Abstract
Hypervirulent Klebsiella pneumoniae (hvKp) is a variant that has been increasingly linked to severe, life-threatening infections including pyogenic liver abscess and bloodstream infections. HvKps belonging to the capsular serotypes K1 and K2 have been reported worldwide, however, very scarce studies are available on their genomics and virulence. In the current study, we report four hypermucoviscous extended-spectrum β-lactamase-producing hvKp clinical strains of capsular serotype K1 and K2 isolated from pus and urine of critically ill patients in tertiary care hospitals in Oman. These strains belong to diverse sequence types (STs), namely ST-23(K1), ST-231(K2), ST-881(K2), and ST-14(K2). To study their virulence, a Galleria mellonella model and resistance to human serum killing were used. The G. mellonella model revealed that the K1/ST-23 isolate was the most virulent, as 50% of the larvae died in the first day, followed by isolate K2/ST-231 and K2/ST-14, for which 75% and 50% of the larvae died in the second day, respectively. Resistance to human serum killing showed there was complete inhibition of bacterial growth of all four isolates by the end of the first hour and up to the third hour. Whole genome sequencing (WGS) revealed that hvKp strains display a unique genetic arrangement of k-loci. Whole-genome single-nucleotide polymorphism-based phylogenetic analysis revealed that these hvKp isolates were phylogenetically distinct, belonging to diverse clades, and belonged to different STs in comparison to global isolates. For ST-23(K1), ST-231(K2), ST-881(K2), and ST-14(K2), there was a gradual decrease in the number of colonies up to the second to third hour, which indicates neutralization of bacterial cells by the serum components. However, this was followed by a sudden increase of bacterial growth, indicating possible resistance of bacteria against human serum bactericidal activity. This is the first report from Oman detailing the WGS of hvKp clinical isolates and assessing their resistance and virulence genomics, which reinforce our understanding of their epidemiology and dissemination in clinical settings.
1. Introduction
Klebsiella pneumoniae is considered to be the most common cause of hospital-acquired infections (HAIs), accounting for 10% of all nosocomial infections worldwide [1]. Immunocompromised patients are at higher risk of HAIs, comprising 8% to 12% of hospitalized patients, particularly ventilator-associated pneumonia [2]. This can result in life-threatening illnesses with mortality rates ranging from 50% to 100% [3,4]. K. pneumoniae have evolved over the years from the classical type into a hypervirulent K. pneumoniae (hvKp), which has been observed in many countries around the globe [4]. However, hvKps can also be acquired from the community and have been shown to infect otherwise healthy as well as immunocompetent individuals [5,6]. Emergence of new clonal lineages of hypervirulent strains with co-occurrence of multidrug resistance genotype has been increasing, which is known as multidrug-resistant hvKp (MDR-hvKp). The most common circulating hvKp serotypes include K1, K2, K20, K54, and K57, with K1 and K2 being the most virulent and accounting for 70% of hvKp isolates [7]. HvKp strains produce hypermucoviscous exopolysaccharide coating capsules, which contribute significantly to the pathogenicity and virulence of the bacteria and have been associated with dissemination of severe infections such as pyogenic liver abscess mainly in K1 and ST-23 serotypes [4,5]. Moreover, hvKps can cause other invasive infections, particularly serotypes K2 and ST-65 [4,8]. However, it is well established that the hypermucoviscosity and hypervirulence are two distinct phenotypes and do not necessarily co-exist in virulent strains [9,10].
In addition to the capsule, other virulence factors in K. pneumoniae can increase the virulence, including the mucoid phenotype regulators (rmpA and rmpA2), siderophores including Yersiniabactin irp2, ybt, and Aerobactin iucA, outer membrane porin (KpnO), and phospholipase D family protein (PLD1) [11]. Furthermore, there are other virulence factors which have not been thoroughly characterized, including efflux pumps, iron transport systems, and genes that are involved in allantoin metabolism [12,13,14]. In addition, some studies have demonstrated that the virulome of the same species could change depending on the host factors [15,16]. Some factors that expose hospitalized individuals to colonization and infection include admission into the intensive care unit (ICU), poor adherence to infection control strategies, prolonged use of invasive devices, and broad-spectrum antibiotics. Moreover, host-related factors such as immunocompromised state, especially diabetics and alcoholics, could increase the risk of colonization of virulent K. pneumoniae [17].
Apart from the intrinsic resistance to ampicillin in hvKps, the antibiotic resistance phenotype in hvKps is still not very prevalent, and the reason is still unclear [4]. However, there has been an increasing trend in resistance in hvKp isolates due to dissemination via mobile genetic elements such as types of New Delhi metallo-β-lactamases (NDMs), oxacillinases β-lactamases (OXA-48), and K. pneumoniae carbapenamases (KPCs) [13,16,17]. Recently, the hvKp serotypes K1 and K2 have been detected from clinical isolates in Oman [8]. Therefore, this study aimed to characterize their virulence and antibiotic resistance genomically and to further study host responses against infection with K. pneumoniae K1 and K2 serotypes circulating in Oman.
2. Results
2.1. Sequence Types of K. pneumoniae Isolates
Initial PCR screening revealed that only 4 out of 129 (3%) K. pneumoniae isolates were hypervirulent capsular strains, namely Kp124 (K1/ST-23), Kp 125 (K2/ST-231), Kp 126 (K2/ST-881), and Kp 83 (K2/ST-14) (Figure 1).
