Comparative Transcriptome Analysis Reveals the Genes and Pathways Related to Wheat Root Hair Length
Abstract
1. Introduction
2. Results
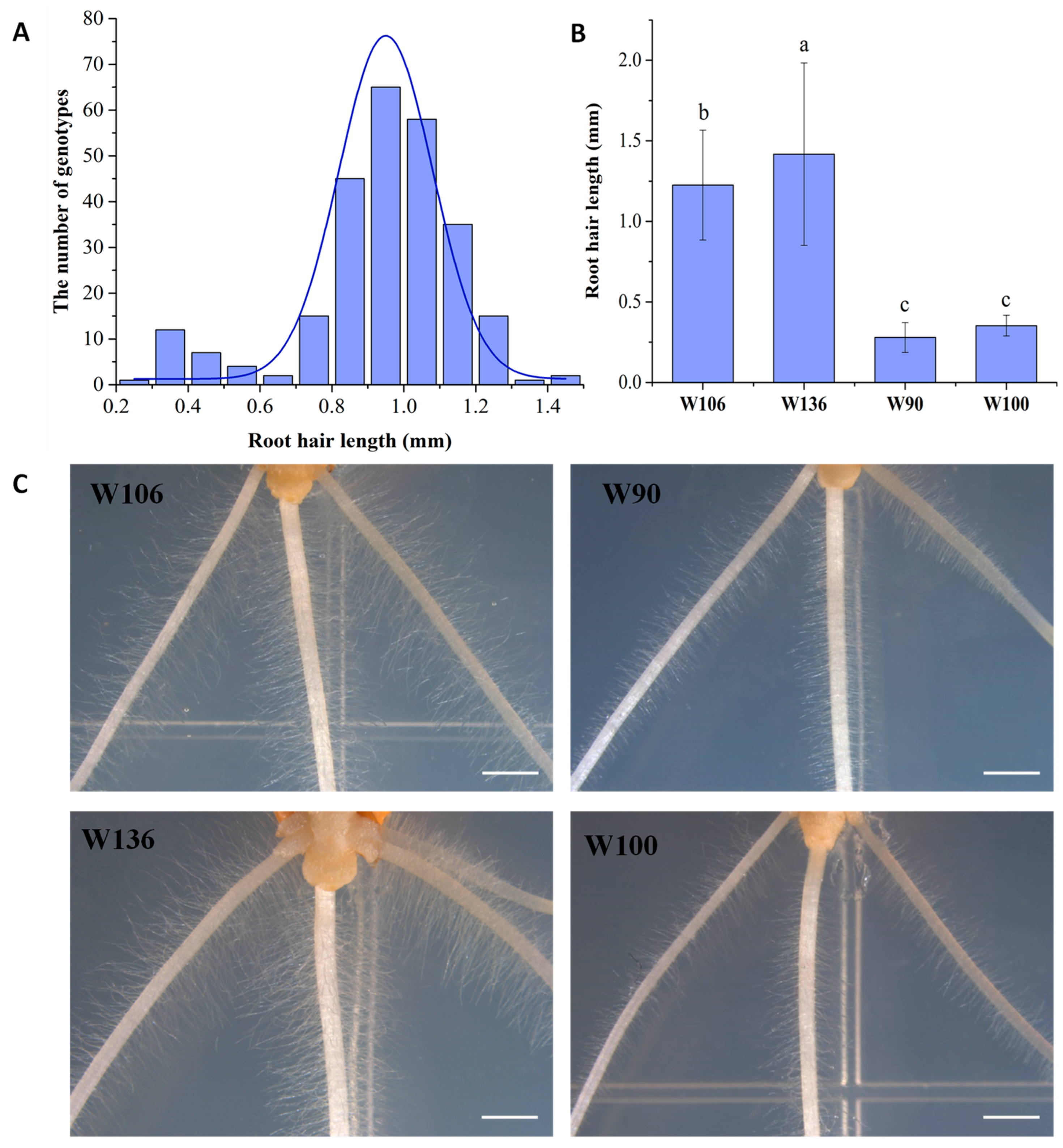
2.1. Measurement of Wheat Root Hair Length and Screening of Long Root Hair Genotypes
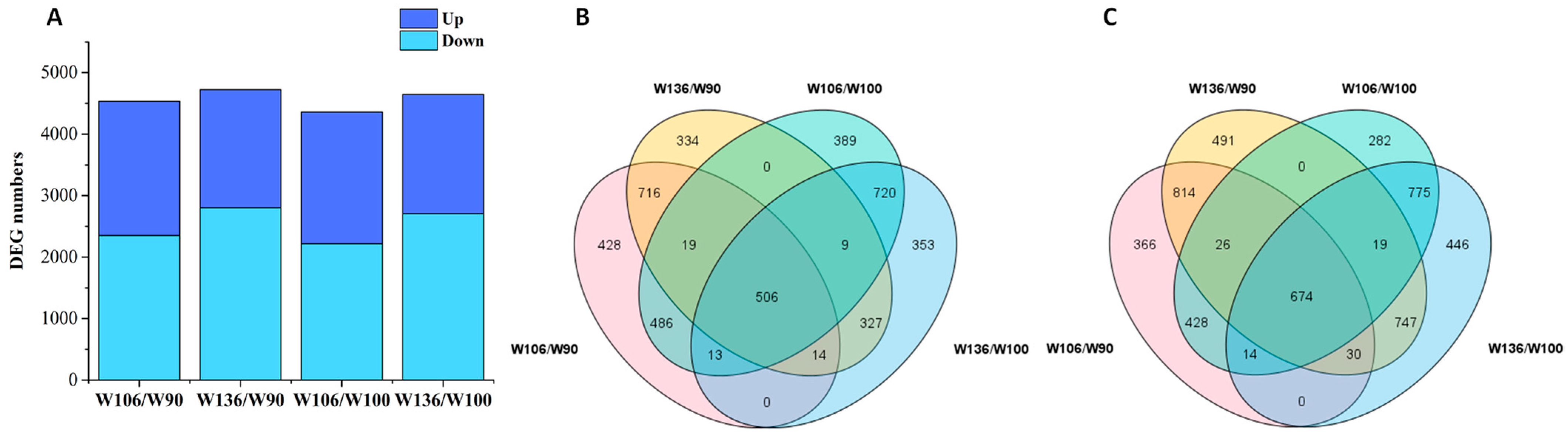
2.2. RNA-Sequencing Data Output and Screening of DEGs
2.3. Validation of DEGs Expression Levels
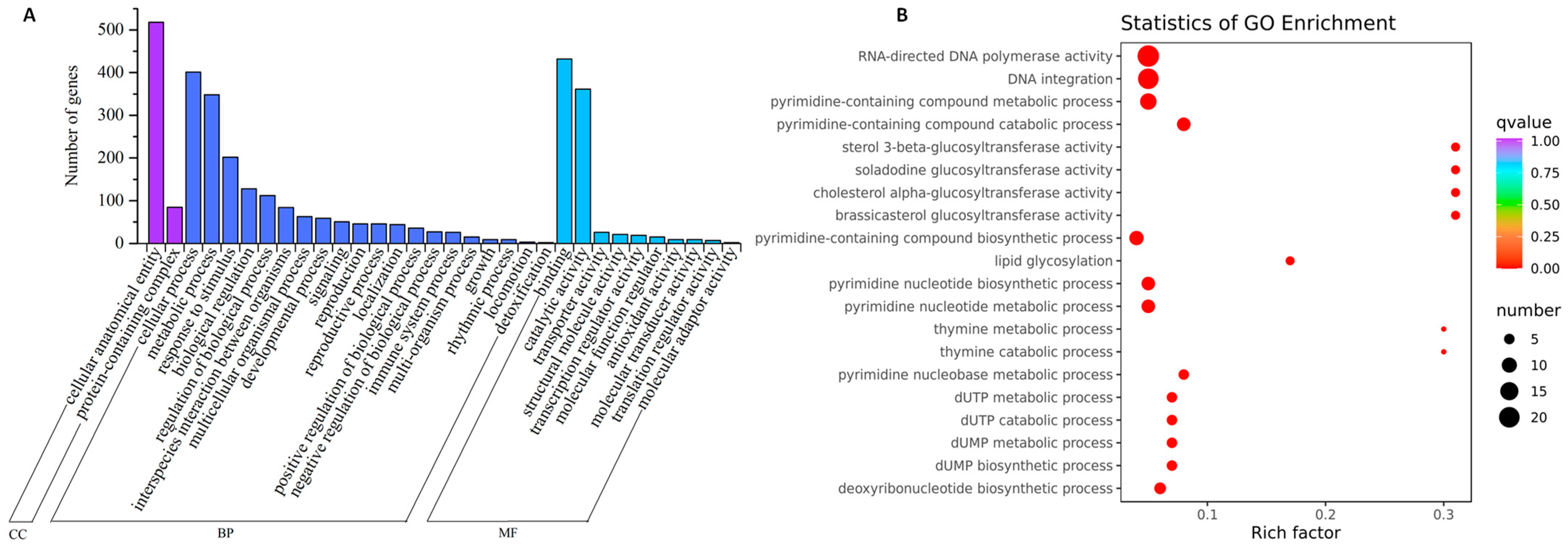
2.4. GO (Gene Ontology) Annotation and Enrichment Analysis of the Shared DEGs in Four Genotypes
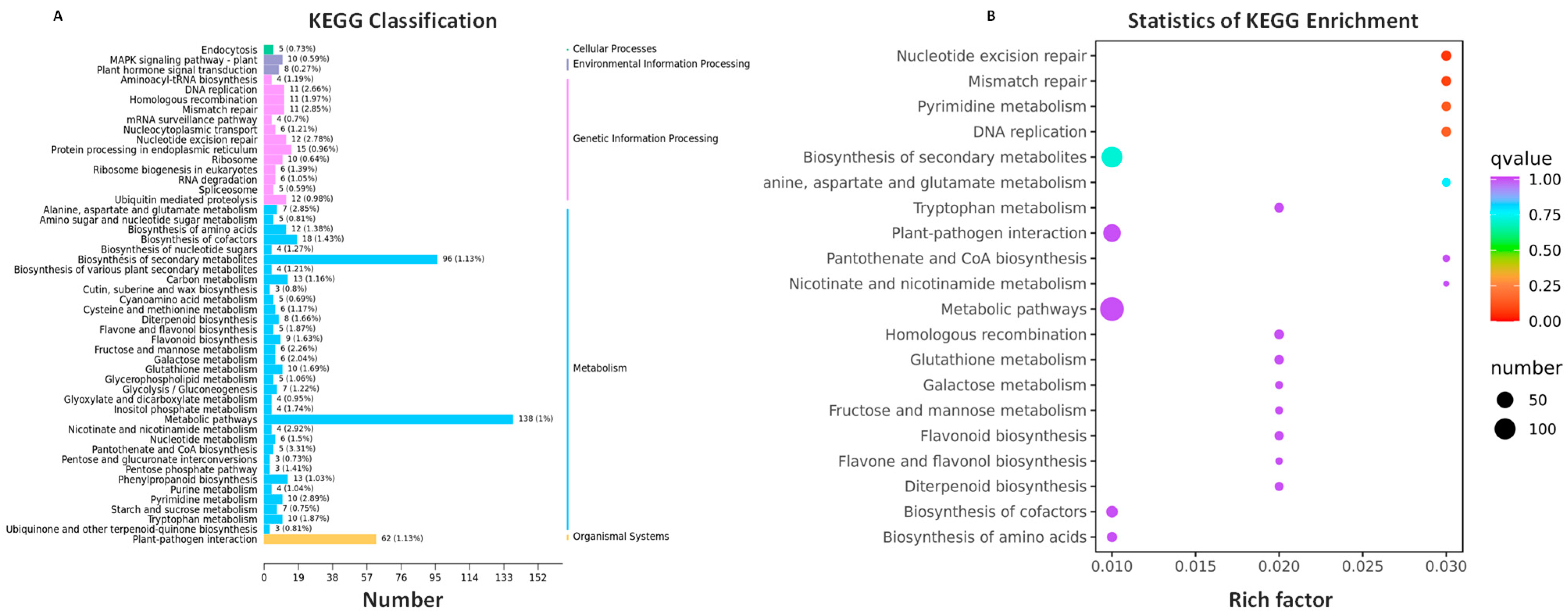
2.5. KEGG Classification and Enrichment Analysis of the Shared DEGs in Four Genotypes
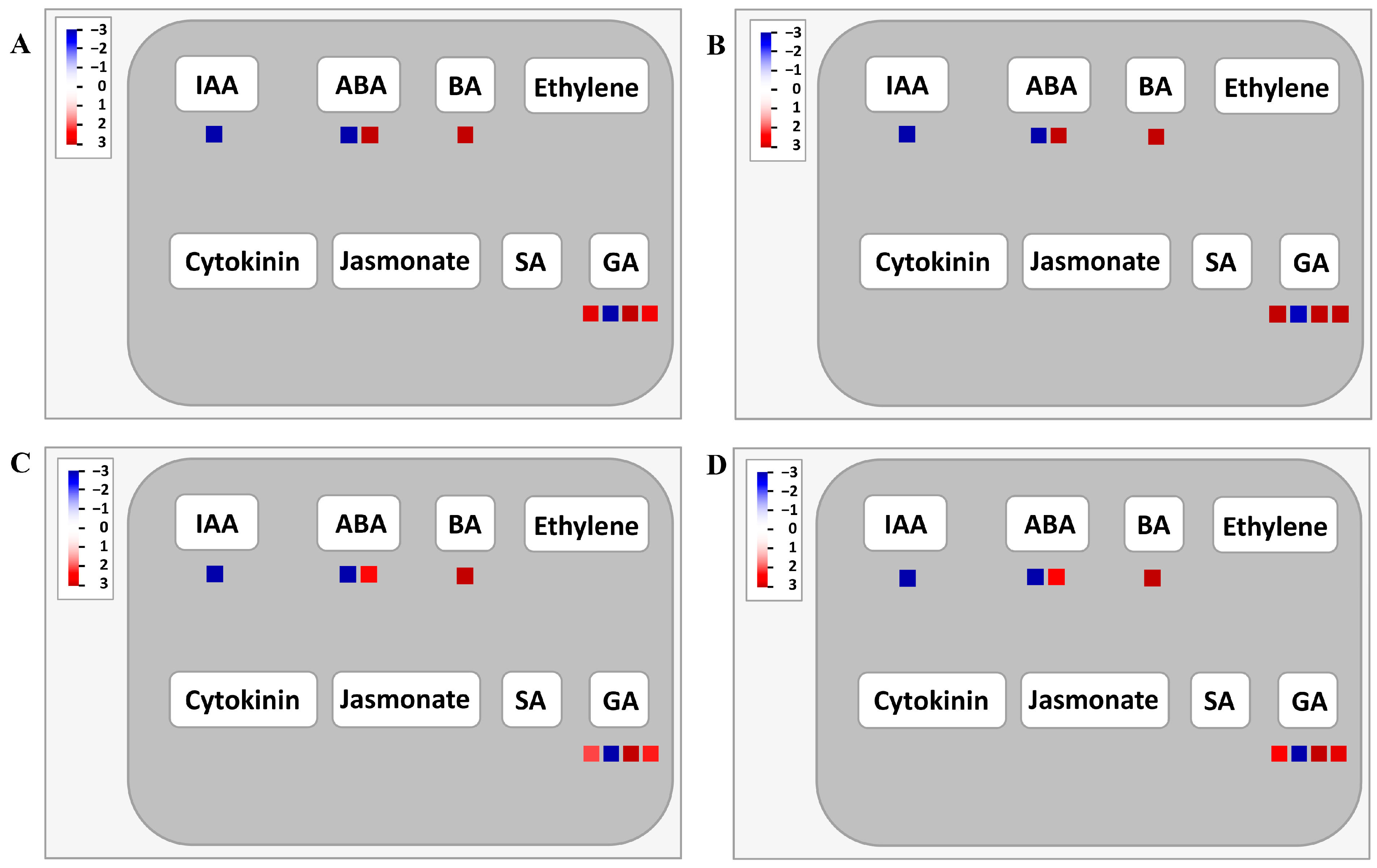
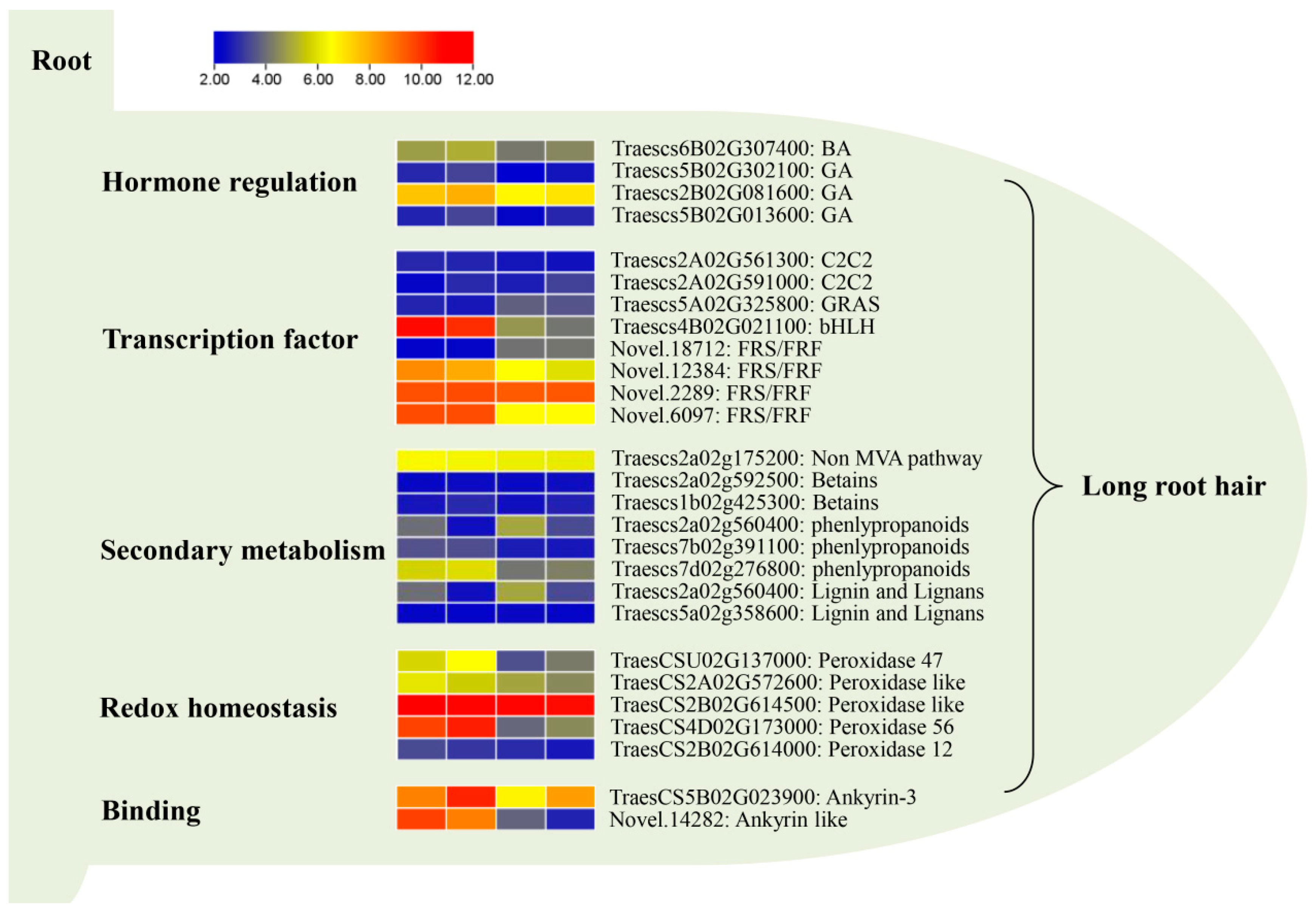
2.6. Pathway Analysis of the Shared DEGs in Four Genotypes Using MapMan
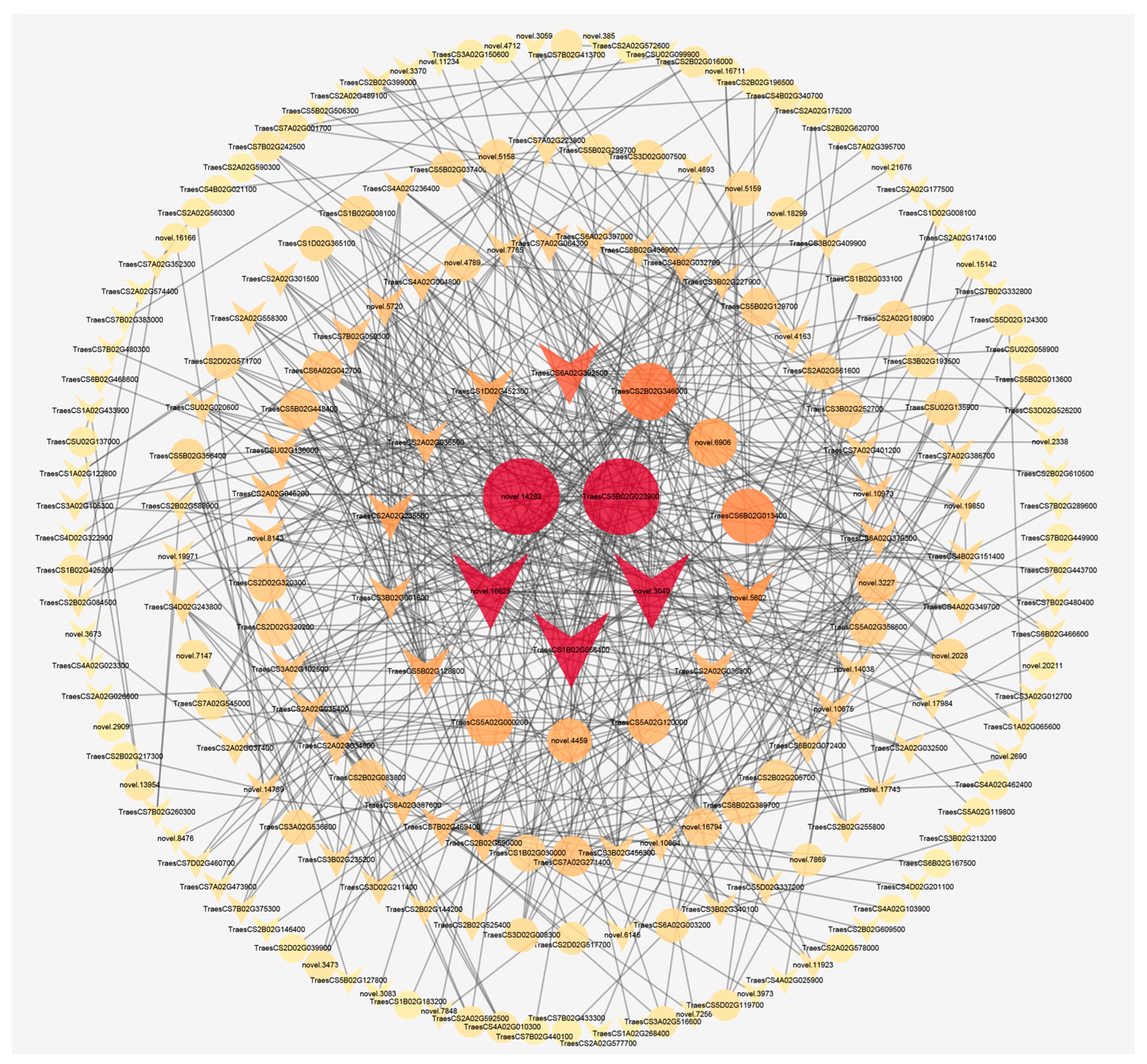
2.7. PPI Network of Shared DEGs Encoding Proteins
3. Discussion
3.1. Root Hairs Play a Key Role in Water and Nutrient Uptake by Plants
3.2. GA and BA Are Key Factors Regulating Root Hair Elongation
3.3. Transcription Factors Such as FRS/FRF and bHLH Are One of the Influencing Factors of Root Hair Length
3.4. Secondary Metabolic Pathway Is an Influencing Factor of Root Hair Length
3.5. Peroxidase Plays a Key Role in the Regulation of Root Hair Growth
3.6. Ankyrin May Be a Key Factor Regulating Wheat Root Hair Growth
4. Materials and Methods
4.1. Plant Materials
4.2. Observation and Length Measurements of Root Hairs
4.3. Sample Preparation and RNA Extraction
4.4. Development of cDNA Library and Transcriptome Sequencing
4.5. Screening and Analyzing Differentially Expressed Genes (DEGs)
4.6. Functional Annotation of DEGs
4.7. MapMan and PPI Analysis
4.8. qRT-PCR Validation
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rongsawat, T.; Peltier, J.B.; Boyer, J.C.; Véry, A.A.; Sentenac, H. Looking for Root Hairs to Overcome Poor Soils. Trends Plant Sci. 2021, 26, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Kato, M.; Tomioka, R.; Kurata, R.; Fukao, Y.; Aoyama, T.; Maeshima, M. Characteristics of a Root Hair-Less Line of Arabidopsis Thaliana under Physiological Stresses. J. Exp. Bot. 2014, 65, 1497–1512. [Google Scholar] [CrossRef] [PubMed]
- Carminati, A.; Passioura, J.B.; Zarebanadkouki, M.; Ahmed, M.A.; Ryan, P.R.; Watt, M.; Delhaize, E. Root Hairs Enable High Transpiration Rates in Drying Soils. New Phytol. 2017, 216, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Grabov, A. Plant KT/KUP/HAK Potassium Transporters: Single Family—Multiple Functions. Ann. Bot. 2007, 99, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Haling, R.E.; Brown, L.K.; Bengough, A.G.; Young, I.M.; Hallett, P.D.; White, P.J.; George, T.S. Root Hairs Improve Root Penetration, Root–Soil Contact, and Phosphorus Acquisition in Soils of Different Strength. J. Exp. Bot. 2013, 64, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Gahoonia, T.S.; Nielsen, N.E. Barley Genotypes with Long Root Hairs Sustain High Grain Yields in Low-P Field. Plant Soil 2004, 262, 55–62. [Google Scholar] [CrossRef]
- Klinsawang, S. Effects of root hair length on potassium acquisition in rice (Oryza sativa L.). Appl. Ecol. Env. Res. 2018, 16, 1609–1620. [Google Scholar] [CrossRef]
