Abstract
Gestational diabetes mellitus (GDM) is a significant pregnancy complication linked to perinatal complications and an elevated risk of future metabolic disorders for both mothers and their children. GDM is diagnosed when women without prior diabetes develop chronic hyperglycemia due to β-cell dysfunction during gestation. Global research focuses on the association between GDM and single nucleotide polymorphisms (SNPs) and aims to enhance our understanding of GDM’s pathogenesis, predict its risk, and guide patient management. This review offers a summary of various SNPs linked to a heightened risk of GDM and explores their biological mechanisms within the tissues implicated in the development of the condition.
1. Introduction
Gestational diabetes mellitus (GDM) refers to diabetes diagnosed in the second or third trimester of pregnancy that was not present before gestation and carries risks for the mother, fetus, and newborn. GDM is often indicative of underlying β-cell dysfunction and increases the risk for later development of diabetes, which is often type 2 diabetes (T2D), in the mother after delivery [1].
The maternal body undergoes a sequence of physiological changes to accommodate the needs of the developing fetus, with insulin sensitivity representing a vital metabolic adaptation during this process. An upsurge in local and placental hormones during mid- to late gestation induces insulin resistance. GDM occurs when pancreatic β-cells cannot adequately respond to the heightened requirement for insulin secretion, leading to the onset of spontaneous hyperglycemia during pregnancy. Both the malfunction of β-cells and the resistance of body tissues to insulin are integral aspects of the pathophysiology of GDM [2].
GDM is a multifactorial disorder that is influenced by interactions between genetic and environmental factors. The interplay between these factors suggests the complexity of the mechanistic pathways underlying GDM. Genome-wide association studies (GWAS) have identified specific genetic variations called single nucleotide polymorphisms (SNPs) associated with the risk of developing GDM. These SNPs not only help researchers understand the causes of GDM for potential drug development, but also have practical applications in clinical settings, which can be used to predict the risk of GDM, guide treatment choices, and manage GDM patients during pregnancy and postpartum, enhancing our understanding and approach to this condition [3].
Several genes, such as the transcription factor 7-like 2 (TCF7L2) gene, the haematopoietically expressed homeobox gene (HHEX), the adiponectin, C1Q, and collagen domain containing (ADIPOQ) gene, and the vascular endothelial growth factor A (VEGFA) gene, are expressed in major tissues contributing to GDM, including the placenta. Their SNPs have been reported to be associated with GDM. This review will explore the biological and physiological effects of these SNPs on the development of GDM. We retrieved the prediction of transcription factors on each SNP by JASPAR CORE 2022 [4] from the UCSC Genome Browser [5]. We also discuss how each tissue contributes to the pathogenesis of GDM, as shown in Figure 1.
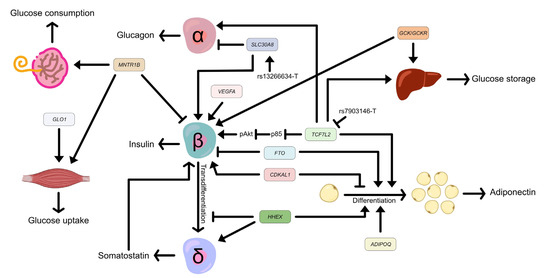
Figure 1.
Effects of associated genes and SNPs in major tissues contribute to gestational diabetes mellitus. TCF7L2 promotes glucose storage in the liver, activates insulin secretion in β-cells by suppressing the expression of p85, and inhibits adipogenesis. The TCFL2 rs7903146 minor allele, T, downregulates the expression of TCF7L2. HHEX increases somatostatin secretion and adipocyte differentiation but inhibits the transdifferentiation of β-cells to δ-cells. SLC30A8 activates insulin secretion and reduces glucagon secretion. The SLC30A8 missense variant rs13266634 allele T improves glucose responsiveness and enhances proinsulin conversion. ADIPOQ activates adipogenesis. FTO inhibits insulin secretion and promotes adipocyte differentiation, and vice versa by CDKAL1. VEGFA maintains β-cell function. MTNR1B increases glucose consumption in the placenta and skeletal muscle, but decreases insulin secretion in pancreatic β-cells. GLO1 activates glucose uptake in skeletal muscle and GCK/GCKR promotes glucose storage in the liver. The transcription factor 7-like 2 (TCF7L2) gene, the haematopoietically expressed homeobox (HHEX) gene, the solute carrier family 30 member 8 gene (SLC30A8), the adiponectin, C1Q, and collagen domain containing (ADIPOQ) gene, the human fat mass and obesity-related (FTO) gene, the CDK5 regulatory subunit associated protein 1 like 1 (CDKAL1) gene, the vascular endothelial growth factor A (VEGFA) gene, the melatonin receptor 1B (MTNR1B) gene, the glyoxalase I (GLO1) gene, the glucokinase (GCK) gene, and the glucokinase regulator (GCKR) gene.
2. TCF7L2
The transcription factor 7-like 2 gene (TCF7L2) is located on chromosome 10q25.3 and contains 18 exons, presenting complex splicing in different tissues. TCF7L2 is a member of the T-cell factor/lymphoid enhancer binding factor family (TCF/LEF), the transcription factor of the Wnt/β-catenin signaling pathway. Mechanistically, TCF7L2 heterodimerizes with β-catenin, then binds to DNA through a high-mobility group (HMG)-box domain to act as either a stimulator or repressor of target gene expression [6].
Several TCF7L2 SNPs have been reported to be associated with a high risk of developing GDM in various populations [7,8,9,10]. Notably, all SNPs are in intronic regions, as shown in Table 1, suggesting the regulatory function of the SNPs in cell-context specificity. Among TCF7L2 variants, rs7903146 (C > T) presented a strong association with GDM risk and increased more than fivefold in the TT genotype in Caucasian women [11]. In contrast, there were no significant differences in the frequencies of the TCF7L2 rs7903146 genotypes between GDM and normal healthy women in the Chinese population; however, this SNP affected glycolipid metabolism in GDM women [9]. The risk allele at SNP rs7903146, identified through fine mapping, coincided with enhanced histone marks in both islets and adipose tissue [12]. This SNP was associated with the offspring’s birth weight [13]. Notably, TCF7L2 SNPs rs7895340 and rs11196205 demonstrated a significant association with T2D in Thai patients similar to the association observed with SNP rs7903146 in Europeans, suggesting the effect of SNPs on the association in different ethnicities [14]. TCF7L2 is expressed in many tissues involved in glucose and lipid metabolism, such as adipose tissue, liver, pancreas, and placenta.

Table 1.
Gestational diabetes mellitus–associated SNPs.
Pancreas-specific and β-cell-specific Tcf7l2 null mice altered glucose homeostasis by decreasing glucose tolerance, impairing insulin secretion, and reducing pancreatic β-cell volume [15,16]. TCF7L2 was bound to phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) promoter to suppress the encoding of p38, resulting in the activation of p-Akt and an increase in insulin secretion [17]. Selective deletion of Tcf7l2 in mice pancreatic α-cells showed a lower α-cell mass and defective counterregulatory response by increasing glucose infusion rates and decreasing plasma glucose concentration. Loss of Tcf7l2 also suppressed the expression of glucagon (Gcg) and MAF BZIP transcription factor B (Mafb) in the islet [18]. However, no effects were found on insulin (Ins) gene expression in insulin-positive cells and β-cell mass [18]. A Tcf7l2 mutant in zebrafish showed hyperglycemia, pancreatic and vascular defects, and decreasing regeneration [19]. RNAseq in zebrafish adult pancreatic cell type revealed that Tcf7l2 exhibited the highest expression in acinar tissue and was expressed in δ-cells but not in β-cells [19].
TCF7L2 plays a role in the development and functioning of adipose tissue. Humans with impaired glucose tolerance and adipocyte insulin resistance [20], humans with obesity [12], and mice with high-fat diet-induced obesity [21] showed low TCF7L2 levels. TCF7L2 was required for Wnt/β-catenin signaling during adipogenesis [20]. Conditional adipocyte Tcf7l2f/f knockout mice induced adipocyte hypertrophy and impaired lipolysis response to fasting [21]. Specifically in mature adipocytes in vivo, disabling the TCF7L2 protein by eliminating the HMG-box DNA-binding domain resulted in overall glucose intolerance and insulin resistance in the liver [20]. Transcriptome-wide profiling indicated that TCF7L2 regulates the extracellular matrix, immune response, and apoptosis in the adipose progenitor [12]. However, the actions of TCF7L2 in adipose tissue remain controversial. Human adipose progenitor had the highest expression of TCF7L2 mRNA but then decreased during differentiation [12]. In contrast, Tcf7l2 was elevated during adipocyte differentiation in murine 3T3-L1 cells [20]. In obesity, the protein level of TCF7L2 is reduced in whole adipose tissue but increased in adipocyte progenitor. Knockdown of TCF7L2 in adipose progenitor showed activation of Wnt/β-catenin signaling in a dose-dependent manner, resulting in impaired proliferation and adipogenesis, while overexpression of TCF7L2 activated adipocyte differentiation [12]. Finally, SNP rs7903146 minor allele T increased insulin accumulation in adipose tissue by decreasing the level of TCF7L2 in adipose progenitor but not in mature adipocytes. A luciferase assay elucidated that the risk allele T abolished the enhancer activity of TCF7L2 [12].
