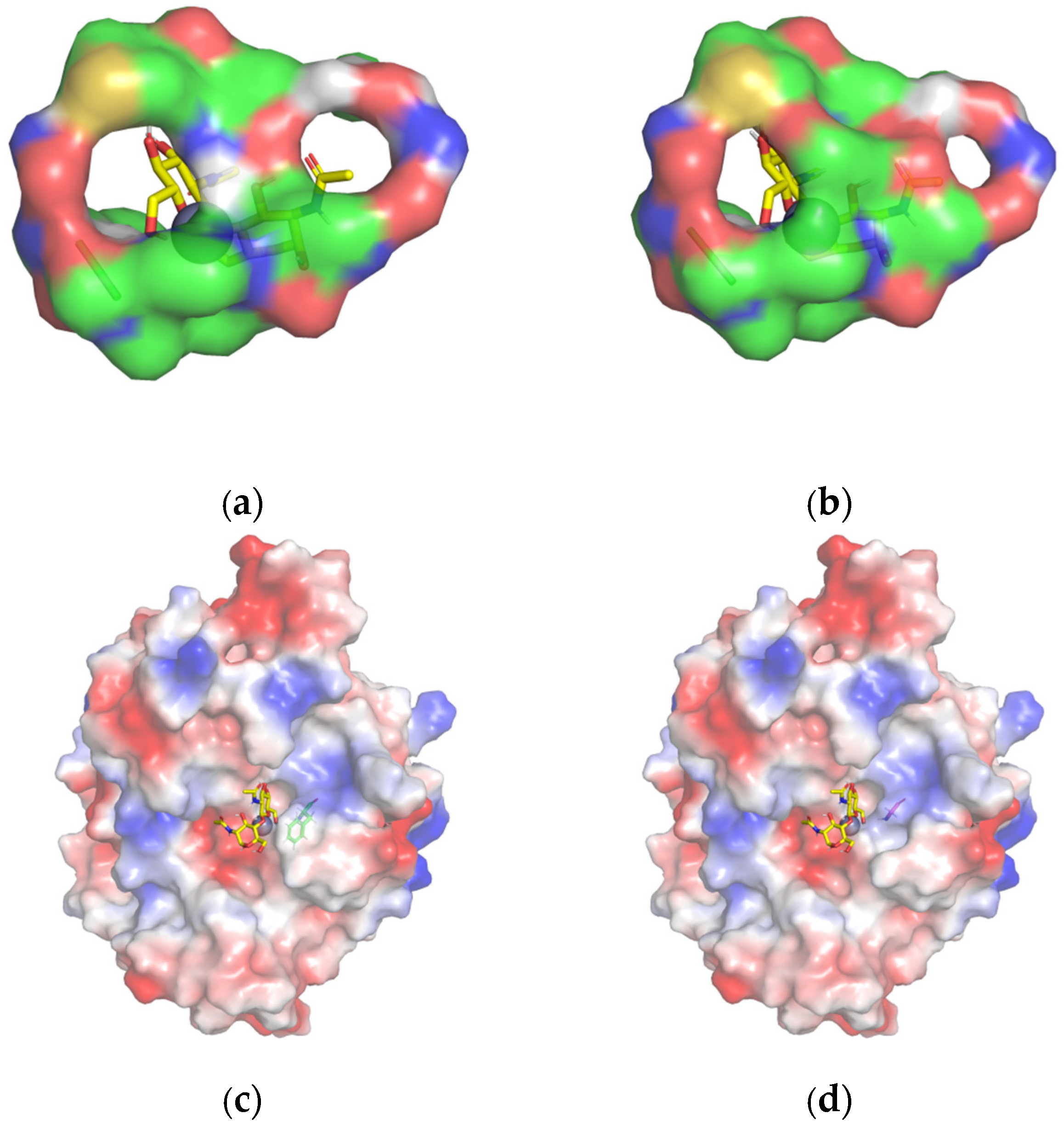
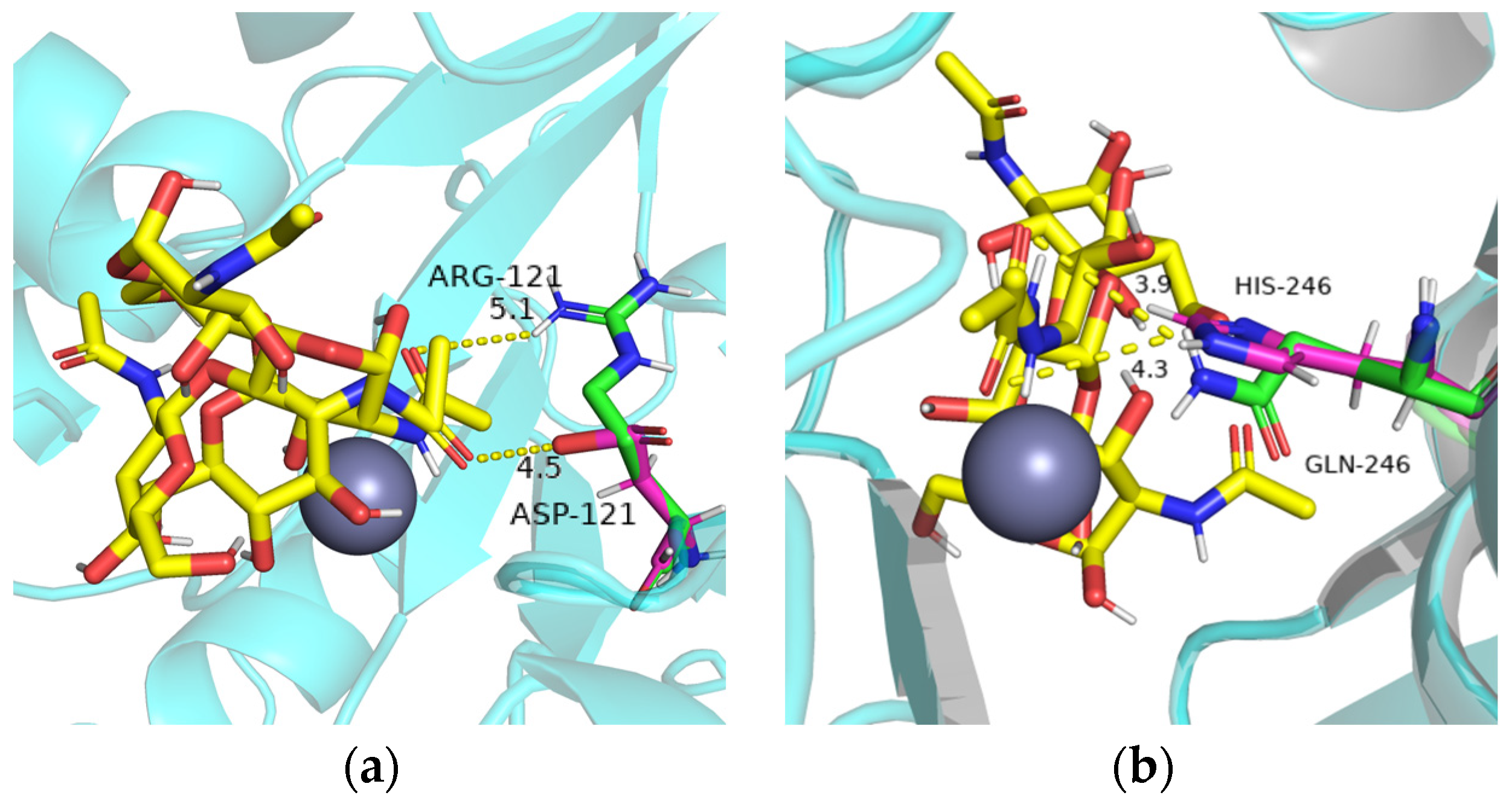
2.5. Site-Directed Mutagenesis and Comparison of Enzyme Activity of Mutants
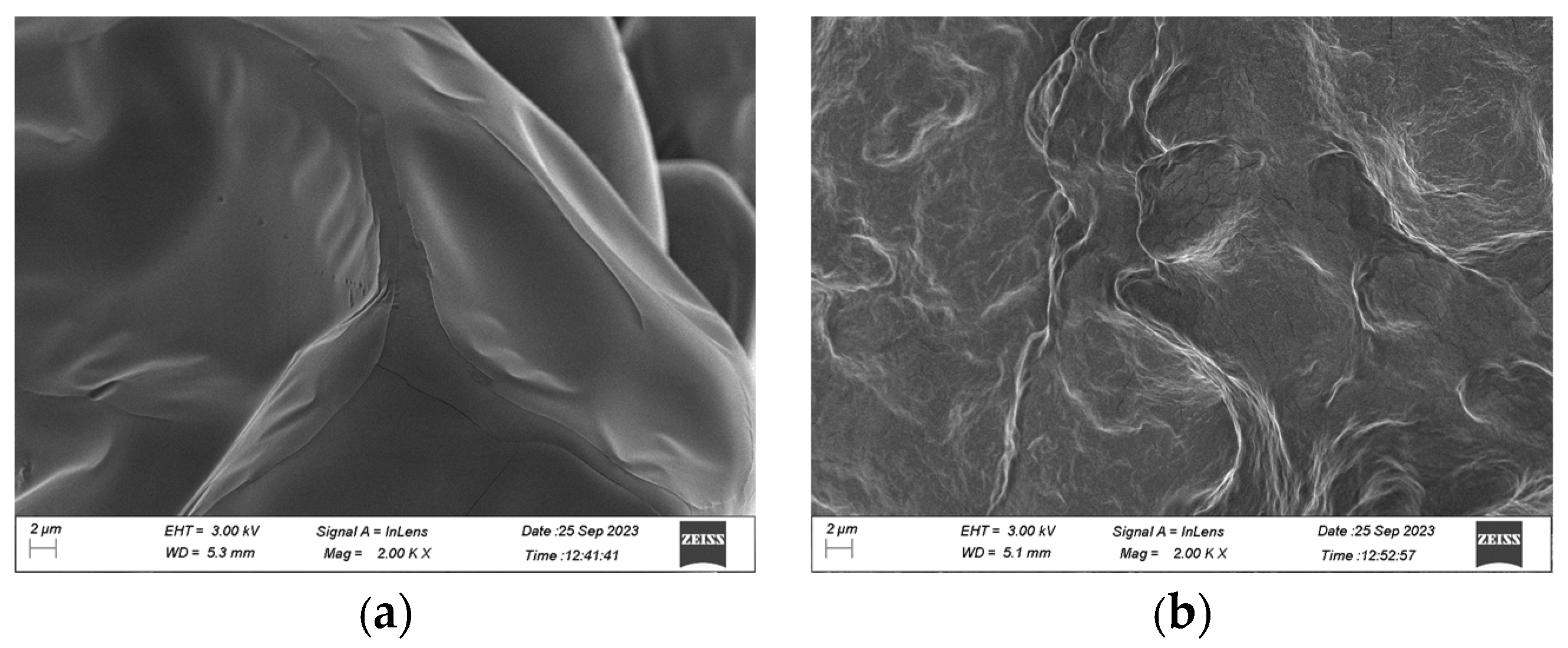
2.6. Scanning Electron Microscope Analysis
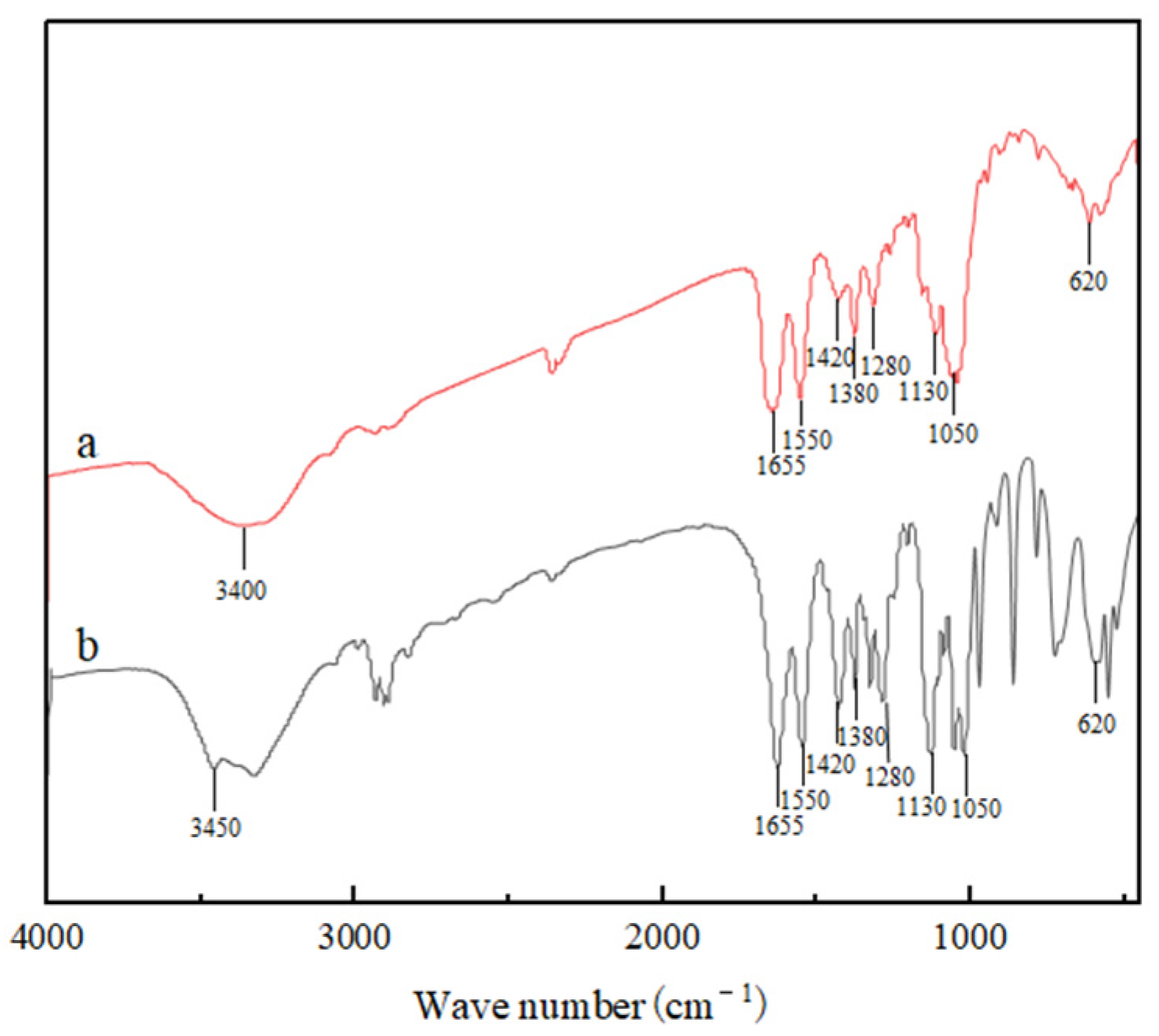
2.7. Fourier Transform-Infrared (FT-IR) Spectroscopy
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Construction of Recombinant Plasmid
4.3. Protein Expression and Purification
4.4. Activity Assay
4.5. Biochemical Characterization
4.6. Homology Modeling and Molecular Docking
4.7. Site-Directed Mutagenesis
4.8. Scanning Electron Microscope Characterization
4.9. Determination of the Degree of Deacetylation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Younes, I.; Rinaudo, M. Chitin and chitosan preparation from marine sources. Structure, properties and applications. Mar. Drugs 2015, 13, 1133–1174. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.Q.; Gozaydin, G.; Yang, H.Y.; Ning, W.; Han, X.; Poon, N.Y. Upcycling chitin-containing waste into organonitrogen chemicals via an integrated process. Proc. Natl. Acad. Sci. USA 2020, 117, 7719–7728. [Google Scholar] [CrossRef] [PubMed]
- Tharanathan, R.N.; Kittur, F.S. Chitin the undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 2003, 43, 61–87. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.L.; Wang, Q.; Sun, J.N.; Mao, X.Z. Expression and Molecular Modification of Chitin Deacetylase from Streptomyces bacillaris. Molecules 2022, 28, 113. [Google Scholar] [CrossRef]
- Sudatta, B.P.; Sugumar, V.; Varma, R.; Nigariga, P. Extraction, characterization and antimicrobial activity of chitosan from pen shell, Pinna bicolor. Int. J. Biol. Macromol. 2020, 163, 423–430. [Google Scholar] [CrossRef]