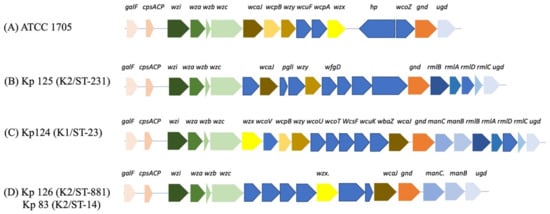
Figure 1.
k-loci gene arrangement of hvKP isolates. (A) ATCC 1705, (B) Kp 125 (K2/ST-231), (C) Kp 124 (K1/ST-23, (D) Kp 126 (K2/ST-881), and Kp 83 (K2/ST-14). Gene names are indicated above the colored arrows, and each group of genes with a shared function or operon are colored with the same colors. Chromosomal genes are conserved in all isolates including capsular synthesis, capsule translocation, and surface assembly genes galF, ORF2, wzi, wza, wzb, wzc, and gnd. wcoZ is uniquely found in the K. pneumoniae ATCC 1705 control strain and one additional isolate from our study (Kp102) (Supplementary Figure S5). GDP-D-mannose synthesis genes manC and manB (C,D), and glucose conversion genes rmlA, rmlB, rmlC, and rmlD (B,C).
Three of these isolates were invasive K. pneumoniae strains with hypermucoviscous phenotype (HMV K. pneumoniae), and one of these (K2/ST-881) was a carbapenemase-producing (CRE) isolate. WGS analysis was performed on these four isolates in addition to the other isolates for determining the ST and k-loci composition. MLST was performed on the isolates from WGS data to determine their sequence type (ST) using the CGE website (Table 1) [18]. One isolate from a pyogenic infection belonged ST-23, similar to global trends associating these features [7]. On the other hand, the three other isolates belonged to three different STs, including ST-14, ST-231, and ST-881 (Table 1). One-third (32%, n = 10) of the isolates belonged to ST-231 and 19% of the isolates (n = 6) belonged to ST-395 (Table 1). Moreover, 6% of the isolates (n = 2) belonged to ST-23 and another 6% of the isolates (n = 2) belonged to ST-405.

Table 1.
Sequence types of local K. pneumoniae clinical isolates (n = 31).
2.2. k-loci Analysis of hvKp Isolates
It was shown that the isolates with unique k-loci genetic arrangements belonged to the hvKp isolates with serotypes K1/ST-23, K2/ST-231, K2/ST-881, and K2/ST-14 (Table 2).

Table 2.
Source and molecular genotype of clinical hypervirulent K. pneumoniae isolates.
The phenotypic antimicrobial susceptibility profiles by disk diffusion of the hvKp isolates are presented in Table 3 and Table 4. All hvKp isolates in this study (Kp 83, Kp 125, and Kp 126) that were K2 positive and Kp 124 that was K1 positive were ESBL producers. Kp 83 (K2/ST-14) showed resistance to AMP, TZP, FEP, CTX, IMP, CAZ, and CIP. Kp 126 (K2/ST-881) was resistant to AMP, FEP, CTX, IMP, CAZ, and CIP. Meanwhile, Kp124 (K1/ST-23) was resistant to AMP, TZP, FEP, CTX, FOX, and CAZ. On the other hand, Kp 125 (K2/ST-231) was a KPC producer with resistance to AMP, TZP, FEP, CTX, and CAZ.

Table 3.
Antimicrobial resistance genotype and phenotype of hypervirulent K. pneumoniae isolates.

Table 4.
Interpretive categories and zone diameter breakpoints according to CLSI guidelines.
2.3. Plasmid Compositions of hvKp K1/ST-23 (Kp 124), K2/ST-231 (Kp 125), K2/ST-881 (Kp 126), and K2/ST-14 (Kp 83)
Whole-genome sequences of K1/ST-23, K2/ST-231, K2/ST-881, and K2/ST-14 isolates comprised 5,569,468 (Kp 125 K2/ST-231) bases, 5,572,070 bases (K1/ST-23), 5,638,661 bases (Kp 126 K2/ST-881), and 5,447,756 bases (Kp 83 K2/ST-14) with 57% GC content in all strains (Supplementary Table S7). The isolate Kp 125 belonging to K1/ST-231 contained seven plasmids: pColKP3, which carried one copy of OXA-181, and KPC-3, conferring carbapenem and cephalosporin resistance by mechanism of antibiotic inactivation. The other plasmids were pIncFII(pAMA11 67-NDM-5), carrying New Delhi metallo-β-lactamase (NDM)-producing enzymes, and pIncFIB(pQil). Isolate belonging to K2/ST-23 carried three plasmids, including pIncFIB(pQil), containing KPC-3, which has been attributed to the global spread of bla-KPC.
In K1/ST-23, Col440II shared 100% identity with the reference plasmid (CP023921). On the other hand, pIncFIB(K) shared 98% identity with the reference plasmids. Isolate belonging to K2/ST-231 had three plasmids, which were IncFIB(pQil), IncFII(pAMA1167-NDM-5), and RepB. pRepB is a megaplasmid (244,152 bp) that is associated with the IncFIB family and has been known to integrate massive accessory modules. In ST-231, pRepB carried a very large composite transposon (15,832 bp), insertion sequences (ISEc27), IS102-like among other mobile elements like integrons, and MDR-conferring genes [19].
Isolate Kp 83 (K2/ST-14) had four plasmids, including pIncFIA (HI1), which carried resistance genes for several antibiotic categories such as tetracyclines, phenicols, folate pathway antagonists, and aminoglycosides. pIncFIB(K) harbored resistance genes against sulfonamides, macrolides, and beta lactams. pIncFIB(pNDM-Mar) carried antimicrobial genes against phenicols, beta lactams, quinolones, and aminoglycosides. Finally, pIncR carried antibiotic genes conferring resistance to beta lactams, quinolones, and aminoglycosides (Table 3).