Mice with liver-specific Tcf7l2 knockout (Alb-Cre; Tcf7l2f/f) displayed elevated fasting glucose levels without alterations in body weight, plasma insulin, or insulin-like growth factor-1 levels. The deficiency of hepatic Tcf7l2 resulted in impaired glucose and insulin tolerance, yet no changes were observed in hepatic insulin signaling or energy expenditure. Interestingly, hepatic Tcf7l2 depletion significantly increased the expression of lipogenic genes in the liver, without affecting genes related to β-oxidation and lipolysis [22].
Notably, TCF7L2 is also exhibited in the placenta [23]; however, the mechanism associated with GDM has never been revealed.
3. HHEX
The haematopoietically expressed homeobox (HHEX) gene encodes a member of the homeobox family of transcription factors. The genomic structure of human HHEX comprises four exons. It is mapped in the 270-kb linkage disequilibrium (LD) block of 10q23.33, containing three genes: insulin degradation enzyme (IDE), kinesin family member 11 (KIF11), and HHEX [24].
HHEX variants rs1111875 (T > C), rs5015480 (T > C), and rs7923837 (A > G) were associated with increased susceptibility to GDM [25,26,27]. All three SNPs are in the intergenic region of the IDE-KIF11-HHEX LD block, in which the closest gene is HHEX [25]. However, in silico studies, such as cis-expression quantitative trait locus (cis-eQTL), which linked SNPs to HHEX, have never been confirmed. Interestingly, the major alleles in all SNPs increased the risk of GDM and are mainly predicted to be the binding sites of forkhead box (FOX) transcription factors, as shown in Table 1.
HHEX plays roles during pancreas and liver embryogenic development, which may affect glucose homeostasis and insulin secretion later in life. The pancreas and liver originate from a shared pool of progenitors, and HHEX plays a crucial role in development by serving as a gatekeeper for pancreatic lineage specification by collaborating with pancreatic transcription factors and endodermal transcription factors like FOXA2 and GATA4 [28]. HHEX participates in the development of the pancreatic endoderm by controlling the expression of genes associated with pancreatic development, including NKX6.1, PTF1A, ONECUT1, and ONECUT3 [29]. Notably, HHEX was not detected in the pancreatic β-cells and α-cells of adult humans and mice, while it was exhibited in adult pancreatic δ-cells and regulated somatostatin secretion [30]. Lysine-specific demethylase-1 targeted the Hhex locus and promoted the repression of Hhex by facilitating methylation at H3K4me1/2, leading to the prevention of the transition of β-cells to δ-cells [31].
Although HHEX is expressed in adipocytes [32] and the placenta [33], studies of mechanism and function remain limited. Knockdown of Hhex in preadipocyte cell line 3T3-L1 suppressed adipogenesis in a dose-dependent manner and led to the suppression of peroxisome proliferator-activated receptor gamma (PPARG) level [32].
4. SLC30A8
The solute carrier family 30 member 8 gene (SLC30A8) encodes zinc transporter 8 (ZnT8) protein, which is highly specifically expressed in insulin-producing β-cells and required for insulin biosynthesis and secretion. SLC30A8 is located on chromosome 8q24.11 and consists of eight exons spanning a length of 37 kb and encoding a 369-amino acid protein [34].
Three SLC30A8 SNPs have been reported to be associated with the risk of GDM (Table 1). SNP rs13266634 (C > T) is a missense variant affecting the amino acid residue 325 in the ZnT8 C-terminal. The major allele CGG at the codon results in the production of arginine (R), whereas the minor allele TGG leads to the generation of tryptophan (W), denoted as R325W. This SNP showed significant associations with GDM and ethnicity specificity. The rs13266634 C allele increased the risk of developing GDM [35,36,37] whereas the T allele was a protective variant against the development of GDM [36,38,39]. Loss of ZnT8 function improved glucose responsiveness and enhanced proinsulin conversion, leading to more effective insulin secretion and preservation of the function of β-cells [40,41]. Mechanistically, the protective T allele enhanced the response to the glucose challenge by increasing insulin secretion and decreasing glucagon secretion in primary islets. Additionally, there was a tendency for the T allele to lower the expression of SLC30A8 (p = 0.053) and genes related to proinsulin processing [42].
The other two SLC30A8 SNPs, rs3802177 (G > A) and rs2466293 (A > G), are both located at 3′ UTR variants. SLC30A8 rs3802177 [7] and rs2466293 [38] were significantly associated with an increased risk of GDM in the Chinese population and were predicted to be binding sites for transcription factors, as shown in Table 1. However, the molecular mechanisms governing how these SNPs affect the expression of SLC30A8 require further investigation.
5. ADIPOQ
The adiponectin, C1Q, and collagen domain containing gene (ADIPOQ) is located on chromosome 3q27.3 and is major expressed in adipose tissue. It encodes adiponectin, a 30 kDa circulating protein in the plasma, which is involved in various physiological mechanisms encompassing energy metabolism and insulin sensitivity [43]. Women with GDM often experience hypoadiponectinemia, leading to insulin resistance and glucose intolerance [44].
SNPs rs17300539 (−11391G > A) and rs266729 (−11377C > G) in the promoter region, rs2241766 (+45T > G) in exon 2, and rs1501299 (+276G > T) in intron 2 have been reported as being associated with the risk of GDM development (Table 1). These four variants are situated within the two ADIPOQ LD blocks. Block 1 includes the promoter sequence spanning from −14,811 to −4120, while block 2 encompasses the exons within the region −450 to +4545 [45].
ADIPOQ rs266729 (C > G) allele G increased the risk of GDM in Asian and European populations [46,47], while reducing it in the American population [48]. The study of Chinese women did not show a significant difference in the genotypes and allele frequencies of rs266729 between GDM patients and the control [49]. The patient’s carrier allele G had a higher adiponectin level [46] and diastolic blood pressure [49] compared to carrier allele C. In contrast, the study of Bulgarian women elucidated that the G allele had a protective effect against GDM [50]. Another SNP in the promoter region, rs17300539 (G > A) allele G, was associated with lower adiponectin [51]. Notably, the variant rs17300539, along with rs266729, has been reported as having no association with GDM in black South African women [52].
SNP rs2241766 (T > G) is a synonymous mutation (Gly15Gly) at exon 2. Although rs2241766 allele G was associated with the risk of GDM [53], the effects of this SNP on adiponectin levels in serum are still controversial in different ethnicities [45,46,54,55]. In Thai women with GDM, the study showed no significant differences in adiponectin levels between different genotypes [46]. However, in Chinese patients, allele G was associated with higher adiponectin levels compared to allele T [55]. Conversely, in Malaysian patients, the trend was reversed, with allele T linked to higher adiponectin levels [54].
Rs1501299 (G > T) showed an association with the risk of T1D, T2D, and GDM [56]; however, there are still few studies on GDM [46,49,53,57]. Tangjittipokin et al. elucidated that patients with minor allele T had higher fasting glucose levels than those with major allele G [46]. GDM patients with the rs1501299 GT genotype had lower diastolic blood pressure than those with the GG genotype [49].
Taken together, the ADIPOQ SNPs are important susceptibility factors for developing GDM. Nevertheless, the molecular mechanisms governing how these SNPs affect the level of adiponectin need to be investigated.
6. FTO
The human fat mass and obesity-related gene (FTO) is mapped on chromosome 16q12.2, including nine exons and eight introns. FTO encodes a 2-oxoglutarate-dependent nucleic acid demethylase, and its main substrate is N6-methyladenosine (m6A) in nuclear RNA, controlling several post-transcriptional regulatory processes [58]. FTO is widely expressed in adipose tissues and skeletal muscles in humans, playing a crucial role in the regulation of body weight and fat mass. FTO is also a critical regulator of adipogenesis that acts in the early stages of adipogenesis, leading to obesity [59]. FTO inhibits insulin secretion and contributes to pancreas islet β-cells dysfunction [60]. However, no study has investigated the role of skeletal muscles in the functioning of the FTO gene in GDM.