- Choi, H.S.; Cho, H.T. Root Hairs Enhance Arabidopsis Seedling Survival upon Soil Disruption. Sci. Rep. 2019, 9, 11181. [Google Scholar] [CrossRef]
- Bengough, A.G.; Loades, K.; McKenzie, B.M. Root Hairs Aid Soil Penetration by Anchoring the Root Surface to Pore Walls. J. Exp. Bot. 2016, 67, 1071–1078. [Google Scholar] [CrossRef]
- Galloway, A.F.; Akhtar, J.; Marcus, S.E.; Fletcher, N.; Field, K.; Knox, P. Cereal Root Exudates Contain Highly Structurally Complex Polysaccharides with Soil-binding Properties. Plant J. 2020, 103, 1666–1678. [Google Scholar] [CrossRef]
- Pang, J.; Ryan, M.H.; Siddique, K.H.M.; Simpson, R.J. Unwrapping the Rhizosheath. Plant Soil 2017, 418, 129–139. [Google Scholar] [CrossRef]
- Delhaize, E.; Rathjen, T.M.; Cavanagh, C.R. The Genetics of Rhizosheath Size in a Multiparent Mapping Population of Wheat. J. Exp. Bot. 2015, 66, 4527–4536. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Shin, R.; Schachtman, D.P. Expression of KT/KUP Genes in Arabidopsis and the Role of Root Hairs in K+ Uptake. Plant Physiol. 2004, 134, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Reintanz, B.; Szyroki, A.; Ivashikina, N.; Ache, P.; Godde, M.; Becker, D.; Palme, K.; Hedrich, R. AtKC1, a Silent Arabidopsis Potassium Channel α-Subunit Modulates Root Hair K + Influx. Proc. Natl. Acad. Sci. USA 2002, 99, 4079–4084. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Shi, X.; Wang, W.; Ryu, K.H.; Schiefelbein, J. Diversification of Root Hair Development Genes in Vascular Plants. Plant Physiol. 2017, 174, 1697–1712. [Google Scholar] [CrossRef] [PubMed]
- Nazoa, P.; Vidmar, J.J.; Tranbarger, T.J.; Mouline, K.; Tillard, P.; Zhuo, D.; Glass, A.D.M.; Touraine, B. Regulation of the Nitrate Transporter Gene AtNRT2.1 in Arabidopsis Thaliana: Responses to Nitrate, Amino Acids and Developmental Stage. Plant Mol. Biol. 2003, 3, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Loqué, D.; Kojima, S.; Rauch, S.; Ishiyama, K.; Inoue, E.; Takahashi, H.; Von Wirén, N. The Organization of High-Affinity Ammonium Uptake in Arabidopsis Roots Depends on the Spatial Arrangement and Biochemical Properties of AMT1-Type Transporters. Plant Cell 2007, 19, 2636–2652. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Deng, M.; Xu, J.; Zhu, X.; Mao, C. Molecular Mechanisms of Phosphate Transport and Signaling in Higher Plants. Semin. Cell Dev. Biol. 2018, 74, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H. Sulfate Transport Systems in Plants: Functional Diversity and Molecular Mechanisms Underlying Regulatory Coordination. J. Exp. Bot. 2019, 70, 4075–4087. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant Growth-Promoting Rhizobacteria and Root System Functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef]
- Holz, M.; Zarebanadkouki, M.; Kuzyakov, Y.; Pausch, J.; Carminati, A. Root Hairs Increase Rhizosphere Extension and Carbon Input to Soil. Ann. Bot. 2018, 121, 61–69. [Google Scholar] [CrossRef]