2.4. Virulence-Associated Genes of hvKp Isolates
WGS of Kp 124 (K1/ST-23), Kp 125 (K2/ST-231), Kp 126 (K2/ST-881), and Kp 83 (K2/ST-14) were compared to the reference genomes of K. pneumoniae strain RJF293 (accession number CP014008.1), which belongs to serotype K2 assigned to ST-374. WGS analysis revealed that genes encoding for pathogenicity were conserved in the analyzed strains at the same relative chromosomal position. There are four main virulence classes in K. pneumoniae, including the capsule, lipopolysaccharide (LPS), siderophores, and fimbriae (Supplementary Table S8). Enterobactin (entB) was detected in all the four hvKp isolates, whereas aerobactin (iucA) was found in Kp 124 (K1/ST-23), Kp 125 (K2/ST-231), and Kp 126 (K2/ST-881) but not Kp 83 (K2/ST-14). Yersiniabactin (irp2 and ybt) and salmochelin (iroB) were missing in all isolates. Capsule biosynthesis gene rcsB was detected in all isolates but not the regulators rmpA and rmpA2. LPS, adhesins, and type 3 fimbriae were detected in all isolates.
2.5. Human Serum Bactericidal Killing Susceptibility and G. mellonella Lethality
In isolate Kp 124 (K1/ST-23) (Figure 2), there was a slight decrease in the growth, indicating the death of bacterial cells, followed by a slight increase in the bacterial growth at T = 3. In isolate Kp 125 (K2/ST-231), in the first hour, there was a decrease in the number of colonies, however, in the second and third hours, there was a significant increase in bacterial growth. In isolate Kp 126 (K2/ST-881), there was a decrease in growth of bacteria in the first hour, but in the second and third hour there was a noticeable increase in the number of colonies, which was higher compared to the previous two isolates. In Kp 83 (K2/ST-14), there was a slight decrease in the number of colonies in the first and second hours. However, after the second hour, and by the third hour, there was a dramatic increase in the bacterial growth. In all replicates of the four tested isolates, the p-value showed statistical significance (p-value of 0.0007).
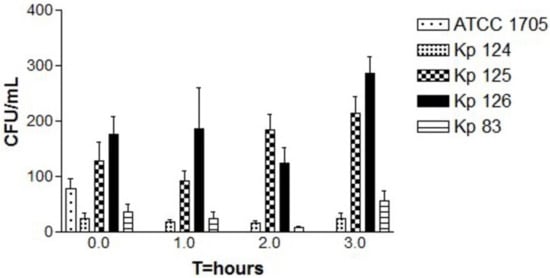
Figure 2.
Bactericidal activity of human serum samples against hvKp clinical strains. The CFU counts of all strains including the control were tested at four time points, T = 0, T = 1, T = 2, and T = 3 (time in hours). Each strain represents the mean for three replicates: K. pneumoniae ATCC 1705 (control), K1/ST-23(Kp 124), K2/ST-231 (Kp 125), K2/ST-881 (Kp 126), and K2/ST-14 (Kp 83). The p-value was 0.0007 for CFU counts, which is statistically significant as shown by one-way ANOVA. Error bars represent SEM. A slight dip in the bacterial growth at T = 2 is followed by a slight to significant increase in the bacterial growth in all three replicates at T = 3 for Kp 125, Kp 126, and Kp 38.
To correlate between resistance to serum killing and virulence, the same strains were injected into G. mellonella larvae. The survival curves of the three replicates were shown separately for comparison (Figure 3). Isolates Kp 124 (K1/ST-23) and Kp 83 (K2/ST-14) seemed to be the most virulent isolates, as most larvae died in the first two days. However, isolates Kp 125 (K2/ST-231) and Kp 126 (K2/ST-881) seemed to be less virulent, as the number of larvae gradually decreased throughout the experiment. In the three replicates (Figure 3B), the Control 1 larvae consisted of the untouched larvae. In the three replicates of the control group, there was a gradual decline in the survival of the larvae from day 2 to 5, as the first larva died in the second day in one of the replicas, while in the other two replicates the larvae died at the end of day 5.
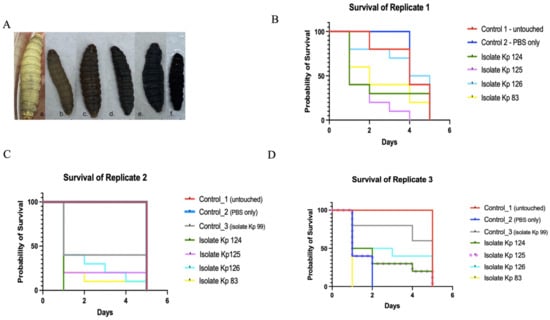
Figure 3.
Kaplan–Meier survival curve of K. pneumoniae clinical isolates. (A) Phenotypic characteristics of G. mellonella larvae pre and post injection and throughout the 5 observation days. a is the healthy larvae at day 0 of the experiment, b–f shows gradual death of the larvae with blackening from day 1-5. (B–D) Survival of G. mellonella larvae infected with K1/ST-23(Kp 124), K2/ST-231 (Kp 125), K2/ST-881 (Kp 126), and K2/ST-14 (Kp 83) in comparison to controls 1 and 2 over the course of 5 days. Control 1—untouched, uninfected larvae are represented in red. Control 2—larvae injected with PBS are represented in blue. A third control, Kp 99 in replicate (C,D), was used, which was a non-hypervirulent K. pneumoniae isolate, just to compare the rate of survival to the hypervirulent strains. Isolate Kp 124 was represented in green, isolate Kp 125 in the purple straight line in (C) and dotted in panel (D) for clarity, isolate Kp 126 in cyan, and isolate Kp 83 in yellow as indicated in the color legend on the right side of each image.