FTO rs1121980 (G > A) minor allele A was found to be significantly associated with a reduced risk of GDM [61]. Additionally, the SNP rs9939609 (T > A) showed a significant association with the risk of developing GDM, and the association was detected only in the Caucasian subgroup by meta-analysis [37]. Individuals carrying the AT genotype of rs9939609 were found to have a higher risk of exceeding excessive gestational weight gain earlier than those with the TT genotype [62]. Notably, multiple meta-analysis studies have reported an inability to detect significant associations between FTO polymorphisms, including rs9939609, rs8050136, rs1421085, and rs1121980, and the risk of developing GDM [63,64,65,66,67].
Although the FTO SNPs rs8050136, rs9939609, and rs1421085 did not emerge as major genetic regulators in the development of GDM, these SNPs were found to be associated with concentrations of adiponectin and TNFα in subjects with GDM [63]. SNP rs8050136 showed no significant association with GDM, but posed an increased risk of GDM in Bangladeshi multigravida women [67]. Interestingly, FTO SNPs are located in intron 1 (as listed in Table 1), suggesting a potential role in regulating the expression of FTO itself and possibly influencing nearby genes [58].
7. VEGFA
The vascular endothelial growth factor A (VEGFA) gene is located at chromosome 6p21.3 and consists of eight exons separated by seven introns with a full molecular length of 14 kb. VEGFA undergoes alternative mRNA splicing, resulting in the generation of different-length isoforms. These isoforms possess distinct biological properties, and each plays a specific role in the differentiation and development of the vascular system [68].
Hypervascularity is normally found with increase in placental weight in GDM [69]. However, the plasma levels of VEGFA are still contradictory. Troncoso et al. showed no difference in VEGFA levels between GMD and the control group, while Meng et al. indicated that GDM patients had lower VEGFA levels [70]. In contrast, other studies showed that GDM patients had elevated VEGFA levels [71,72]. VEGFA is also expressed in β-cells and controls development and function. The protein level of VEGFA remained constant in β-cells throughout pregnancy in mice [73]. However, when VEGFA signaling was disrupted in β-cells of pregnant mice, it led to a decrease in the maintenance of islet vessels and induced transient glucose intolerance without affecting the expansion of the β-cell mass [74].
Although the expression of VEGFA seems important in the pathogenesis of GDM, there is only one study on the association of VEFGA polymorphisms with GDM in Chinese women [71]. Five SNPs of the VEGFA gene were selected: rs2146323 (C > A), rs2010963 (G > C), rs3025039 (C > T), rs3025010 (T > C), and rs833069 (G > A) (Table 1). SNPs rs2146323 allele A (intron 2) and rs3025039 allele T (exon 7) showed a dramatically increased risk of GDM. The patients who carried the rs3025039 CT+TT genotype had a higher level of VEGFA than those who carried the CC genotype. Other SNPs showed no difference in the distributions of genotypes and alleles between GDM and control. Interestingly, the frequency of haplotypes CAAC, CAAT, CACC, CACT, GACT, and GGCT for the rs2010963–rs833069–rs2146323–rs3025010 combination was found to be statistically different between women with GDM and healthy women [71].
An imbalance in the expression of various other vascular endothelial growth factors (VEGFs) and their receptors (VEGFRs) also influences the pathophysiology of the GDM [75], future studies examining susceptibility variants and the underlying biological mechanisms will likely unveil associations within this growth factor family with GDM.
8. CDKAL1
The CDK5 regulatory subunit associated protein 1-like 1 gene (CDKAL1) is in chromosome 6p22.3. This gene encodes a 65-kDas CDKAL1 protein, functioning as a methylthiotransferase. It acts as a tRNA modification enzyme, playing a role in enhancing the translation of various transcripts in pancreatic β-cells, including that of proinsulin. CDKAL1 also serves as an endogenous inhibitor of cyclin-dependent kinase 5 (CDK5). The inhibition of CDK5 by CDKAL1 plays a crucial role in preventing the translocation of PDX1, a transcription factor responsible for regulating the insulin gene. This disruption in the translocation process leads to an interruption in insulin production [76]. In adipocytes, CDKAL1 negatively regulates the adipocyte-specific genes in response to hyperglycemic and hyperlipidemic conditions [77]. CDKAL1 regulates the differentiation of adipocytes in murine 3T3-L1 cells through the Wnt/β-catenin signaling pathway [78]. Additionally, in adipose-specific Cdkal1 knockout mice, CDKAL1 is involved in controlling mitochondrial function [79].
The GDM-associated SNPs fall within an intronic 5 region of the CDKAL1 locus (Table 1) and have recently been the hot spot for studying the risk with GDM. SNP rs7754840 (G > C) allele C significantly correlated with GDM risk in meta-analysis [64,80,81]; however, the associations varied across different ethnicities. No association between variations and GDM was found in Egyptian and Chinese populations [82,83]. Meanwhile, Caucasian women showed an association, and Bangladeshi women had a significant association only after adjusting for gravidity and family history of diabetes [39,84]. Meta-analysis showed that SNP rs7756992 (A > G) minor allele G increased the risk of GDM in Bangladeshi women [64,80,84]. The allele G of this SNP also showed a strong association with impaired glucose metabolism, low birth weight, and a decreased insulin secretion index later in life [85]. Interestingly, SNP rs7747752 (G > C) was reported as being associated with GDM risk without the previous report on association with T2D. The studies conducted on Chinese women elucidated that SNP rs7747752 allele C was genetically associated with elevated GDM risk and with interaction with other factors [86,87,88]. Another SNP, rs9368222 minor allele A, which is in the same block as rs7747752, was also associated with the risk of GDM in the Hispanic population but not in the Caucasian population [89]. For SNP rs10946398 (A > C), well known to be associated with T2D, it was recently reported that allele C was associated with GDM in Pakistani women [90]. In contrast, the CC genotype was not associated with GDM, but was associated with the need for insulin therapy in Caucasian women [91]. Notably, the CDKAL1 locus stands out as a prominent example of ancestry-correlated heterogeneity, as multi-ancestry meta-analyses showed that the SNPs at this locus exhibited more pronounced effects on GDM in GWAS conducted on individuals of East Asian ancestry compared to other populations [92].
9. MTNR1B
The melatonin receptor 1B gene (MTNR1B), located in chromosome 11q14.3, encodes melatonin receptor MT2, which is expressed in the human brain, pancreatic β-cells, and skeletal muscle. MT2 not only regulates circadian rhythm, but is also involved in glucose metabolism. Melatonin binds with MT2 and suppresses cyclic guanosine monophosphate (cGMP) signaling, which results in a decrease in insulin secretion in pancreatic β-cells. In the skeletal muscle, the binding of melatonin to MT2 stimulates glucose transport into cells via the insulin receptor substrate-1 (IRS-1)/phosphoinositide 3-kinase (PI-3-kinase) pathway [93]. MTNR1B is also expressed in the placenta, and GDM women have higher levels of MT2 compared to normal women [94]. A study of trophoblast HTR-8/SVneo cells showed that melatonin upregulates GLU4, PPARG, and MT2 expression, leading to elevated glucose consumption [94].
Several MTNR1B polymorphisms were shown to be associated with GDM (Table 1). SNP rs10830963 (C > G), within the single 11.5-kb intron, is the most popular variant, and allele G showed susceptibility to increased risk of GDM from meta-analysis and different ethnicities [7,13,35,39,66,89,94,95,96,97]. The placental expression level of MT2 in GDM women carrying the GG and GC genotypes was higher than in those carrying the CC genotype [95]. SNPs rs1387153 (C > T) allele T and rs10830962 (C > G) allele G are both located in the intergenic region over 2 kb upstream from MTNR1B and demonstrated a significant association with an increased risk of GDM [96,98,99,100,101]. Notably, maternal rs10830963 (C > G) had an association with offspring birth weight [13,102]. SNP rs4753426 (T > C), located in the MTNR1B promoter, still has contradictory reports. No statistically significant differences were observed in the distribution of MTNR1B rs4753426 genotypes and alleles between women with GDM and healthy pregnant women in a study conducted on the Caucasian population [66]. However, rs4753426 minor allele C was associated with susceptibility to GDM [96,98], and another study demonstrated that major allele T was a protective variant for GDM development [99].
10. GLO1
The glyoxalase I gene (GLO1) is located on chromosome 6p21.2 and is predominantly expressed in the human skeleton. GLO1 serves as a protective enzyme against dicarbonyl stress. Mechanistically, methylglyoxal (MG) is a byproduct of glycolysis, and GLO1 plays a crucial role in detoxifying MG to D-lactate. Elevated levels of MG can directly inhibit insulin signaling by binding to IRS-1 within the skeletal muscle and by binding to circulating insulin. Conditions associated with metabolic impairment, such as obesity, insulin resistance, and T2D, have been observed to suppress the expression of GLO1, disrupting the MG/GLO1 axis in skeletal muscle metabolism [103].