- Robertson-Albertyn, S.; Alegria Terrazas, R.; Balbirnie, K.; Blank, M.; Janiak, A.; Szarejko, I.; Chmielewska, B.; Karcz, J.; Morris, J.; Hedley, P.E.; et al. Root Hair Mutations Displace the Barley Rhizosphere Microbiota. Front. Plant Sci. 2017, 8, 1094. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC Transporter AtPDR8 Is a Cadmium Extrusion Pump Conferring Heavy Metal Resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D.; Downie, J.A. Coordinating Nodule Morphogenesis with Rhizobial Infection in Legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Vissenberg, K.; Claeijs, N.; Balcerowicz, D.; Schoenaers, S. Hormonal Regulation of Root Hair Growth and Responses to the Environment in Arabidopsis. J. Exp. Bot. 2020, 71, 2412–2427. [Google Scholar] [CrossRef]
- Haling, R.E.; Brown, L.K.; Bengough, A.G.; Valentine, T.A.; White, P.J.; Young, I.M.; George, T.S. Root Hair Length and Rhizosheath Mass Depend on Soil Porosity, Strength and Water Content in Barley Genotypes. Planta 2014, 239, 643–651. [Google Scholar] [CrossRef]
- Zhang, C.; Simpson, R.J.; Kim, C.M.; Warthmann, N.; Delhaize, E.; Dolan, L.; Byrne, M.E.; Wu, Y.; Ryan, P.R. Do Longer Root Hairs Improve Phosphorus Uptake? Testing the Hypothesis with Transgenic Brachypodium Distachyon Lines Overexpressing Endogenous RSL Genes. New Phytol. 2018, 217, 1654–1666. [Google Scholar] [CrossRef]
- Müller, M.; Schmidt, W. Environmentally Induced Plasticity of Root Hair Development in Arabidopsis. Plant Physiol. 2004, 134, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Zhou, D.-X.; Zhao, Y. WUSCHEL -Related Homeobox Gene WOX11 Increases Rice Drought Resistance by Controlling Root Hair Formation and Root System Development. Plant Signal. Behav. 2016, 11, e1130198. [Google Scholar] [CrossRef]
- Zhang, X.; Mi, Y.; Mao, H.; Liu, S.; Chen, L.; Qin, F. Genetic Variation in ZmTIP1 Contributes to Root Hair Elongation and Drought Tolerance in Maize. Plant Biotechnol. J. 2020, 18, 1271–1283. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Li, K.; Sun, F.; Han, C.; Wang, Y.; Li, X. Salt-Induced Plasticity of Root Hair Development Is Caused by Ion Disequilibrium in Arabidopsis Thaliana. J. Plant Res. 2008, 121, 87–96. [Google Scholar] [CrossRef]
- Robin, A.H.K.; Matthew, C.; Uddin, M.J.; Bayazid, K.N. Salinity-Induced Reduction in Root Surface Area and Changes in Major Root and Shoot Traits at the Phytomer Level in Wheat. J. Exp. Bot. 2016, 67, 3719–3729. [Google Scholar] [CrossRef]
- Stéger, A.; Palmgren, M. Root Hair Growth from the pH Point of View. Front. Plant Sci. 2022, 13, 949672. [Google Scholar] [CrossRef]
- Eljebbawi, A.; Guerrero, Y.D.C.R.; Dunand, C.; Estevez, J.M. Highlighting Reactive Oxygen Species as Multitaskers in Root Development. iScience 2021, 24, 101978. [Google Scholar] [CrossRef]
- Brown, L.K.; George, T.S.; Neugebauer, K.; White, P.J. The Rhizosheath—A Potential Trait for Future Agricultural Sustainability Occurs in Orders throughout the Angiosperms. Plant Soil 2017, 418, 115–128. [Google Scholar] [CrossRef]
- Marin, M.; Feeney, D.S.; Brown, L.K.; Naveed, M.; Ruiz, S.; Koebernick, N.; Bengough, A.G.; Hallett, P.D.; Roose, T.; Puértolas, J.; et al. Significance of Root Hairs for Plant Performance under Contrasting Field Conditions and Water Deficit. Ann. Bot. 2021, 128, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Sugimoto, K. A Gene Regulatory Network for Root Hair Development. J. Plant Res. 2019, 132, 301–309. [Google Scholar] [CrossRef]