In Control 2, larvae that received only PBS, there was also a gradual decline in the survival of the larvae throughout the 5 days in replicate one, however, no death was noticed in the second replicate until the larvae slowed down in motility as cocoon formation commenced. However, in the third replicate, there was 100% death, which could suggest that the batch contained mature larvae in that control group. In Control 3 larvae, consisting of larvae injected with isolate Kp 99 that is negative for all hypervirulent capsular serotypes, on the first day, about 20–50% of the larvae died, with no death in the remaining 50–60% of larvae until the end of the experiment as observed in the three replicates. In the strain Kp 124 (K1/ST-23) test group, more than 50% in two replicates and 100% in the other replicate of the larvae died on the first day of the experiment. The remainder of larvae were dead by the fourth day. For the strain Kp 125 (K2/ST-231) test group, about 50–75% of the larvae died on the second day, and by the fourth day, all larvae were dead in all three replicates. In the isolate Kp 126 (K2/ST-881) test group, the death ranged between 25% and 60% of the larvae on the first day, but overall, the survival rate declined gradually throughout the 5 observation days in the three replicates. In isolate Kp 83 (K2/ST-14), more than 50–100% of larvae died by the second day of the experiment in the three replicates.
2.6. Genetic Relatedness to Global Isolates
A whole-genome SNPs calling phylogenetic tree of 131 isolates including 30 strains in this study (from CPHL and Kp 83, represented in blue) as well as 101 strains from Genbank global strains (represented in black) [17] were plotted to compare genetic relatedness (Figure 4).

Figure 4.
A whole-genome single-nucleotide polymorphisms (SNPs) calling phylogenetic tree of K. pneumoniae isolates (A) (n = 131). A maximum-likelihood tree showing genetic relatedness between K. pneumoniae strains based on WGS data. The strains are labeled with serial numbers and grouped according to their STs. K. pneumoniae RJF293 (GenBank accession number CP014008) was used as a reference. The isolates starting with NZ and CP (in black) are global published isolates retrieved from Genbank and were used for comparison. The isolates in blue are strains from this study. The hypervirulent isolates are pointed at with arrows: red for K2/ST-881 (Kp126), green for K2/ST-231 (Kp125), purple for K1/ST-23 (Kp 124), and pink for K2/ST-14 (Kp83). iTOL was used to visualize and annotate the tree. Clades are indicated in (A–D). This figure compares the local strains in this study (represented in blue) as well as global strains, which clearly demonstrates the diverse clonality of clinical isolates from Oman, belonging to various STs.
A separate phylogenetic tree of 31 isolates for a close-up inspection, including the hypervirulent isolates of this study Kp125 (K2/ST-231), Kp 126 (K2/ST-881), and Kp 83 (K2/ST-14), as well as 27 other non-hypervirulent K. pneumoniae local clinical isolates (Figure 5), was created. The unique STs were highlighted in different colors. Group A in blue is representing isolates with ST-395, group B in yellow belongs to ST-405, group C in green belongs to ST-23, group D in red represents ST-231, and finally the unhighlighted isolates belong to miscellaneous STs. The hypervirulent isolates Kp 124 (K1/ST-23), Kp 125 (K2/ST-231), Kp 126 (K2/ST-881), and Kp 83 (K2/ST-14) are indicated with magenta arrows. Hypervirulent Kp 83 (K2/ST-14) and Kp 124 (K1/ST-23) are branching from the same clade but belong to two different clusters, C and D, respectively. Following construction of the k-loci in Kaptive, it was evident from the phylogenetic tree that the clinical strains with similar STs have high similarity in the genetic arrangement of k-loci (Figure 1 and Figures S3–S30). The hypervirulent serotypes positive for K2, namely K2/ST-231 (Kp 125), K2/ST-881 (Kp 126), and K2/ST-14 (Kp 83), exhibit diversity in their STs, resulting in distinct k-loci gene compositions.
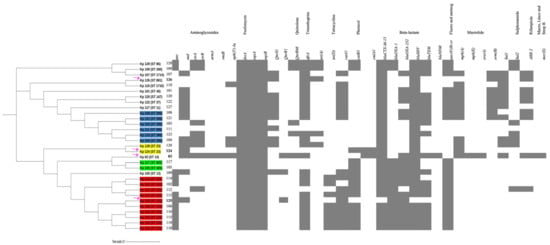
Figure 5.
A whole-genome single-nucleotide polymorphisms (SNPs) calling phylogenetic tree of K. pneumoniae isolates (n = 31) and heat map of resistance-encoding genes distribution and their antimicrobial classes. A maximum-likelihood tree showing genetic relatedness between K. pneumoniae strains based on WGS results. The isolates are grouped and highlighted according to their STs (blue = ST 395, red = ST 231, green = ST 405, yellow = ST 23, unhighlighted = miscellaneous STs). The hypervirulent isolates K1/ST-23 (Kp 124, K2/ST-231 (Kp 125), K2/ST-881 (Kp 126), and K2/ST-14 (Kp 83)) are pointed at using magenta arrows. K. pneumoniae RJF293 (GenBank accession number CP014008) reference was used. iTOL was used to visualize and annotate the tree. Hypervirulent Kp 83 (K2/ST-14) and Kp 124 (K1/ST-23) are branching from the same clade but belong to two different clusters, C and D, respectively. The relation between their STs and resistance phenotype indicates the diversity in the acquired antimicrobial genes in these hypervirulent strains.