- Ding, Z.W.; Ahmed, S.; Hang, J.H.; Mi, H.; Hou, X.Y.; Yang, G. Rationally engineered chitin deacetylase from Arthrobacter sp. AW19M34-1 with improved catalytic activity toward crystalline chitin. Carbohydr. Polym. 2021, 274, 118637. [Google Scholar] [CrossRef]
- Assis, C.F.; Araujo, N.K.; Pagnoncelli, M.G.; Silva Pedrini, M.R.; Macedo, G.R.; Santos, E.S. Chitooligosaccharides enzymatic production by Metarhizium anisopliae. Bioprocess Biosyst. Eng. 2010, 33, 893–899. [Google Scholar] [CrossRef]
- Liaqat, F.; Eltem, R. Chitooligosaccharides and their biological activities: A comprehensive review. Carbohydr. Polym. 2018, 184, 243–259. [Google Scholar] [CrossRef]
- Chai, J.L.; Hang, J.B.; Zhang, C.G.; Yang, J.; Wang, S.J.; Liu, S.; Fang, Y.W. Purification and characterization of chitin deacetylase active on insoluble chitin from Nitratireductor aquimarinus MCDA3-3. Int. J. Biol. Macromol. 2020, 152, 922–929. [Google Scholar] [CrossRef]
- Chen, X.; Yang, H.Y.; Zhong, Z.Y.; Yan, N. Base-catalysed, one-step mechanochemical conversion of chitin and shrimp shells into low molecular weight chitosan. Green Chem. 2017, 19, 2783–2792. [Google Scholar] [CrossRef]
- Kaczmarek, M.B.; Struszczyk-Swita, K.; Li, X.; Szczesna-Antczak, M.; Daroch, M. Enzymatic Modifications of Chitin, Chitosan, and Chitooligosaccharides. Front. Bioeng. Biotechnol. 2019, 7, 243. [Google Scholar] [CrossRef]
- Pawaskar, G.M.; Pangannaya, S.; Raval, K.; Trivedi, D.R.; Raval, R. Screening of chitin deacetylase producing microbes from marine source using a novel receptor on agar plate. Int. J. Biol. Macromol. 2019, 131, 716–720. [Google Scholar] [CrossRef]