- Dugasa, M.T.; Feng, X.; Wang, N.-H.; Wang, J.; Wu, F. Comparative Transcriptome and Tolerance Mechanism Analysis in the Two Contrasting Wheat (Triticum aestivum L.) Cultivars in Response to Drought and Salinity Stresses. Plant Growth Regul. 2021, 94, 101–114. [Google Scholar] [CrossRef]
- He, X.; Zeng, J.; Cao, F.; Ahmed, I.M.; Zhang, G.; Vincze, E.; Wu, F. HvEXPB7, a Novel β-Expansin Gene Revealed by the Root Hair Transcriptome of Tibetan Wild Barley, Improves Root Hair Growth under Drought Stress. J. Exp. Bot. 2015, 66, 7405–7419. [Google Scholar] [CrossRef]
- Paez-Garcia, A.; Motes, C.; Scheible, W.-R.; Chen, R.; Blancaflor, E.; Monteros, M. Root Traits and Phenotyping Strategies for Plant Improvement. Plants 2015, 4, 334–355. [Google Scholar] [CrossRef] [PubMed]
- Kohli, P.S.; Maurya, K.; Thakur, J.K.; Bhosale, R.; Giri, J. Significance of Root Hairs in Developing Stress-resilient Plants for Sustainable Crop Production. Plant Cell Environ. 2022, 45, 677–694. [Google Scholar] [CrossRef]
- Wendrich, J.R.; Yang, B.; Vandamme, N.; Verstaen, K.; Smet, W.; Van De Velde, C.; Minne, M.; Wybouw, B.; Mor, E.; Arents, H.E.; et al. Vascular Transcription Factors Guide Plant Epidermal Responses to Limiting Phosphate Conditions. Science 2020, 370, eaay4970. [Google Scholar] [CrossRef]
- Zhu, L.; Liu, L.; Sun, H.; Zhang, Y.; Liu, X.; Wang, N.; Chen, J.; Zhang, K.; Bai, Z.; Wang, G.; et al. The Responses of Lateral Roots and Root Hairs to Nitrogen Stress in Cotton Based on Daily Root Measurements. J. Agro. Crop Sci. 2022, 208, 89–105. [Google Scholar] [CrossRef]
- Huang, L.; Jiang, Q.; Wu, J.; An, L.; Zhou, Z.; Wong, C.; Wu, M.; Yu, H.; Gan, Y. Zinc Finger Protein 5 (ZFP5) Associates with Ethylene Signaling to Regulate the Phosphate and Potassium Deficiency-Induced Root Hair Development in Arabidopsis. Plant Mol. Biol. 2020, 102, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Liu, L.; Zhang, Y.; Sun, H.; Zhang, K.; Bai, Z.; Dong, H.; Li, C. Fine Root and Root Hair Morphology of Cotton under Drought Stress Revealed with RhizoPot. J. Agro. Crop Sci. 2020, 206, 679–693. [Google Scholar] [CrossRef]
- Ober, E.S.; Alahmad, S.; Cockram, J.; Forestan, C.; Hickey, L.T.; Kant, J.; Maccaferri, M.; Marr, E.; Milner, M.; Pinto, F.; et al. Wheat Root Systems as a Breeding Target for Climate Resilience. Theor. Appl. Genet. 2021, 134, 1645–1662. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.-M.; Achard, P. Gibberellin Signaling in Plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Devlin, R.M.; Brown, D.P. Effect of Gibberellic Acid on the Elongation Rate of Agrostis Alba Root Hairs. Physiol. Plant. 1969, 22, 759–763. [Google Scholar] [CrossRef]
- Ohkawa, H.; Kamada, H.; Sudo, H.; Harada, H. Effects of Gibberellic Acid on Hairy Root Growth in Datura Innoxia. J. Plant Physiol. 1989, 134, 633–636. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, L.; Yan, A.; Liu, Y.; Liu, B.; Yu, C.; Zhang, A.; Schiefelbein, J.; Gan, Y. Multiple Phytohormones Promote Root Hair Elongation by Regulating a Similar Set of Genes in the Root Epidermis in Arabidopsis. J. Exp. Bot. 2016, 67, 6363–6372. [Google Scholar] [CrossRef]