The association between GLO1 polymorphisms and GDM has only one report from Zeng et al., who recently showed the effects in the Chinese population [104]. SNP rs1130534 (T > A) is a synonymous mutation at codon 124 (Gly124Gly). The AA genotype of rs1130534 was identified as a protective factor against GDM. The TA genotype increased fasting glucose levels and the risk of GDM. Furthermore, newborn weight was notably lower in individuals with the TA genotype than in those with the TT genotype. SNP rs4746 (T > G) is a missense variant affecting the amino acid residue 111 in exon 4. The codon major allele GAG generates a glutamic acid (E), while the minor allele GCG instead generates an alanine (A) (E111A). The GG genotype and the G allele were associated with a lower risk of GDM [104]. Another GLO1 variant, rs1781735, is in the promoter, which significantly affects the transcriptional activity of GLO1 [105]. This SNP showed no association with the risk of GDM, but the GG genotype was more associated with cumulative neonatal weight and MG levels than the TG or TT genotypes. Also, individuals with the GG genotype exhibited significantly higher MG levels than individuals with the TT genotype. The combined effects of multiple SNPs, specifically the rs1781735–rs4746–rs1130534 (T–G–T) haplotypes, were found to significantly reduce the risk of developing GDM [104]. Notably, none of the GLO1 variants is predicted to be the binding site of the transcription factor, as shown in Table 1.
11. GCK and GCKR
The glucokinase gene (GCK) is situated on chromosome 7p15.3, composed of 10 coding exons, and is expressed in pancreatic β-cells, liver, and brain. On the other hand, the glucokinase regulator gene (GCKR) is located on chromosome 2p23.3 and encodes a glucokinase regulatory protein (GKRP), responsible for controlling the activity of GCK. GCK, in turn, plays a key role in glucose storage and disposal in the liver and modulates insulin secretion in the pancreas. Mechanistically, GKRP forms an inactive complex with GCK at basal glucose concentrations. Together, GCK and GKRP work in tandem, maintaining blood glucose homeostasis. Variants in the GCK and GCKR genes can disrupt the balance of the GCK/GKRP complex, leading to abnormal glucose concentrations and hyperglycemia [106,107]. Indeed, heterozygous inactivating mutations in GCK can lead to a form of diabetes known as maturity-onset diabetes of the young (GCK-MODY). This condition presents a challenge in diagnosis, especially during pregnancy, as it can easily be confused with GDM [108].
GCK SNPs rs1799884 (C > T) and rs4607517 (G > A) were both reported to be associated with T2D; however, the association was different in GDM. SNP rs1799884 (C > T) is in the GCK promoter without any study and prediction to be the binding site of transcription factor, as shown in Table 1. The association of this variant with GDM is still contradicted in Asian and Caucasian populations. GCK rs179988 minor allele T was significantly associated with GDM [39,109,110] but no association was reported [61,86,91]. GDM women showed a predominance of the T allele and more commonly required insulin treatment [91]. SNP rs4607517 (G > A), an intergenic variant between GCK and YKT6, had no report to show an association with GDM [61,86,109], except for the interaction between this SNP and sweets consumption on GDM [111]. However, fasting blood glucose was significantly lower in individuals with the rs4607517 GA genotype than in those with the GG genotype [109]. Notably, the distribution of rs4607517 was not in Hardy–Weinberg equilibrium in the Filipinos [35].
SNP rs1260326 (C > T) is a missense variant at GCKR exon 15, which substitutes proline to leucine at position 446 (P446L). Nevertheless, there was no difference in genotypes between cases and controls [11] and no association of rs1260326 with GDM risk was found from the meta-analysis [37,64]. Interestingly, the association between rs1260326 major allele C and GDM was shown in Chinese and Euro-Brazilian women [49,112], among whom GDM women with the CC genotype had a higher 1-h oral glucose tolerance test (OGTT) level compared to those with the TT genotype [49]. In contrast, the GCKR intronic rs780094 (T > C) variant was the risk factor for GDM from the meta-analysis [37,64]. However, the case-control studies showed no difference in genotypes between control and GDM women [91], and no association of the SNP with GDM risk was observed [109].
12. Conclusions
The potential to comprehend the pathophysiology of GMD through genetic polymorphisms (Figure 2) holds promise, providing insights into predicting and managing GDM patients’ risks. However, the incorporation of associated SNPs into clinical practice remains a challenge, influenced by such factors as GDM detection criteria, sample size, and ethnicity in GWAS analysis. Additionally, the shared pathogenesis with T2D complicates the identification of unique GDM variants. Given the multicomplex nature of GDM involving various tissues, it is crucial to uncover the underlying mechanism of each genetic variant. Furthermore, genetic information must be considered alongside factors such as diet, lifestyle, and gut microbes that influence individual susceptibility. The continuous exploration of new SNPs, improving the genetic background of GDM, and integrating machine learning into polygenic risk score (PRS) models may have the potential to refine our understanding and offer valuable assistance in clinical practice.
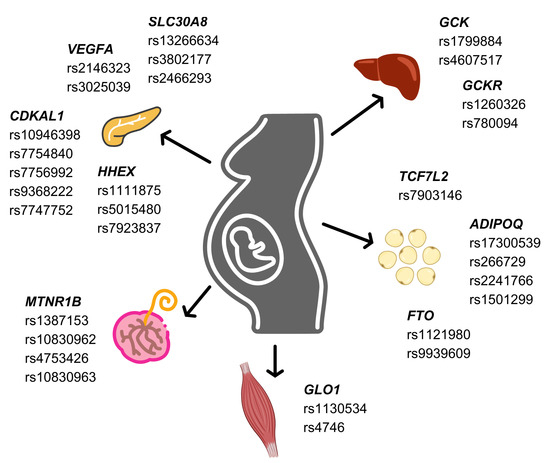
Figure 2.
Associated single nucleotide polymorphisms on gestational diabetes mellitus risk in pancreas, liver, skeletal muscle, adipose tissue, and placenta.
Author Contributions
S.S. primarily wrote the manuscript. W.T. provided supervisory and editorial assistance. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Faculty of Medicine Siriraj Hospital, Mahidol University and by a Health Systems Research Institute (HSRI) (grant No. 66-138) to W.T.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Hilliard, M.E.; Isaacs, D.; Johnson, E.L.; et al. 2. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes-2023. Diabetes Care 2023, 46 (Suppl. S1), S19–S40. [Google Scholar] [CrossRef]
- Plows, J.F.; Stanley, J.L.; Baker, P.N.; Reynolds, C.M.; Vickers, M.H. The Pathophysiology of Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2018, 19, 3342. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Li, P. Novel single nucleotide polymorphisms in gestational diabetes mellitus. Clin. Chim. Acta 2023, 538, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Cheneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Del Bosque-Plata, L.; Martinez-Martinez, E.; Espinoza-Camacho, M.A.; Gragnoli, C. The Role of TCF7L2 in Type 2 Diabetes. Diabetes 2021, 70, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Chavarro, J.; Olsen, S.; Lin, Y.; Ley, S.H.; Bao, W.; Rawal, S.; Grunnet, L.G.; Thuesen, A.C.B.; Mills, J.L.; et al. Genetic variants of gestational diabetes mellitus: A study of 112 SNPs among 8722 women in two independent populations. Diabetologia 2018, 61, 1758–1768. [Google Scholar] [CrossRef]
- Chang, S.; Wang, Z.; Wu, L.; Lu, X.; Shangguan, S.; Xin, Y.; Li, L.; Wang, L. Association between TCF7L2 polymorphisms and gestational diabetes mellitus: A meta-analysis. J. Diabetes Investig. 2017, 8, 560–570. [Google Scholar] [CrossRef]