- Hu, Z.; Lu, S.T.; Cheng, Y.; Kong, S.Z.; Li, S.D.; Li, C.P.; Yang, L. Investigation of the Effects of Molecular Parameters on the Hemostatic Properties of Chitosan. Molecules 2018, 23, 3147. [Google Scholar] [CrossRef]
- Raval, R.; Simsa, R.; Raval, K. Expression studies of Bacillus licheniformis chitin deacetylase in E. coli Rosetta cells. Int. J. Biol. Macromol. 2017, 104, 1692–1696. [Google Scholar] [CrossRef]
- Landwehr, S.; Melcher, R.L.; Kolkenbrock, S.; Moerschbacher, B.M. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 2016, 6, 38018. [Google Scholar] [CrossRef] [PubMed]
- Aktuganov, G.E.; Melent’ev, A.I. Specific features of chitosan depolymerization by chitinases, chitosanases, and nonspecific enzymes in the production of bioactive chitooligosaccharides. Appl. Biochem. Microbiol. 2017, 53, 611–627. [Google Scholar] [CrossRef]
- Ma, Q.Y.; Gao, X.Z.; Bi, X.Y.; Tu, L.N.; Xia, M.L.; Shen, Y.B.; Wang, M. Isolation, characterisation, and genome sequencing of Rhodococcus equi: A novel strain producing chitin deacetylase. Sci. Rep. 2020, 10, 4329. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Ito, E. A pathway of chitosan formation in Mucor rouxii: Enzymatic deacetylation of chitin. Biochem. Biophys. Res. Commun. 1974, 56, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.D.; Katsumoto, T.; Onodera, K. Purification and Characterization of Chitin Deacetylase from Absidia coerulea. J. Biochem. 1995, 117, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, B.; Blondeau, K.; Stevens, W.F.; Hegarat, F.L. Expression of chitin deacetylase from Colletotrichum lindemuthianum in Pichia pastoris: Purification and characterization. Protein Expr. Purif. 