- Ma, L.; Li, G. Far1-related sequence (FRS) and FRS-related factor (FRF) family proteins in Arabidopsis growth and development. Front. Plant Sci. 2018, 9, 692. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, Y.; Ma, M.; Zhou, Q.; Zhao, Y.; Zhao, B.; Wang, B.; Wei, H.; Wang, H. Arabidopsis FHY3 and FAR1 Integrate Light and Strigolactone Signaling to Regulate Branching. Nat. Commun. 2020, 11, 1955. [Google Scholar] [CrossRef]
- Li, D.; Fu, X.; Guo, L.; Huang, Z.; Li, Y.; Liu, Y.; He, Z.; Cao, X.; Ma, X.; Zhao, M.; et al. FAR-RED ELONGATED HYPOCOTYL3 Activates SEPALLATA2 but Inhibits CLAVATA3 to Regulate Meristem Determinacy and Maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 9375–9380. [Google Scholar] [CrossRef]
- Lin, Q.; Ohashi, Y.; Kato, M.; Tsuge, T.; Gu, H.; Qu, L.-J.; Aoyama, T. GLABRA2 Directly Suppresses Basic Helix-Loop-Helix Transcription Factor Genes with Diverse Functions in Root Hair Development. Plant Cell 2015, 27, 2894–2906. [Google Scholar] [CrossRef]
- Yi, K.; Menand, B.; Bell, E.; Dolan, L. A Basic Helix-Loop-Helix Transcription Factor Controls Cell Growth and Size in Root Hairs. Nat. Genet. 2010, 42, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, V.; Kakkar, M.; Kumari, P.; Zinta, G.; Gahlaut, V.; Kumar, S. Multifaceted Roles of GRAS Transcription Factors in Growth and Stress Responses in Plants. iScience 2022, 25, 105026. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Xia, Y.; Jiang, W.; Shen, G.; Pang, Y. GbMYBR1 from Ginkgo Biloba Represses Phenylpropanoid Biosynthesis and Trichome Development in Arabidopsis. Planta 2020, 252, 68. [Google Scholar] [CrossRef] [PubMed]
- Thwe, A.A.; Kim, J.K.; Li, X.; Bok Kim, Y.; Romij Uddin, M.; Kim, S.J.; Suzuki, T.; Park, N.I.; Park, S.U. Metabolomic Analysis and Phenylpropanoid Biosynthesis in Hairy Root Culture of Tartary Buckwheat Cultivars. PLoS ONE 2013, 8, e65349. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.-Y.; Li, C.-X.; He, Y.; Hou, X.-Y.; Ma, X.-R. The Regulation of Adaptation to Cold and Drought Stresses in Poa Crymophila Keng Revealed by Integrative Transcriptomics and Metabolomics Analysis. Front. Plant Sci. 2021, 12, 631117. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.X.; Son, S.; Lee, S.; Jung, E.; Lee, Y.; Sung, J.; Lee, C. Combined Effects of Nutrients × Water × Light on Metabolite Composition in Tomato Fruits (Solanum lycopersicum L.). Plants 2021, 10, 1437. [Google Scholar] [CrossRef]
- Mnich, E.; Bjarnholt, N.; Eudes, A.; Harholt, J.; Holland, C.; Jørgensen, B.; Larsen, F.H.; Liu, M.; Manat, R.; Meyer, A.S.; et al. Phenolic Cross-Links: Building and de-Constructing the Plant Cell Wall. Nat. Prod. Rep. 2020, 37, 919–961. [Google Scholar] [CrossRef]
- Holbein, J.; Franke, R.B.; Marhavý, P.; Fujita, S.; Górecka, M.; Sobczak, M.; Geldner, N.; Schreiber, L.; Grundler, F.M.W.; Siddique, S. Root Endodermal Barrier System Contributes to Defence against Plant-parasitic Cyst and Root-knot Nematodes. Plant J. 2019, 100, 221–236. [Google Scholar] [CrossRef]
- Kärkönen, A.; Koutaniemi, S. Lignin Biosynthesis Studies in Plant Tissue Cultures. J. Integr. Plant Biol. 2010, 52, 176–185. [Google Scholar] [CrossRef]