The K. pneumoniae capsular synthesis loci, k-loci, in all our isolates generally consisted of the following conserved genes, including galF, ORF2 (cpsACP), wzi, wza, wzb, wzc, and gnd, which are chromosomally encoded core and similar to what has been reported in the literature [18,19] (Figure 1), as these are necessary for capsular synthesis along with other genes. Moreover, galF, ORF2, and gnd are involved in carbohydrates metabolism, and wzi (orfX), wza, wzb, and wzc are responsible for the capsule translocation and surface assembly [20,21]. In this study, the ICEfinder showed that Kp 125 (K2/ST-231), Kp 126 (K2/ST-881), and Kp 83 (K2/ST-14) had 7, 3, 2, and 1 putative type 4 secretion systems with putative ICEs, respectively (Supplementary Figure S33).
The hypervirulent strains carried a wide range of acquired antimicrobial genes (Figure 5). Kp 83 (K2/ST-14) carried a variety of resistance-conferring genes as compared to the other isolates, except for qnrS1, blaOXA-2, blaAP, and tet(A) genes. Kp 124 (K1/ST-23) carried the least number of resistance-conferring genes and harbored no resistance genes for some antibiotic classes including folate pathway antagonist, quaternary ammonium compounds, streptogramin b, rifamycin, aminocyclitol, macrolide, lincosamide, and tetracycline. Moreover, isolate Kp 125 (K2/ST-231) carried resistance genes for all antibiotic classes in Figure 4, except glycosamide and tetracycline. In addition, Kp 126 (K2/ST-881) seemed to lack resistance-conferring genes to aminoglycosides, amphenicol, streptogramin b, rifamycin, aminocyclitol, macrolide, and lincosamide.
3. Discussion
The hvKp are known to be associated with increased virulence as well as with severe human infections [22]. Only 3% of the clinical isolates tested positive for K1 and K2 during the same period in 2019 at Sultan Qaboos University Hospital (SQUH), from urine and pus samples, and one K2 case was reported in 2017 from a urine specimen. This finding is consistent with several studies, which showed that the most commonly circulating hypervirulent capsular serotypes are K1 and K2, which has been attributed to disruption of epithelial and mucosal barriers such as endotracheal tubes, catheters, and surgical wounds in hospital settings [21,23,24,25,26]. Phenotypically, all the hypervirulent isolates in this study were ESBL producers, which are known to be associated with increased length of stay and mortality rates. In contrast to previous studies, the prevalence rate of ESBL producers was higher in non-hypermucoviscous K. pneumoniae isolates [20,21,23,24,25,26]. There is a high possibility of opportunistic infections in our patients included in this study, as the majority are immunocompromised.
The k-loci of the capsular operon share similar genetic organization, in which major operons are conserved in most isolates with few exceptions. MLST analysis showed that most of the isolates with similar STs had similar gene arrangements with a similar reference genome in Kaptive. Moreover, these strains were in the same cluster in the whole-genome SNP calling phylogenetic tree. The remaining strains belonged to miscellaneous STs and, therefore, variable k-loci genetic arrangement figures were observed. To the best of our knowledge, this observation has not been previously reported, making this study the first in Oman to indicate a significant correlation between genetic composition of the k-loci and their STs.
In addition to the conserved genes in the k-loci, there are other genes that are commonly present in the operon, including manC and manB, which are involved in the synthesis of GDP-D-mannose as well as RmlA, RmlB, RmlC, and RmlD proteins involved in the conversion of glucose 1-phosphate to dTDP-L-rhamnose [27]. Moreover, additional virulence determinants such as aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth and survival of hvKp ex vivo and in vivo [4]. The presence of a wide variety of virulence-conferring genes in the isolates of this study indicates that these hvKp strains were well equipped with the virulence machinery, which could, under the right conditions, enable a hypervirulent phenotype.
Previous studies proved that hypervirulent K. pneumoniae strains are associated with increased antibiotic resistance, therefore, recognizing their global dissemination is an urgent priority [5,28,29,30,31]. There has been an increase in reporting MDR in hvKp clinical strains over the last few years, therefore designated as MDR-hvKp. This was attributed to the horizontal gene transfer mediated by plasmids (type I and type II) [5,22,32,33]. For example, type I plasmids were reported to carry blaNDM-1 and blaKPC-3 [20,21,23,24]. In silico detection of plasmids using plasmidFinder demonstrated that the plasmids in these strains carry mobilizable pathogenicity and resistance-conferring genes [14,15] (Table 3). While most of these plasmids share common sequences, there are wide areas of mosaic genetic structures that might indicate multiple recombination events (Supplementary Figures S31 and S32).