2004, 38, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Martinou, A.; Koutsioulis, D.; Bouriotis, V. Expression, purification, and characterization of a cobalt-activated chitin deacetylase (Cda2p) from Saccharomyces cerevisiae. Protein Expr. Purif. 2002, 24, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, J.H.; Du, Y.M.; Fan, L.H.; Qiu, Y.L.; Li, J.; Kennedy, J.F. Purification and characterization of chitin deacetylase from Scopulariopsis brevicaulis. Carbohydr. Polym. 2006, 65, 211–217. [Google Scholar] [CrossRef]
- Gauthier, C.; Clerisse, F.; Dommes, J.; Jaspar, M.F. Characterization and cloning of chitin deacetylases from Rhizopus circinans. Protein Expr. Purif. 2008, 59, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Zhao, Y.; Oh, K.T.; Nguyen, V.N.; Park, R.D. Enzymatic deacetylation of chitin by extracellular chitin deacetylase from a newly screened Mortierella sp. DY-52. J. Microbiol. Biotechnol. 2008, 18, 759–766. [Google Scholar] [PubMed]
- Yamada, M.; Kurano, M.; Inatomi, S.; Taguchi, G.; Okazaki, M.; Shimosaka, M. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol. Lett. 2008, 289, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, K.P.; Panes, V.A. Molecular cloning and expression of chitin deacetylase 1 gene from the gills of Penaeus monodon (black tiger shrimp). Fish Shellfish Immunol. 2016, 55, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Y.; Zhang, J.Q.; Xiang, J.H. Immune function against bacteria of chitin deacetylase 1 (EcCDA1) from Exopalaemon carinicauda. Fish Shellfish. Immunol. 2018, 75, 115–123. [Google Scholar] [CrossRef]
- Li, X.G.; Diao, P.Y.; Chu, J.D.; Zhou, G.; Zhou, J.; Lin, H.; Chen, J.H.; Zeng, Q.F. Molecular characterization and function of chitin deacetylase-like from the Chinese mitten crab, Eriocheir sinensis. Comp. Biochem. Physiol. 2021, 256, 110612. [Google Scholar] [CrossRef]