- Nestler, J.; Schütz, W.; Hochholdinger, F. Conserved and Unique Features of the Maize (Zea mays L.) Root Hair Proteome. J. Proteome Res. 2011, 10, 2525–2537. [Google Scholar] [CrossRef] [PubMed]
- Orman-Ligeza, B.; Parizot, B.; De Rycke, R.; Fernandez, A.; Himschoot, E.; Van Breusegem, F.; Bennett, M.J.; Périlleux, C.; Beeckman, T.; Draye, X. RBOH-Mediated ROS Production Facilitates Lateral Root Emergence in Arabidopsis. Development 2016, 143, 3328–3339. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the Paradoxical: How Plant Peroxidases Modify the Cell Wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Dunand, C.; Crèvecoeur, M.; Penel, C. Distribution of Superoxide and Hydrogen Peroxide in Arabidopsis Root and Their Influence on Root Development: Possible Interaction with Peroxidases. New Phytol. 2007, 174, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Marzol, E.; Borassi, C.; Carignani Sardoy, M.; Ranocha, P.; Aptekmann, A.A.; Bringas, M.; Pennington, J.; Paez-Valencia, J.; Martínez Pacheco, J.; Rodríguez-Garcia, D.R.; et al. Class III Peroxidases PRX01, PRX44, and PRX73 Control Root Hair Growth in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 5375. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, J.M.; Ranocha, P.; Kasulin, L.; Fusari, C.M.; Servi, L.; Aptekmann, A.A.; Gabarain, V.B.; Peralta, J.M.; Borassi, C.; Marzol, E.; et al. Apoplastic Class III Peroxidases PRX62 and PRX69 Promote Arabidopsis Root Hair Growth at Low Temperature. Nat. Commun. 2022, 13, 1310. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, S.G.; Smerdon, S.J. The Ankyrin Repeat: A Diversity of Interactions on a Common Structural Framework. Trends Biochem. Sci. 1999, 24, 311–316. [Google Scholar] [CrossRef]
- Vo, K.T.X.; Kim, C.-Y.; Chandran, A.K.N.; Jung, K.-H.; An, G.; Jeon, J.-S. Molecular Insights into the Function of Ankyrin Proteins in Plants. J. Plant Biol. 2015, 58, 271–284. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; Ding, P.; Johnson, K.; Li, X.; Zhang, Y. The Ankyrin-Repeat Transmembrane Protein BDA1 Functions Downstream of the Receptor-Like Protein SNC2 to Regulate Plant Immunity. Plant Physiol. 2012, 159, 1857–1865. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.E.; Schofield, A.; Lyzenga, W.; Liu, H.; Stone, S.L. Arabidopsis RING E3 Ligase XBAT32 Regulates Lateral Root Production through Its Role in Ethylene Biosynthesis. Plant Physiol. 2010, 153, 1587–1596. [Google Scholar] [CrossRef]
- Huang, J.; Chen, F.; Del Casino, C.; Autino, A.; Shen, M.; Yuan, S.; Peng, J.; Shi, H.; Wang, C.; Cresti, M.; et al. An Ankyrin Repeat-Containing Protein, Characterized as a Ubiquitin Ligase, Is Closely Associated with Membrane-Enclosed Organelles and Required for Pollen Germination and Pollen Tube Growth in Lily. Plant Physiol. 2006, 140, 1374–1383. [Google Scholar] [CrossRef]
- Kim, D.H.; Lee, J.-E.; Xu, Z.-Y.; Geem, K.R.; Kwon, Y.; Park, J.W.; Hwang, I. Cytosolic Targeting Factor AKR2A Captures Chloroplast Outer Membrane-Localized Client Proteins at the Ribosome during Translation. Nat. Commun. 2015, 6, 6843. [Google Scholar] [CrossRef] [PubMed]