- Fang, C.; Wu, S.; Zhang, J.; Tian, Q.; Zhang, Z.; Wu, L. Impaired glucolipid metabolism in gestational diabetes mellitus with T variation of TCF7L2 rs7903146: A case–control study. Int. J. Diabetes Dev. Ctries. 2023, 1–8. [Google Scholar] [CrossRef]
- Zhang, P.; Deng, M.; Li, W.; Dai, Q.; He, H.; Zheng, W.; She, L.; Xiang, B.; Zeng, J.; Zhou, F.; et al. The correlation between transcription factor 7-like 2 gene polymorphisms and susceptibility of gestational diabetes mellitus in the population of central China: A case-control study. Front. Endocrinol. 2022, 13, 916590. [Google Scholar] [CrossRef]
- Franzago, M.; Fraticelli, F.; Nicolucci, A.; Celentano, C.; Liberati, M.; Stuppia, L.; Vitacolonna, E. Molecular Analysis of a Genetic Variants Panel Related to Nutrients and Metabolism: Association with Susceptibility to Gestational Diabetes and Cardiometabolic Risk in Affected Women. J. Diabetes Res. 2017, 2017, 4612623. [Google Scholar] [CrossRef]
- Verma, M.; Loh, N.Y.; Sabaratnam, R.; Vasan, S.K.; van Dam, A.D.; Todorcevic, M.; Neville, M.J.; Toledo, E.; Karpe, F.; Christodoulides, C. TCF7L2 plays a complex role in human adipose progenitor biology, which might contribute to genetic susceptibility to type 2 diabetes. Metabolism 2022, 133, 155240. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, R.N.; Warrington, N.M.; Cavadino, A.; Tyrrell, J.; Nodzenski, M.; Horikoshi, M.; Geller, F.; Myhre, R.; Richmond, R.C.; Paternoster, L.; et al. Genome-wide association study of offspring birth weight in 86 577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Hum. Mol. Genet. 2018, 27, 742–756. [Google Scholar] [CrossRef]
- Tangjittipokin, W.; Chongjarean, N.; Plengvidhya, N.; Homsanit, M.; Yenchitsomanus, P.T. Transcription factor 7-like 2 (TCF7L2) variations associated with earlier age-onset of type 2 diabetes in Thai patients. J. Genet. 2012, 91, 251–255. [Google Scholar] [CrossRef] [PubMed]
- da Silva Xavier, G.; Mondragon, A.; Sun, G.; Chen, L.; McGinty, J.A.; French, P.M.; Rutter, G.A. Abnormal glucose tolerance and insulin secretion in pancreas-specific Tcf7l2-null mice. Diabetologia 2012, 55, 2667–2676. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.K.; Mondragon, A.; Chen, L.; McGinty, J.A.; French, P.M.; Ferrer, J.; Thorens, B.; Hodson, D.J.; Rutter, G.A.; Da Silva Xavier, G. Selective disruption of Tcf7l2 in the pancreatic beta cell impairs secretory function and lowers beta cell mass. Hum. Mol. Genet. 2015, 24, 1390–1399. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.H.; Li, Y.L.; Liu, N.J.; Yang, Z.; Tao, X.M.; Du, Y.P.; Wang, X.C.; Lu, B.; Zhang, Z.Y.; Hu, R.M.; et al. TCF7L2 regulates pancreatic beta-cell function through PI3K/AKT signal pathway. Diabetol. Metab. Syndr. 2019, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- da Silva Xavier, G.; Mondragon, A.; Mourougavelou, V.; Cruciani-Guglielmacci, C.; Denom, J.; Herrera, P.L.; Magnan, C.; Rutter, G.A. Pancreatic alpha cell-selective deletion of Tcf7l2 impairs glucagon secretion and counter-regulatory responses to hypoglycaemia in mice. Diabetologia 2017, 60, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Facchinello, N.; Tarifeno-Saldivia, E.; Grisan, E.; Schiavone, M.; Peron, M.; Mongera, A.; Ek, O.; Schmitner, N.; Meyer, D.; Peers, B.; et al. Tcf7l2 plays pleiotropic roles in the control of glucose homeostasis, pancreas morphology, vascularization and regeneration. Sci. Rep. 2017, 7, 9605. [Google Scholar] [CrossRef]
- Chen, X.; Ayala, I.; Shannon, C.; Fourcaudot, M.; Acharya, N.K.; Jenkinson, C.P.; Heikkinen, S.; Norton, L. The Diabetes Gene and Wnt Pathway Effector TCF7L2 Regulates Adipocyte Development and Function. Diabetes 2018, 67, 554–568. [Google Scholar] [CrossRef]
- Geoghegan, G.; Simcox, J.; Seldin, M.M.; Parnell, T.J.; Stubben, C.; Just, S.; Begaye, L.; Lusis, A.J.; Villanueva, C.J. Targeted deletion of Tcf7l2 in adipocytes promotes adipocyte hypertrophy and impaired glucose metabolism. Mol. Metab. 2019, 24, 44–63. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; An, T.H.; Kim, H.; Jung, E.; Kim, G.; Oh, S.Y.; Kim, J.S.; Chun, H.J.; Jung, J.; Lee, E.W.; et al. Tcf7l2 in hepatocytes regulates de novo lipogenesis in diet-induced non-alcoholic fatty liver disease in mice. Diabetologia 2023, 66, 931–954. [Google Scholar] [CrossRef] [PubMed]
- Kuzmicki, M.; Telejko, B.; Wawrusiewicz-Kurylonek, N.; Kalejta, K.; Lemancewicz, A.; Zdrodowski, M.; Grabiec, M.; Pryszczepko-Wawreszuk, A.M.; Kretowski, A.; Gorska, M.; et al. The expression of transcription factor 7-like 2 (TCF7L2) in fat and placental tissue from women with gestational diabetes. Diabetes Res. Clin. Pract. 2011, 94, e43–e46. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Lu, F.; Dong, M.; Lin, Y.; Li, H.; Chen, J.; Shen, C.; Jin, G.; Hu, Z.; Shen, H. Genetic variants of IDE-KIF11-HHEX at 10q23.33 associated with type 2 diabetes risk: A fine-mapping study in Chinese population. PLoS ONE 2012, 7, e35060. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Zhang, L.; Wang, J.; Wang, Y. Association of HHEX and SLC30A8 Gene Polymorphisms with Gestational Diabetes Mellitus Susceptibility: A Meta-analysis. Biochem. Genet. 2023, 61, 2203–2221. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ding, Y.; Zhang, X.; Rao, J.; Yu, H.; Pan, H. The association between HHEX single-nucleotide polymorphism rs5015480 and gestational diabetes mellitus: A meta-analysis. Medicine 2020, 99, e19478. [Google Scholar] [CrossRef] [PubMed]
- Tarnowski, M.; Malinowski, D.; Safranow, K.; Dziedziejko, V.; Czerewaty, M.; Pawlik, A. Hematopoietically expressed homeobox (HHEX) gene polymorphism (rs5015480) is associated with increased risk of gestational diabetes mellitus. Clin. Genet. 2017, 91, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Cho, H.; Tayyebi, Z.; Shukla, A.; Luo, R.; Dixon, G.; Ursu, V.; Stransky, S.; Tremmel, D.M.; Sackett, S.D.; et al. CRISPR screening uncovers a central requirement for HHEX in pancreatic lineage commitment and plasticity restriction. Nat. Cell Biol. 2022, 24, 1064–1076. [Google Scholar] [CrossRef]
- Ito, R.; Kimura, A.; Hirose, Y.; Hatano, Y.; Mima, A.; Mae, S.I.; Keidai, Y.; Nakamura, T.; Fujikura, J.; Nishi, Y.; et al. Elucidation of HHEX in pancreatic endoderm differentiation using a human iPSC differentiation model. Sci. Rep. 2023, 13, 8659. [Google Scholar] [CrossRef]
- Zhang, J.; McKenna, L.B.; Bogue, C.W.; Kaestner, K.H. The diabetes gene Hhex maintains delta-cell differentiation and islet function. Genes Dev. 2014, 28, 829–834. [Google Scholar] [CrossRef]
- Liang, X.; Duan, H.; Mao, Y.; Koestner, U.; Wei, Y.; Deng, F.; Zhuang, J.; Li, H.; Wang, C.; Hernandez-Miranda, L.R.; et al. The SNAG Domain of Insm1 Regulates Pancreatic Endocrine Cell Differentiation and Represses beta- to delta-Cell Transdifferentiation. Diabetes 2021, 70, 1084–1097. [Google Scholar] [CrossRef]
- Evseeva, M.N.; Dyikanov, D.T.; Karagyaur, M.N.; Prikazchikova, T.A.; Sheptulina, A.F.; Balashova, M.S.; Zatsepin, T.S.; Rubtsov, Y.P.; Kulebyakin, K.Y. Hematopoietically-expressed homeobox protein HHEX regulates adipogenesis in preadipocytes. Biochimie 2021, 185, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, G.; Murthi, P.; Quinn, L.; Brennecke, S.P.; Kalionis, B. Homeodomain protein HLX is expressed primarily in cytotrophoblast cell types in the early pregnancy human placenta. Reprod. Fertil. Dev. 2008, 20, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Chimienti, F.; Devergnas, S.; Favier, A.; Seve, M. Identification and cloning of a beta-cell-specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes 2004, 53, 2330–2337. [Google Scholar] [CrossRef] [PubMed]