- Wang, L.Z.; Xue, C.H.; Wang, Y.M.; Yang, B. Extraction of proteins with low fluoride level from Antarctic krill (Euphausia superba) and their composition analysis. J. Agric. Food Chem. 2011, 59, 6108–6112. [Google Scholar] [CrossRef]
- Schaafsma, F.L.; David, C.L.; Kohlbach, D.; Ehrlich, J.; Castellani, G.; Lange, B.A. Allometric relationships of ecologically important Antarctic and Arctic zooplankton and fish species. Polar Biol. 2022, 45, 203–224. [Google Scholar] [CrossRef]
- Cavan, E.L.; Belcher, A.; Atkinson, A.; Hill, S.L.; Kawaguchi, S.; McCormack, S.; Meyer, B.; Nicol, S.; Ratnarajah, L.; Schmidt, K.; et al. The importance of Antarctic krill in biogeochemical cycles. Nat. Commun. 2019, 10, 4742. [Google Scholar] [CrossRef] [PubMed]
- Pauly, D.; Christensen, V.J. Primary production required to sustain global fisheries. Nature 1995, 374, 255–257. [Google Scholar] [CrossRef]
- Wang, R.; Song, P.; Li, Y. An Integrated, Size-Structured Stock Assessment of Antarctic Krill, Euphausia superba. Front. Mar. Sci. 2021, 8, 710544. [Google Scholar] [CrossRef]
- Bondad, M.G.; Subasinghe, R.P.; Josupeit, H.; Cai, J.N.; Zhou, X.W. The role of crustacean fisheries and aquaculture in global food security: Past, present and future. J. Invertebr. Pathol. 2012, 110, 158–165. [Google Scholar] [CrossRef]
- Bao, J.Q.; Chen, L.; Liu, T.T. Dandelion polysaccharide suppresses lipid oxidation in Antarctic krill (Euphausia superba). Int. J. Biol. Macromol. 2019, 133, 1164–1167. [Google Scholar] [CrossRef]
- Alfonso, C.; Nuero, O.M.; Santamaría, F.; Reyes, F. Purification of a heat-stable chitin deacetylase from Aspergillus nidulans and its role in cell wall degradation. Curr. Microbiol. 1995, 30, 49–54. [Google Scholar] [CrossRef]