- Garcion, C.; Guilleminot, J.; Kroj, T.; Parcy, F.; Giraudat, J.; Devic, M. AKRP and EMB506 Are Two Ankyrin Repeat Proteins Essential for Plastid Differentiation and Plant Development in Arabidopsis. Plant J. 2006, 48, 895–906. [Google Scholar] [CrossRef]
- Wang, H.; Zou, S.; Li, Y.; Lin, F.; Tang, D. An Ankyrin-Repeat and WRKY-Domain-Containing Immune Receptor Confers Stripe Rust Resistance in Wheat. Nat. Commun. 2020, 11, 1353. [Google Scholar] [CrossRef]
- Zhang, X.; Li, D.; Zhang, H.; Wang, X.; Zheng, Z.; Song, F. Molecular Characterization of Rice OsBIANK1, Encoding a Plasma Membrane-Anchored Ankyrin Repeat Protein, and Its Inducible Expression in Defense Responses. Mol. Biol. Rep. 2010, 37, 653–660. [Google Scholar] [CrossRef]
- Zhao, J.-Y.; Lu, Z.-W.; Sun, Y.; Fang, Z.-W.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S.; Min, D.-H. The Ankyrin-Repeat Gene GmANK114 Confers Drought and Salt Tolerance in Arabidopsis and Soybean. Front. Plant Sci. 2020, 11, 584167. [Google Scholar] [CrossRef]
- Chen, L.; Hu, W.; Mishra, N.; Wei, J.; Lu, H.; Hou, Y.; Qiu, X.; Yu, S.; Wang, C.; Zhang, H.; et al. AKR2A Interacts with KCS1 to Improve VLCFAs Contents and Chilling Tolerance of Arabidopsis Thaliana. Plant J. 2020, 103, 1575–1589. [Google Scholar] [CrossRef]
- Paeng, S.K.; Kang, C.H.; Chi, Y.H.; Chae, H.B.; Lee, E.S.; Park, J.H.; Wi, S.D.; Bae, S.B.; Phan, K.A.T.; Lee, S.Y. AtTPR10 Containing Multiple ANK and TPR Domains Exhibits Chaperone Activity and Heat-Shock Dependent Structural Switching. Appl. Sci. 2020, 10, 1265. [Google Scholar] [CrossRef]
- Cao, Y.R.; Chen, H.W.; Li, Z.G.; Tao, J.J.; Ma, B.; Zhang, W.-K.; Chen, S.-Y.; Zhang, J.-S. Tobacco Ankyrin Protein NEIP2 Interacts with Ethylene Receptor NTHK1 and Regulates Plant Growth and Stress Responses. Plant Cell Physiol. 2015, 56, 803–818. [Google Scholar] [CrossRef]
- Zhang, F.; Xie, Y.; Jiang, H.; Wang, X.; Hao, Y.-J.; Zhang, Z.; You, C.X. The Ankyrin Repeat-Containing Protein MdANK2B Regulates Salt Tolerance and ABA Sensitivity in Malus Domestica. Plant Cell Rep. 2021, 40, 405–419. [Google Scholar] [CrossRef]
- Lu, H.; Rate, D.N.; Song, J.T.; Greenberg, J.T. ACD6, a Novel Ankyrin Protein, Is a Regulator and an Effector of Salicylic Acid Signaling in the Arabidopsis Defense Response. Plant Cell 2003, 15, 2408–2420. [Google Scholar] [CrossRef]
- Hemsley, P.A.; Kemp, A.C.; Grierson, C.S. The TIP GROWTH DEFECTIVE1 S-Acyl Transferase Regulates Plant Cell Growth in Arabidopsis. Plant Cell 2005, 17, 2554–2563. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Barber, R.F. Multiple Testing with the Structure-Adaptive Benjamini–Hochberg Algorithm. J. R. Stat. Soc. B 2019, 81, 45–74. [Google Scholar] [CrossRef]
- Rivals, I.; Personnaz, L.; Taing, L.; Potier, M.-C. Enrichment or Depletion of a GO Category within a Class of Genes: Which Test? Bioinformatics 2007, 23, 401–407. [Google Scholar] [CrossRef] [PubMed]


| Sample | Raw Reads | Clean Reads | Reads Mapped | Unique Mapped | Multi Mapped | ‘+’ Mapped | ‘−’ Mapped | Exon Mapped | Expressed Gene Number |
|---|---|---|---|---|---|---|---|---|---|
| W90-1 | 65,362,590 | 59,353,078 | 53,608,182 (90.32%) | 49,153,912 (82.82%) | 4,454,270 (7.50%) | 24,573,452 (41.40%) | 24,580,460 (41.41%) | 8,363,394 (91.22%) | 88,280 |