- Benny, P.; Ahn, H.J.; Burlingame, J.; Lee, M.J.; Miller, C.; Chen, J.; Urschitz, J. Genetic risk factors associated with gestational diabetes in a multi-ethnic population. PLoS ONE 2021, 16, e0261137. [Google Scholar] [CrossRef] [PubMed]
- Dereke, J.; Palmqvist, S.; Nilsson, C.; Landin-Olsson, M.; Hillman, M. The prevalence and predictive value of the SLC30A8 R325W polymorphism and zinc transporter 8 autoantibodies in the development of GDM and postpartum type 1 diabetes. Endocrine 2016, 53, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wang, Y.; Zhang, B.; Jin, Z. Association of type 2 diabetes susceptible genes GCKR, SLC30A8, and FTO polymorphisms with gestational diabetes mellitus risk: A meta-analysis. Endocrine 2018, 62, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Tan, B.; Han, F.; Huang, X.; Huang, J.; Wei, Y.; Guo, R. Association of solute carrier family 30 A8 zinc transporter gene variations with gestational diabetes mellitus risk in a Chinese population. Front. Endocrinol. 2023, 14, 1159714. [Google Scholar] [CrossRef] [PubMed]
- Rosta, K.; Al-Aissa, Z.; Hadarits, O.; Harreiter, J.; Nadasdi, A.; Kelemen, F.; Bancher-Todesca, D.; Komlosi, Z.; Nemeth, L.; Rigo, J.; et al. Association Study with 77 SNPs Confirms the Robust Role for the rs10830963/G of MTNR1B Variant and Identifies Two Novel Associations in Gestational Diabetes Mellitus Development. PLoS ONE 2017, 12, e0169781. [Google Scholar] [CrossRef]
- Zhang, Y.; Jian, W.; He, L.; Wu, J. ZnT8 in T2D: A novel therapeutic target for maintaining insulin secretion capacity. Acta Biochim. Biophys. Sin. 2020, 52, 1050–1051. [Google Scholar] [CrossRef]
- Ma, Q.; Xiao, Y.; Xu, W.; Wang, M.; Li, S.; Yang, Z.; Xu, M.; Zhang, T.; Zhang, Z.N.; Hu, R.; et al. ZnT8 loss-of-function accelerates functional maturation of hESC-derived beta cells and resists metabolic stress in diabetes. Nat. Commun. 2022, 13, 4142. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, O.P.; Lehtovirta, M.; Hastoy, B.; Chandra, V.; Krentz, N.A.J.; Kleiner, S.; Jain, D.; Richard, A.M.; Abaitua, F.; Beer, N.L.; et al. Loss of ZnT8 function protects against diabetes by enhanced insulin secretion. Nat. Genet. 2019, 51, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Moyce Gruber, B.L.; Dolinsky, V.W. The Role of Adiponectin during Pregnancy and Gestational Diabetes. Life 2023, 13, 301. [Google Scholar] [CrossRef] [PubMed]
- Tangjittipokin, W.; Thanatummatis, B.; Wardati, F.; Narkdontri, T.; Teerawattanapong, N.; Boriboonhirunsarn, D. The genetic polymorphisms and levels of adipokines and adipocytokines that influence the risk of developing gestational diabetes mellitus in Thai pregnant women. Gene 2023, 860, 147228. [Google Scholar] [CrossRef] [PubMed]
- Takhshid, M.A.; Haem, Z.; Aboualizadeh, F. The association of circulating adiponectin and +45 T/G polymorphism of adiponectin gene with gestational diabetes mellitus in Iranian population. J. Diabetes Metab. Disord. 2015, 14, 30. [Google Scholar] [CrossRef] [PubMed]
- Tangjittipokin, W.; Narkdontri, T.; Teerawattanapong, N.; Thanatummatis, B.; Wardati, F.; Sunsaneevithayakul, P.; Boriboonhirunsarn, D. The Variants in ADIPOQ are Associated with Maternal Circulating Adipokine Profile in Gestational Diabetes Mellitus. J. Multidiscip. Healthc. 2023, 16, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, A.; Teler, J.; Maciejewska, A.; Sawczuk, M.; Safranow, K.; Dziedziejko, V. Adiponectin and leptin gene polymorphisms in women with gestational diabetes mellitus. J. Assist. Reprod. Genet. 2017, 34, 511–516. [Google Scholar] [CrossRef]
- Bai, Y.; Tang, L.; Li, L.; Li, L. The roles of ADIPOQ rs266729 and MTNR1B rs10830963 polymorphisms in patients with gestational diabetes mellitus: A meta-analysis. Gene 2020, 730, 144302. [Google Scholar] [CrossRef]
- Zhu, M.; Lv, Y.; Peng, Y.; Wu, Y.; Feng, Y.; Jia, T.; Xu, S.; Li, S.; Wang, W.; Tian, J.; et al. GCKR and ADIPOQ gene polymorphisms in women with gestational diabetes mellitus. Acta Diabetol. 2023, 60, 1709–1718. [Google Scholar] [CrossRef]
- Beltcheva, O.; Boyadzhieva, M.; Angelova, O.; Mitev, V.; Kaneva, R.; Atanasova, I. The rs266729 single-nucleotide polymorphism in the adiponectin gene shows association with gestational diabetes. Arch. Gynecol. Obstet. 2014, 289, 743–748. [Google Scholar] [CrossRef]
- Hivert, M.F.; Scholtens, D.M.; Allard, C.; Nodzenski, M.; Bouchard, L.; Brisson, D.; Lowe, L.P.; McDowell, I.; Reddy, T.; Dastani, Z.; et al. Genetic determinants of adiponectin regulation revealed by pregnancy. Obesity 2017, 25, 935–944. [Google Scholar] [CrossRef]
- Dias, S.; Adam, S.; Rheeder, P.; Pheiffer, C. No Association Between ADIPOQ or MTHFR Polymorphisms and Gestational Diabetes Mellitus in South African Women. Diabetes Metab. Syndr. Obes. 2021, 14, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.T.; Wu, S.L.; Liao, X.; Ma, S.J.; Tan, H.Z. Adiponectin gene polymorphisms and risk of gestational diabetes mellitus: A meta-analysis. World J. Clin. Cases 2019, 7, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Low, C.F.; Mohd Tohit, E.R.; Chong, P.P.; Idris, F. Adiponectin SNP45TG is associated with gestational diabetes mellitus. Arch. Gynecol. Obstet. 2011, 283, 1255–1260. [Google Scholar] [CrossRef]
- Feng, Y.; Jiang, C.D.; Chang, A.M.; Shi, Y.; Gao, J.; Zhu, L.; Zhang, Z. Interactions among insulin resistance, inflammation factors, obesity-related gene polymorphisms, environmental risk factors, and diet in the development of gestational diabetes mellitus. J. Matern. Fetal. Neonatal. Med. 2019, 32, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Howlader, M.; Sultana, M.I.; Akter, F.; Hossain, M.M. Adiponectin gene polymorphisms associated with diabetes mellitus: A descriptive review. Heliyon 2021, 7, e07851. [Google Scholar] [CrossRef] [PubMed]
- Cakina, S.; Ulu, S.; Beyazit, F.; Özen, E.; Postacı, E.S. Association between adiponectin and gene polymorphism in gestational diabetes mellitus patients. Rev. Romana De Med. De Lab. 2023, 31, 119–124. [Google Scholar]
- Lan, N.; Lu, Y.; Zhang, Y.; Pu, S.; Xi, H.; Nie, X.; Liu, J.; Yuan, W. FTO—A Common Genetic Basis for Obesity and Cancer. Front. Genet. 2020, 11, 559138. [Google Scholar] [CrossRef]
- Merkestein, M.; Laber, S.; McMurray, F.; Andrew, D.; Sachse, G.; Sanderson, J.; Li, M.; Usher, S.; Sellayah, D.; Ashcroft, F.M.; et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat. Commun. 2015, 6, 6792. [Google Scholar] [CrossRef]
- Fan, H.Q.; He, W.; Xu, K.F.; Wang, Z.X.; Xu, X.Y.; Chen, H. FTO Inhibits Insulin Secretion and Promotes NF-kappaB Activation through Positively Regulating ROS Production in Pancreatic beta cells. PLoS ONE 2015, 10, e0127705. [Google Scholar]
- Cao, M.; Zhang, L.; Chen, T.; Shi, A.; Xie, K.; Li, Z.; Xu, J.; Chen, Z.; Ji, C.; Wen, J. Genetic Susceptibility to Gestational Diabetes Mellitus in a Chinese Population. Front. Endocrinol. 2020, 11, 247. [Google Scholar] [CrossRef]