- Grasso, G.; Satriano, C.; Milardi, D.J. A neglected modulator of insulin-degrading enzyme activity and conformation: The pH. Biophys. Chem. 2015, 203, 33–40. [Google Scholar] [CrossRef]
- Nahar, P.; Ghormade, V.; Deshpande, M.V. The extracellular constitutive production of chitin deacetylase in Metarhizium anisopliae: Possible edge to entomopathogenic fungi in the biological control of insect pests. J. Invertebr. Pathol. 2004, 85, 80–88. [Google Scholar] [CrossRef]
- Caufrier, F.; Martinou, A.; Dupont, C.; Bouriotis, V.J. Carbohydrate esterase family 4 enzymes: Substrate specificity. Carbohydr. Res. 2003, 338, 687–692. [Google Scholar] [CrossRef]
- Pareek, N.; Vivekanand, V.; Saroj, S. Purification and characterization of chitin deacetylase from Penicillium oxalicum SAEM-51. Carbohydr. Polym. 2012, 87, 1091–1097. [Google Scholar] [CrossRef]
- Roman, D.L.; Roman, M.; Sletta, H.; Ostafe, V.; Isvoran, A.J. Assessment of the properties of chitin deacetylases showing different enzymatic action patterns. J. Mol. Graphics Modell. 2019, 88, 41–48. [Google Scholar] [CrossRef]
- El-Sherbiny, I.J. Synthesis, characterization and metal uptake capacity of a new carboxymethyl chitosan derivative. Eur. Polym. J. 2009, 45, 199–210. [Google Scholar] [CrossRef]
- Velde, K.V.; Kiekens, P.J. Structure analysis and degree of substitution of chitin, chitosan and dibutyrylchitin by FT-IR spectroscopy and solid state 13C NMR. Carbohydr. Polym. 2004, 58, 409–416. [Google Scholar] [CrossRef]