According to ResFinder and CARD online tools findings, the hypervirulent strains in this study including Kp 124 (K1/ST-23), Kp 125 (K2/ST-231), Kp 126 (K2/ST-881), and Kp 83 (K2/ST-14) were in fact highly resistant, carrying a wide variety of antibiotic resistance genes for multiple antimicrobial classes. All these strains carried incompatibility group (Inc) FIB plasmids that are known to encode both virulence and antimicrobial resistance genes in a wide variety of intestinal bacteria including E. coli, Salmonella, and Klebsiella spp. including IncFIA and IncFIB that represent one of the most common plasmid types that plays a major role in the dissemination of AMR in Enterobacteriaceae [34,35,36,37,38,39]. Furthermore, IncFIB (K) was carried by both Kp 83 (K2/ST-14) and Kp 126 (K2/ST-881), which suggested the likelihood of HGT of these plasmids between different bacteria [40]. Moreover, Kp 125 (K2/ST-231) carried a ColKP3 plasmid that is known to harbor OXA-232 carbapenemase responsible for the rapidly increasing carbapenem resistance in K. pneumoniae strains worldwide [41]. Moreover, Kp 125 (K2/ST-231) and Kp 124 (K1/ST-23) both carried IncFIB(pQil), which is known to be harbored by K. pneumoniae and other pathogens responsible for nosocomial infections [42]. This finding suggests the likelihood of HGT of this plasmid amongst K. pneumoniae bacterial isolates since both Kp 125 (K2/ST-231) and Kp 124 (K1/ST-23) were isolated in the same period in 2019 in SQUH.
SNP calling phylogenetic trees performed on WGS data show that K. pneumoniae strains isolated from different geographical regions (n = 131) were arranged with distinct clustering [40,41,42,43]. The strains in our study were found to be phylogenetically diverse, belonging to various STs. The hvKps Kp 124 (K1/ST-23) and Kp 125 (K2/ST-231) are present in a clade A, whereas Kp126 (K2/ST-881) is in clade B. The close-up tree showed that our strains were grouped in four main clusters, highlighted with different colors, indicating the diverse groups of strains circulating in our local hospitals. However, there was clustering in these four main groups, indicating a possibility of dissemination at a possibly larger scale [43,44]. Isolates belonging to ST-231 have been associated with MDR spread in Europe, with the potential to cause a future epidemic, posing a public threat. These strains produce OXA-232 as well as RmtF, which are associated with high-level carbapenem resistance [45,46]. Furthermore, K. pneumoniae belonging to ST-395 are associated with the production of OXA-48, which is the most common enzyme causing healthcare-associated infections globally. The OXA-48 enzyme hydrolyses carbapenems and shows weak activity against extended-spectrum cephalosporins such as cefepime and ceftazidime, making it difficult to treat these infections [47,48]. This clone of K. pneumoniae has been detected in many countries of the Middle East, including Oman, Yemen, and Saudi Arabia [49]. Furthermore, Kp 117 and Kp 105 belong to ST-405 (blue cluster) and have also been associated with the production of OXA-48 just like clones belonging to ST 395 and thus have been responsible for many hospital-associated outbreaks and war-related outbreaks (in Germany), and some of which are resistant to colistin [50,51,52].
Several studies have shown that infection of G. mellonella with K. pneumoniae has resulted in host responses similar to the innate immune responses in murine models including cell death, inhibition of phagocytosis, and antimicrobial peptide production. Furthermore, G. mellonella can differentiate between pathogenic and non-pathogenic Klebsiella strains and showed that the more virulent strains were associated with increased survival and host cellular damage as compared to avirulent strains [53].
In the present study, it was shown that Kp 124 (K1/ST-23) was the most virulent, as 50% of the larvae died on the first day, followed by K2/ST-231, where 75% of the larvae died on the second day, and Kp 83 (K2/ST-14), where 50% of the larvae died on the second day. Similarly, in Europe, K1-positive K. pneumoniae belonging to ST-23 have been associated with severe and fatal infections associated with liver abscesses [3,25].
However, in Kp 126 (K2/ST-881), there was a gradual decline in the number of larvae over the course of 5 days, perhaps making it the least virulent, as the death was slower and steadier. It was expected for Kp 126 (K2/ST-881), although it was a hypervirulent strain, that it belonged to ST-881, which, according to the literature, has not been associated with severe human infections, as compared to the other STs of Kp 83 (K2/ST-14), Kp 124 (K1/ST-23), and Kp 125 (K2/ST-231). The G. mellonella experiment was repeated three times to allow for comparison and to check how the quality of larvae might affect the results. The results of the experiments indicated the higher virulence of hypervirulent K1 and K2 capsular serotype isolates as compared to non-hypervirulent strains [1,54]. However, the main limitation in this assay was due to shipping and delivery restrictions caused by the COVID-19 pandemic, as the larvae used in this study were subjected to variations in temperatures and storing conditions.
Human serum resistance experiments were performed for the hypervirulent strains Kp 124 (K1/ST-23), Kp 125 (K2/ST-231), Kp 126 (K2/ST-881), and Kp 83 (K2/ST-14). For each isolate, three replicates were performed for accurate comparison and reproducibility. There was complete inhibition of bacterial growth in all four isolates by the end of the first hour and up to the third hour. This was due to the presence of complement proteins and lysozyme, which have a bactericidal effect [55]. Furthermore, an unexpected pattern of bacterial growth was observed in all replicas for all four strains. For Kp 124 (K1/ST-23), there was a gradual decrease in the number of colonies up to the second hour, which indicates neutralization of bacterial cells by the serum components; however, in the third hour, there was a sudden splurge of bacterial growth. For Kp 125 (K2/ST-231), the same decrease in bacterial growth was noticed by the end of the first hour, but there was a dramatic increase in the bacterial growth starting from the second hour, unlike in Kp 124 (K1/ST-23). Furthermore, for Kp 126 (K2/ST-881) and Kp 83 (K2/ST-14), similar patterns of sudden growth of bacterial cells commenced in the third hour. Therefore, the sudden increase in the bacterial growth despite the unfavorable serum environment for growth indicates possible resistance of bacteria against human serum bactericidal activity [56]. Human serum, unlike blood, contains antibodies and complement proteins that contribute to the neutralization of bacteria but does not contain immune cells such as phagocytes. Previous studies discuss the bactericidal activity that human serum possesses against Gram-negative bacteria including K. pneumoniae. Moreover, they highlight that the resistance against human serum may be a significant virulence factor. Therefore, the strains used in this assay might have acquired the resistance against human serum as one of their virulence factors [55]. Furthermore, earlier studies have shown that K. pneumoniae LPS O antigen and the capsule polysaccharide contribute strongly to the resistance against human serum antibodies and complement proteins. The isolates tested in this experiment were hypervirulent and possessed a thick capsule, therefore, their resistance to human serum could be explained by this theory [55,56,57,58]. In future studies, it would be worthy to test the hypervirulent strains of this study in human blood that contains immune cells such as phagocytes to investigate how it compares to human serum. In previous studies, K. pneumoniae has shown its ability to resist killing by phagocytosis in human blood killing assays [59].