| W90-2 | 61,327,388 | 58,045,598 | 53,142,825 (91.55%) | 49,217,176 (84.79%) | 3,925,649 (6.76%) | 24,608,886 (42.40%) | 24,608,290 (42.39%) | 8,332,918 (91.49%) | 88,250 |
| W90-3 | 73,110,780 | 68,218,444 | 62,857,440 (92.14%) | 58,143,204 (85.23%) | 4,714,236 (6.91%) | 29,071,891 (42.62%) | 29,071,313 (42.62%) | 9,882,370 (91.59%) | 89,285 |
| W100-1 | 63,157,218 | 57,594,580 | 51,858,044 (90.04%) | 47,556,819 (82.57%) | 4,301,225 (7.47%) | 23,781,711 (41.29%) | 23,775,108 (41.28%) | 7,891,593 (91.27%) | 88,124 |
| W100-2 | 55,525,334 | 46,352,918 | 43,341,067 (93.50%) | 40,140,499 (86.60%) | 3,200,568 (6.90%) | 20,075,390 (43.31%) | 20,065,109 (43.29%) | 6,675,681 (92.42%) | 85,594 |
| W100-3 | 63,619,112 | 59,787,134 | 54,564,001 (91.26%) | 50,472,705 (84.42%) | 4,091,296 (6.84%) | 25,241,384 (42.22%) | 25,231,321 (42.20%) | 8,325,128 (91.57%) | 89,752 |
| W106-1 | 67,132,016 | 62,597,994 | 58,960,629 (94.19%) | 54,803,393 (87.55%) | 4,157,236 (6.64%) | 27,400,681 (43.77%) | 27,402,712 (43.78%) | 9,420,269 (92.63%) | 87,203 |
| W106-2 | 78,750,774 | 73,059,016 | 68,344,337 (93.55%) | 63,408,836 (86.79%) | 4,935,501 (6.76%) | 31,702,820 (43.39%) | 31,706,016 (43.40%) | 10,861,159 (92.35%) | 88,279 |
| W106-3 | 71,615,688 | 65,291,454 | 61,261,231 (93.83%) | 56,812,593 (87.01%) | 4,448,638 (6.81%) | 28,405,411 (43.51%) | 28,407,182 (43.51%) | 9,774,520 (92.51%) | 86,945 |
| W136-1 | 96,589,012 | 90,374,388 | 83,210,404 (92.07%) | 76,814,622 (85.00%) | 6,395,782 (7.08%) | 38,404,283 (42.49%) | 38,410,339 (42.50%) | 12,749,176 (91.69%) | 88,522 |
| W136-2 | 103,797,240 | 97,690,332 | 90,367,433 (92.50%) | 83,612,384 (85.59%) | 6,755,049 (6.91%) | 41,805,273 (42.79%) | 41,807,111 (42.80%) | 13,902,102 (91.92%) | 88,771 |
| W136-3 | 92,550,006 | 87,161,176 | 81,051,648 (92.99%) | 75,268,049 (86.36%) | 5,783,599 (6.64%) | 37,630,806 (43.17%) | 37,637,243 (43.18%) | 12,523,229 (92.16%) | 88,330 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, J.; Wang, Y.; Wu, G.; Sun, Q.; He, X.; Zhang, X.; Sun, X.; Zhao, Y.; Liu, W.; Xu, D.; et al. Comparative Transcriptome Analysis Reveals the Genes and Pathways Related to Wheat Root Hair Length. Int. J. Mol. Sci. 2024, 25, 2069. https://doi.org/10.3390/ijms25042069
Zeng J, Wang Y, Wu G, Sun Q, He X, Zhang X, Sun X, Zhao Y, Liu W, Xu D, et al. Comparative Transcriptome Analysis Reveals the Genes and Pathways Related to Wheat Root Hair Length. International Journal of Molecular Sciences. 2024; 25(4):2069. https://doi.org/10.3390/ijms25042069
Chicago/Turabian StyleZeng, Jianbin, Yongmei Wang, Gang Wu, Qingyi Sun, Xiaoyan He, Xinyi Zhang, Xuelian Sun, Yan Zhao, Wenxing Liu, Dengan Xu, and et al. 2024. "Comparative Transcriptome Analysis Reveals the Genes and Pathways Related to Wheat Root Hair Length" International Journal of Molecular Sciences 25, no. 4: 2069. https://doi.org/10.3390/ijms25042069
APA StyleZeng, J., Wang, Y., Wu, G., Sun, Q., He, X., Zhang, X., Sun, X., Zhao, Y., Liu, W., Xu, D., Dai, X., & Ma, W. (2024). Comparative Transcriptome Analysis Reveals the Genes and Pathways Related to Wheat Root Hair Length. International Journal of Molecular Sciences, 25(4), 2069. https://doi.org/10.3390/ijms25042069