- Ngo, T.H.; Ngo, D. Effects of low–frequency ultrasound on heterogenous deacetylation of chitin. Int. J. Biol. Macromol. 2017, 104, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Y.; Zhao, Y.; Zhang, H.D.; Wang, W.X.; Cong, H.H.; Yin, H. Characterization of the specific mode of action of a chitin deacetylase and separation of the partially acetylated chitosan oligosaccharides. Mar. Drugs 2019, 17, 74. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Niu, X.; Guo, X.L. Heterologous expression, characterization and possible functions of the chitin deacetylases, Cda1 and Cda2, from Mushroom Coprinopsis cinerea. Glycobiology 2018, 28, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gay, L.M.; Tuveng, T.R.; Agger, J.W. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017, 7, 1746. [Google Scholar] [CrossRef]
- Andrés, E.; Albesa-Jové, D.; Biarnés, X.; Guerin, M.E.; Planas, A. Structural basis of chitin oligosaccharide deacetylation. Angew. Chem. 2014, 126, 7002–7007. [Google Scholar] [CrossRef]
- Deng, J.J.; Shi, D.; Mao, H.H.; Li, Z.W.; Liang, S.; Ke, Y.; Luo, X.C. Heterologous expression and characterization of an antifungal chitinase (Chit46) from Trichoderma harzianum GIM 3.442 and its application in colloidal chitin conversion. Int. J. Biol. Macromol. 2019, 134, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhou, Y.; Qu, M.; Qiu, Y.; Guo, X.; Zhang, Y. Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family. J. Biol. Chem. 2019, 294, 5774–5783. [Google Scholar] [CrossRef]
- Seeliger, D.; Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Ling, K.S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes 2007, 34, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Rath, P.; Kumar, A.S.; Tiwari, T.N. Extraction and characterization of chitin and chitosan from fishery waste by chemical method. Environ. Technol. Innov. 2015, 3, 77–85. [Google Scholar] [CrossRef]