- Santos, K.D.; Rosado, E.L.; da Fonseca, A.C.P.; Belfort, G.P.; da Silva, L.B.G.; Ribeiro-Alves, M.; Zembrzuski, V.M.; Martinez, J.A.; Saunders, C. FTO and ADRB2 Genetic Polymorphisms Are Risk Factors for Earlier Excessive Gestational Weight Gain in Pregnant Women with Pregestational Diabetes Mellitus: Results of a Randomized Nutrigenetic Trial. Nutrients 2022, 14, 1050. [Google Scholar] [CrossRef] [PubMed]
- Saucedo, R.; Valencia, J.; Gutierrez, C.; Basurto, L.; Hernandez, M.; Puello, E.; Rico, G.; Vega, G.; Zarate, A. Gene variants in the FTO gene are associated with adiponectin and TNF-alpha levels in gestational diabetes mellitus. Diabetol. Metab. Syndr. 2017, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Long, W.; Zhou, W.; Zhang, B.; Liu, J.; Yu, B. FTO, GCKR, CDKAL1 and CDKN2A/B gene polymorphisms and the risk of gestational diabetes mellitus: A meta-analysis. Arch. Gynecol. Obstet. 2018, 298, 705–715. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Cao, W.T.; Zeng, Y.H.; Huang, Z.Q.; Du, W.R.; Guan, N.D.; Zhao, Y.Z.; Wei, B.R.; Liu, Y.H.; Jing, C.X.; et al. Lack of associations between the FTO polymorphisms and gestational diabetes: A meta-analysis and trial sequential analysis. Gene 2018, 677, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Tarnowski, M.; Bujak, J.; Kopytko, P.; Majcher, S.; Ustianowski, P.; Dziedziejko, V.; Safranow, K.; Pawlik, A. Effect of FTO and IGF2BP2 gene polymorphisms on duration of pregnancy and Apgar scores in women with gestational diabetes. J. Obstet. Gynaecol. 2019, 39, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Amin, U.S.M.; Rahman, T.A.; Hasan, M.; Tofail, T.; Hasanat, M.A.; Seraj, Z.I.; Salimullah, M. Type 2 diabetes linked FTO gene variant rs8050136 is significantly associated with gravidity in gestational diabetes in a sample of Bangladeshi women: Meta-analysis and case-control study. PLoS ONE 2023, 18, e0288318. [Google Scholar] [CrossRef] [PubMed]
- Melincovici, C.S.; Bosca, A.B.; Susman, S.; Marginean, M.; Mihu, C.; Istrate, M.; Moldovan, I.M.; Roman, A.L.; Mihu, C.M. Vascular endothelial growth factor (VEGF)—Key factor in normal and pathological angiogenesis. Rom. J. Morphol. Embryol. 2018, 59, 455–467. [Google Scholar]
- Troncoso, F.; Acurio, J.; Herlitz, K.; Aguayo, C.; Bertoglia, P.; Guzman-Gutierrez, E.; Loyola, M.; Gonzalez, M.; Rezgaoui, M.; Desoye, G.; et al. Gestational diabetes mellitus is associated with increased pro-migratory activation of vascular endothelial growth factor receptor 2 and reduced expression of vascular endothelial growth factor receptor 1. PLoS ONE 2017, 12, e0182509. [Google Scholar] [CrossRef]
- Meng, Q.; Shao, L.; Luo, X.; Mu, Y.; Xu, W.; Gao, L.; Xu, H.; Cui, Y. Expressions of VEGF-A and VEGFR-2 in placentae from GDM pregnancies. Reprod. Biol. Endocrinol. 2016, 14, 61. [Google Scholar] [CrossRef]
- Dong, P.P. Association of vascular endothelial growth factor expression and polymorphisms with the risk of gestational diabetes mellitus. J. Clin. Lab. Anal. 2019, 33, e22686. [Google Scholar] [CrossRef]
- Sirico, A.; Rossi, E.D.; Degennaro, V.A.; Arena, V.; Rizzi, A.; Tartaglione, L.; Di Leo, M.; Pitocco, D.; Lanzone, A. Placental diabesity: Placental VEGF and CD31 expression according to pregestational BMI and gestational weight gain in women with gestational diabetes. Arch. Gynecol. Obstet. 2023, 307, 1823–1831. [Google Scholar] [CrossRef]
- Yang, W.; Jiang, Y.; Wang, Y.; Zhang, T.; Liu, Q.; Wang, C.; Swisher, G.; Wu, N.; Chao, C.; Prasadan, K.; et al. Placental growth factor in beta cells plays an essential role in gestational beta-cell growth. BMJ Open Diabetes Res. Care 2020, 8, e000921. [Google Scholar] [CrossRef] [PubMed]
- Staels, W.; Heremans, Y.; Leuckx, G.; Van Gassen, N.; Salinno, C.; De Groef, S.; Cools, M.; Keshet, E.; Dor, Y.; Heimberg, H.; et al. Conditional islet hypovascularisation does not preclude beta cell expansion during pregnancy in mice. Diabetologia 2017, 60, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Bolatai, A.; He, Y.; Wu, N. Vascular endothelial growth factor and its receptors regulation in gestational diabetes mellitus and eclampsia. J. Transl. Med. 2022, 20, 400. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, C.; Das, N.; Saha, S.; Kundu, T.; Sircar, D.; Roy, P. Involvement of Cdkal1 in the etiology of type 2 diabetes mellitus and microvascular diabetic complications: A review. J. Diabetes Metab. Disord. 2022, 21, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Okamura, T.; Yanobu-Takanashi, R.; Takeuchi, F.; Isono, M.; Akiyama, K.; Shimizu, Y.; Goto, M.; Liang, Y.Q.; Yamamoto, K.; Katsuya, T.; et al. Deletion of CDKAL1 affects high-fat diet-induced fat accumulation and glucose-stimulated insulin secretion in mice, indicating relevance to diabetes. PLoS ONE 2012, 7, e49055. [Google Scholar] [CrossRef] [PubMed]
- Take, K.; Waki, H.; Sun, W.; Wada, T.; Yu, J.; Nakamura, M.; Aoyama, T.; Yamauchi, T.; Kadowaki, T. CDK5 Regulatory Subunit-Associated Protein 1-like 1 Negatively Regulates Adipocyte Differentiation through Activation of Wnt Signaling Pathway. Sci. Rep. 2017, 7, 7326. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.J.; Bruckner, R.J.; Paulo, J.A.; Kazak, L.; Long, J.Z.; Mina, A.I.; Deng, Z.; LeClair, K.B.; Hall, J.A.; Hong, S.; et al. Cdkal1, a type 2 diabetes susceptibility gene, regulates mitochondrial function in adipose tissue. Mol. Metab. 2017, 6, 1212–1225. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Y.; Song, L.P.; Wei, S.D.; Wen, X.L.; Liu, D.B. CDK5 Regulatory Subunit-Associated Protein 1-Like 1 Gene Polymorphisms and Gestational Diabetes Mellitus Risk: A Trial Sequential Meta-Analysis of 13,306 Subjects. Front. Endocrinol. 2021, 12, 722674. [Google Scholar] [CrossRef]
- Mahdizade, A.H.; Bahreiny, S.S.; Bastani, M.-N.; Dabbagh, M.R.; Aghaei, M.; Ali Malayeri, F.; YousefiFard, A.; Taghizadeh, E. The influence of CDKAL1 (rs7754840) gene polymorphism on susceptibility to gestational diabetes mellitus in pregnant women: A systematic review and meta-analysis. Int. J. Diabetes Dev. Ctries. 2023, 1–10. [Google Scholar] [CrossRef]
- Noury, A.E.; Azmy, O.; Alsharnoubi, J.; Salama, S.; Okasha, A.; Gouda, W. Variants of CDKAL1 rs7754840 (G/C) and CDKN2A/2B rs10811661 (C/T) with gestational diabetes: Insignificant association. BMC Res. Notes 2018, 11, 181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nie, M.; Li, W.; Ping, F.; Hu, Y.; Ma, L.; Gao, J.; Liu, J. Association of six single nucleotide polymorphisms with gestational diabetes mellitus in a Chinese population. PLoS ONE 2011, 6, e26953. [Google Scholar] [CrossRef] [PubMed]
- Amin, U.S.M.; Parvez, N.; Rahman, T.A.; Hasan, M.R.; Das, K.C.; Jahan, S.; Hasanat, M.A.; Seraj, Z.I.; Salimullah, M. CDKAL1 gene rs7756992 A/G and rs7754840 G/C polymorphisms are associated with gestational diabetes mellitus in a sample of Bangladeshi population: Implication for future T2DM prophylaxis. Diabetol. Metab. Syndr. 2022, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.F.; Xiao, X.H.; Zhang, Z.X.; Liu, Y.; Xu, T.; Zhu, X.L.; Zhang, Y.; Wu, X.P.; Li, W.H.; Zhang, H.B.; et al. Positive Association Between Type 2 Diabetes Risk Alleles Near CDKAL1 and Reduced Birthweight in Chinese Han Individuals. Chin. Med. J. 2015, 128, 1873–1878. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, Q.; Feng, Y.; Yang, H.; Wu, W.; Zhang, P.; Wang, Y.; Ko, J.; Zhao, F.; Du, W.; et al. Single Nucleotide Polymorphisms in CDKAL1 Gene Are Associated with Risk of Gestational Diabetes Mellitus in Chinese Population. J. Diabetes Res. 2019, 2019, 3618103. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, W.; Liu, J.; Leng, J.; Li, W.; Yu, Z.; Li, J.; Ma, R.C.W.; Hu, G.; Fang, Z.; et al. Serum concentrations of SFAs and CDKAL1 single-nucleotide polymorphism rs7747752 are related to an increased risk of gestational diabetes mellitus. Am. J. Clin. Nutr. 2021, 114, 1698–1707. [Google Scholar] [CrossRef]