4. Materials and Methods
4.1. Bacterial Isolates
This study was commenced after obtaining approval by the Medical Ethics Research committee (MREC#1896), College of Medicine and Health Sciences, Sultan Qaboos University, Muscat, Oman. Initially, we screened 129 K. pneumoniae clinical isolates for K1, K2, K20, K54, and K57 from various samples (urine, wound, tracheal aspirate, blood, sputum) from patients from Central Public Health Laboratories (CPHL) (Supplementary Figures S1 and S2) representing various areas from Oman collected between 2015 and 2020. Another 30 isolates were screened from Sultan Qaboos University Hospital (SQUH), Muscat, Oman, collected between 2019 and 2021 (Supplementary Table S1). PCR preliminary data are presented in Supplementary Tables S5 and S6, including PCR cycling conditions and primers. Colonies were collected from purity plates of Cystine Lactose Electrolyte-Deficient (CLED) agar (Oxoid, Basingstoke Hampshire, UK). These colonies were used to make frozen stock in beads-containing cryotubes and processed according to the manufacturer’s instructions (Mast Diagnostics, Derby, UK). The samples were frozen at −80 °C for future use.
4.2. String Test for Hypermucoviscosity
An inoculation loop was used to stretch a colony on a CLED agar plate (Oxoid, Basingstoke Hampshire, UK), and the immediate formation of a viscous string of >5 mm in length was indicative of a hypermucoviscous strain of K. pneumoniae [60].
4.3. Antibiotic Susceptibility Testing
Antimicrobial susceptibility profiles of the isolates were carried out using the disk diffusion method [61]. Firstly, from an overnight pure culture, three to five colonies were suspended in 5 mL of normal saline (Fisher Scientific, Loughborough, UK). The suspension was adjusted to 0.5 McFarland (approximately 1–2 × 108 CFU/mL) using a CrystalSpec nephlometer (BD Diagnostics, Baltimore, MD, USA) following the manufacturer’s protocol. The antibiotic disks included ampicillin (AMP 10 mg), piperacillin/tazobactam (TZP 110 mg), cefepime (FEP 30 mg), cefotaxime (CTX 30 mg), cefoxitin (FOX 30 mg), ceftazidime (CAZ 30 mg), imipenem (IPM 10 mg), meropenem (MEM 10 mg), amikacin (AK 30 mg), gentamicin (CN 10 mg), and ciprofloxacin (CIP 5 mg) (Liofilchem, Roseto degli Abruzzi, Italy and BioMérieux, Voie Romaine, Craponne, France). The susceptibility assay was carried out according to the CLSI standards [62], placed on the nutrient agar plates (Oxoid, Hampshire, UK), and inoculated using sterile forceps. The plates were then incubated for 18–24 h at 37 °C. E. coli (ATCC 25922) and P. aeruginosa (ATCC 27853) were used as susceptible control strains. The readings were taken after 18–24 h, and the interpretive categories and zone diameter breakpoints are listed in Table 4.
4.4. PCR Screening
Initial PCR screening of MDR K. pneumoniae was performed for the purpose of assessing the prevalence of hvKp capsular serotypes (Supplementary Figure S1). Initial screening of MDR K. pneumoniae revealed that four isolates displayed hvKp phenotype. These four isolates were isolated mainly from urine and one from a wound swab, which were collected during possible cross-transmission episodes in SQUH, Muscat, Oman between 2019 and 2020. We conducted a comparison of the k-loci in these isolates to known K. pneumoniae references in the database using the Kaptive 2.0 online tool to analyze similarities and variabilities [9]. The genetic arrangement of different k-loci in each isolate as well as percentage of similarity between the isolate to known references showed that most of the screened K. pneumoniae clinical isolates have similar genetic arrangement (Supplementary Figures S3–S30). The matches of “Good” or “very good” confidence were reported based on the 90% to 95% identity match.
4.5. DNA Extraction and WGS
DNA was extracted from an overnight culture using the QIAamp DNA Mini Kit (Qiagen, Germany) for whole-genome sequencing. The manufacturer’s protocol was followed with slight modifications. For highly mucoid K. pneumoniae isolates, a pre-lysis step was performed, which includes the preparation of a pre-lysis buffer consisting of 100 μL of Tris//EDTA (TE) buffer (ThermoFisher Scientific, Waltham, MA, USA), 1.00 μL lysozyme 10 mg/mL (final concentration 0.1 mg/mL) (Sigma-Aldrich, St. Louis, MO, USA), 0.20 μL lysostaphin 10 mg/mL (final concentration 0.02 mg/mL) (ThermoFisher Scientific, USA), and 0.10 μL RNAse A 100 mg/mL (final concentration 0.1 mg/mL) (Qiagen, Hilden, Germany). The bacterial suspension from an overnight culture was centrifuged at 8000× g for 2 min, and the pelleted bacterial cells were resuspended in 100 μL of the pre-lysis buffer by pipetting up and down several times with a P200 pipette. The suspension was then incubated for 60 min. After incubation, 1.00 μL of Proteinase K (Sigma-Aldrich, St. Louis, MO, USA) was added as a final step in the pre-lysis stage. The sample was then ready for the first step of the Qiagen DNA extraction protocol as discussed in the previous section.