- Kasaai, M.R. A review of several reported procedures to determine the degree of N-acetylation for chitin and chitosan using infrared spectroscopy. Carbohydr. Polym. 2008, 71, 497–508. [Google Scholar] [CrossRef]



| Total Activity (U) | Protein Concentration (mg/mL) | Specific Activity (U/mg) | Purification Fold | Activity Yield (100%) | |
|---|---|---|---|---|---|
| Crude enzyme | 161.7 | 8.6 | 18.8 | 1 | 100 |
| Ni2+-NTA | 137.8 | 5.7 | 24.2 | 1.3 | 85.2 |
| Enzyme Activity (U/mg) | Relative Enzyme Activity | |
|---|---|---|
| Wild-type | 24.2 ± 0.22 | 100% |
| F18G | 38.7 ± 0.64 | 160% |
| R121D | 30.1 ± 0.67 | 124% |
| F124G | 34.2 ± 0.42 | 141% |
| Q246H | 32.6 ± 0.86 | 135% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Tan, J.; Zou, H.; Wang, F.; Xu, J. Cloning and Characterization of Chitin Deacetylase from Euphausia superba. Int. J. Mol. Sci. 2024, 25, 2075. https://doi.org/10.3390/ijms25042075
Wang X, Tan J, Zou H, Wang F, Xu J. Cloning and Characterization of Chitin Deacetylase from Euphausia superba. International Journal of Molecular Sciences. 2024; 25(4):2075. https://doi.org/10.3390/ijms25042075
Chicago/Turabian StyleWang, Xutong, Jiahao Tan, Huaying Zou, Fang Wang, and Jiakun Xu. 2024. "Cloning and Characterization of Chitin Deacetylase from Euphausia superba" International Journal of Molecular Sciences 25, no. 4: 2075. https://doi.org/10.3390/ijms25042075
APA StyleWang, X., Tan, J., Zou, H., Wang, F., & Xu, J. (2024). Cloning and Characterization of Chitin Deacetylase from Euphausia superba. International Journal of Molecular Sciences, 25(4), 2075. https://doi.org/10.3390/ijms25042075