- Wang, H.; Li, J.; Leng, J.; Li, W.; Liu, J.; Yan, X.; Yu, Z.; Hu, G.; Ma, R.C.W.; Fang, Z.; et al. The CDKAL1 rs7747752-Bile Acids Interaction Increased Risk of Gestational Diabetes Mellitus: A Nested Case-Control Study. Front. Endocrinol. 2022, 13, 808956. [Google Scholar] [CrossRef]
- Ramos-Levi, A.; Barabash, A.; Valerio, J.; Garcia de la Torre, N.; Mendizabal, L.; Zulueta, M.; de Miguel, M.P.; Diaz, A.; Duran, A.; Familiar, C.; et al. Genetic variants for prediction of gestational diabetes mellitus and modulation of susceptibility by a nutritional intervention based on a Mediterranean diet. Front. Endocrinol. 2022, 13, 1036088. [Google Scholar] [CrossRef]
- Asghar, A.; Firasat, S.; Afshan, K.; Naz, S. Association of CDKAL1 gene polymorphism (rs10946398) with gestational diabetes mellitus in Pakistani population. Mol. Biol. Rep. 2023, 50, 57–64. [Google Scholar] [CrossRef]
- Tarnowski, M.; Malinowski, D.; Pawlak, K.; Dziedziejko, V.; Safranow, K.; Pawlik, A. GCK, GCKR, FADS1, DGKB/TMEM195 and CDKAL1 Gene Polymorphisms in Women with Gestational Diabetes. Can. J. Diabetes 2017, 41, 372–379. [Google Scholar] [CrossRef]
- Pervjakova, N.; Moen, G.H.; Borges, M.C.; Ferreira, T.; Cook, J.P.; Allard, C.; Beaumont, R.N.; Canouil, M.; Hatem, G.; Heiskala, A.; et al. Multi-ancestry genome-wide association study of gestational diabetes mellitus highlights genetic links with type 2 diabetes. Hum. Mol. Genet. 2022, 31, 3377–3391. [Google Scholar] [CrossRef]
- Zhu, H.; Zhao, Z.J.; Liu, H.Y.; Cai, J.; Lu, Q.K.; Ji, L.D.; Xu, J. The melatonin receptor 1B gene links circadian rhythms and type 2 diabetes mellitus: An evolutionary story. Ann. Med. 2023, 55, 1262–1286. [Google Scholar] [CrossRef]
- Wei, L.; Jiang, Y.; Gao, P.; Zhang, J.; Zhou, X.; Zhu, S.; Chen, Y.; Zhang, H.; Du, Y.; Fang, C.; et al. Role of melatonin receptor 1B gene polymorphism and its effect on the regulation of glucose transport in gestational diabetes mellitus. J. Zhejiang Univ. Sci. B 2023, 24, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhou, Y.; Qiao, B.; Xu, L.; Li, Y.; Li, C. Association Between a Melatonin Receptor 1B Genetic Polymorphism and Its Protein Expression in Gestational Diabetes Mellitus. Reprod. Sci. 2019, 26, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Shen, Y.; Shi, X.; Gu, X.; Zhang, P.; Liu, Y.; Zhu, A.; Jiang, L. MTNR1B gene on susceptibility to gestational diabetes mellitus: A two-stage hospital-based study in Southern China. Mol. Genet. Genom. 2020, 295, 1369–1378. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Fei, X.; Li, M.; Zhang, Z.; Zhu, W.; Zhang, M.; Chen, X.; Xu, J.; Zhang, M.; Shen, Y.; et al. Associations of the MTNR1B rs10830963 and PPARG rs1801282 variants with gestational diabetes mellitus: A meta-analysis. Int. J. Diabetes Dev. Ctries. 2023, 43, 1029–1042. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, K.; Jin, L.; Zhou, Y.; Shang, X.; Wang, X.; Yu, H. MTNR1B gene variations and high pre-pregnancy BMI increase gestational diabetes mellitus risk in Chinese women. Gene 2023, 894, 148023. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, W.; Liu, B.; Dai, A.; Wang, Y.; She, L.; Zhang, P.; Zheng, W.; Dai, Q.; Yang, M. Melatonin Receptor 1B Genetic Variants on Susceptibility to Gestational Diabetes Mellitus: A Hospital-Based Case-Control Study in Wuhan, Central China. Diabetes Metab. Syndr. Obes. 2022, 15, 1207–1216. [Google Scholar] [CrossRef]
- Shan, D.; Wang, A.; Yi, K. MTNR1B rs1387153 Polymorphism and Risk of Gestational Diabetes Mellitus: Meta-Analysis and Trial Sequential Analysis. Public Health Genom. 2023, 26, 201–211. [Google Scholar] [CrossRef]
- Xie, K.; Chen, T.; Zhang, Y.; Wen, J.; Cui, X.; You, L.; Zhu, L.; Xu, B.; Ji, C.; Guo, X. Association of rs10830962 polymorphism with gestational diabetes mellitus risk in a Chinese population. Sci. Rep. 2019, 9, 5357. [Google Scholar] [CrossRef]
- Liang, Z.; Liu, H.; Wang, L.; Chen, Y.; Zhou, T.; Heianza, Y.; Li, W.; Leng, J.; Wang, J.; Gao, R.; et al. Maternal MTNR1B genotype, maternal gestational weight gain, and childhood obesity. Am. J. Clin. Nutr. 2020, 111, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Mey, J.T.; Haus, J.M. Dicarbonyl Stress and Glyoxalase-1 in Skeletal Muscle: Implications for Insulin Resistance and Type 2 Diabetes. Front. Cardiovasc. Med. 2018, 5, 117. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Yang, T.; Wei, W.; Zou, D.; Wei, Y.; Han, F.; He, J.; Huang, J.; Guo, R. Association between GLO1 variants and gestational diabetes mellitus susceptibility in a Chinese population: A preliminary study. Front. Endocrinol. 2023, 14, 1235581. [Google Scholar] [CrossRef] [PubMed]
- Gale, C.P.; Futers, T.S.; Summers, L.K. Common polymorphisms in the glyoxalase-1 gene and their association with pro-thrombotic factors. Diab. Vasc. Dis. Res. 2004, 1, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Matschinsky, F.M.; Magnuson, M.A.; Zelent, D.; Jetton, T.L.; Doliba, N.; Han, Y.; Taub, R.; Grimsby, J. The network of glucokinase-expressing cells in glucose homeostasis and the potential of glucokinase activators for diabetes therapy. Diabetes 2006, 55, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Matschinsky, F.M.; Wilson, D.F. The Central Role of Glucokinase in Glucose Homeostasis: A Perspective 50 Years After Demonstrating the Presence of the Enzyme in Islets of Langerhans. Front. Physiol. 2019, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Chakera, A.J.; Steele, A.M.; Gloyn, A.L.; Shepherd, M.H.; Shields, B.; Ellard, S.; Hattersley, A.T. Recognition and Management of Individuals With Hyperglycemia Because of a Heterozygous Glucokinase Mutation. Diabetes Care 2015, 38, 1383–1392. [Google Scholar] [CrossRef]
- She, L.; Li, W.; Guo, Y.; Zhou, J.; Liu, J.; Zheng, W.; Dai, A.; Chen, X.; Wang, P.; He, H.; et al. Association of glucokinase gene and glucokinase regulatory protein gene polymorphisms with gestational diabetes mellitus: A case-control study. Gene 2022, 824, 146378. [Google Scholar] [CrossRef]
- Popova, P.V.; Klyushina, A.A.; Vasilyeva, L.B.; Tkachuk, A.S.; Bolotko, Y.A.; Gerasimov, A.S.; Pustozerov, E.A.; Kravchuk, E.N.; Predeus, A.; Kostareva, A.A.; et al. Effect of gene-lifestyle interaction on gestational diabetes risk. Oncotarget 2017, 8, 112024–112035. [Google Scholar] [CrossRef]
- Ao, D.; Zhao, Q.; Song, J.Y.; Liu, Z.; Wang, Y.; Wang, H.J.; Yang, H.X. The association of the glucokinase rs4607517 polymorphism with gestational diabetes mellitus and its interaction with sweets consumption in Chinese women. Public Health Nutr. 2021, 24, 2563–2569. [Google Scholar] [CrossRef]
- Anghebem-Oliveira, M.I.; Webber, S.; Alberton, D.; de Souza, E.M.; Klassen, G.; Picheth, G.; Rego, F.G. The GCKR Gene Polymorphism rs780094 is a Risk Factor for Gestational Diabetes in a Brazilian Population. J. Clin. Lab. Anal. 2017, 31, e22035. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).