After DNA quantification by NanoDrop (ThermoFisher Scientific 1000 NanoDrop Spectrophotometer), the samples were sent to microbesNG in the UKfor WGS by Illumina next-generation sequencing (https://microbesng.co.uk, Birmingham, UK, accessed on 30 June 2021) [60]. The extracted DNA was prepared following the manufacturer’s protocol as described on the company’s website as follows: ILLUMINA SEQUENCING (SGS and EGS) Genomic DNA libraries are prepared using the Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s protocol with the following modifications: input DNA is increased 2-fold, and PCR elongation time is increased to 45 s. DNA quantification and library preparation are carried out on a Hamilton Microlab STAR automated liquid handling system (Hamilton Bonaduz AG, Bonaduz, Switzerland). Pooled libraries are quantified using the Kapa Biosystems Library Quantification Kit for Illumina. Libraries are sequenced using Illumina sequencers (HiSeq/NovaSeq) using a 250 bp paired-end protocol. Reads are adapter trimmed using Trimmomatic 0.30 with a sliding window quality cutoff of Q15 [63]. De novo assembly is performed on samples using SPAdes version 3.7 [64], and contigs are annotated using Prokka 1.11 [65].
4.6. Bioinformatics Analysis
For visualization of the K. pneumoniae contigs as well as the investigation of main virulence genes SnapGene Viewer 4.1 (https://snapgene.com, accessed on 12 January 2021) software was used. Moreover, Artemis was used to identify Klebsiella capsule synthesis loci in the different isolates [66]. Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/blast.cgi accessed on 22 September 2021) was used to identify the similarities between the loci against a reference K. pneumoniae RJF293 (GenBank accession number CP014008) (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 3 June 2021) [67]. In addition, Genbank was used to search for similar DNA sequences or genes from the DNA database to the sequenced genomes of this study. Kaptive was used for locus typing and variant analysis on the pre-assembled sequences against a K. pneumoniae database (https://kaptive-web.erc.monash.edu, accessed on 13 September 2021) [9]. The Klebsiella capsule synthesis loci (k-loci) of each of the sequenced genomes were generated and were used for comparison between the different isolates (Supplementary Figures S3–S30). Kleborate v2.1.0 was used to investigate other virulence factors including yersiniabactin, colibactin, aerobactin, and salmochelin [68]. The ICEfinder v2.0 online tool was used to detect integrative and conjugative elements (https://bioinfo-mml.sjtu.edu.cn/ICEfinder/ICEfinder.html, accessed on 26 May 2021) [69]. Multilocus sequence typing (MLST) v2.0.9 was determined using the MLST tool in the Center for Genomic Epidemiology (CGE) (http://cge.cbs.dtu.dk/services, accessed on 14 June 2021). PlasmidFinder and ResFinder in the CGE website were used to identify plasmids and acquired antimicrobial resistance genes, respectively [70]. Additionally, the Comprehensive Antibiotic Resistance Database v3.2.8 (CARD) (https://card.mcmaster.ca/, accessed on 30 April 2021) was used to detect putative antimicrobial resistance genes using the Resistance Gene Identifier (RGI) tool v6.0.3. This tool identifies the antibiotic resistome(s) as well as point mutations within the resistance-conferring genes [71]. Furthermore, the phylogeny tool in the CGE server was used to investigate the genetic relatedness between the different sequences by identifying single nucleotide polymorphisms (SNPs) [72]. The tree files were visualized using Mega7 v7.0, which performs automatic sequence alignment and infers phylogenetic trees [73]. In addition, a combined phylogenetic tree of 131 isolates that includes the isolates in this study as well as published global isolates was created using R v4.3.2, a statistical programming software for comparison and visualization of genetic similarities or differences between the project’s isolates and the published ones [74]. iTOL v6, an online tool, was used to display, annotate, and manage the phylogenetic tree [75]. A whole-genome SNP alignment was generated using Snippy v4.4.5 [75,76], and the AE015929 genome was used as a reference. Then, iqtree v1.6.12, using model finder and ultrafast bootstrap [77], was used to produce a maximum likelihood phylogenetic tree. A phylogenetic tree of the whole-genome SNP was constructed and linked to the gene analysis heat map using the R platform. Lists of clinical isolate serial numbers of phylogenetic trees in Figure 4 and Figure 5 are presented in Supplementary Table S4. For statistical analysis, one-way ANOVA was used to analyze variations in bactericidal activity of human serum against K. pneumoniae clinical isolates using GraphPad Prism software (10.0.3). A p-value of <0.05 was considered as statistically significant. Observation tables are presented in Supplementary Table S3.
4.7. G. mellonella Virulence Assays
Virulence assays were performed using the larvae of G. mellonella as a model of infection. The protocol was modified from previous published studies [78]. To ensure quality, the larvae were manually picked through local beekeepers in Oman and were kept at room temperature (25 °C) in the dark, and wood shavings were provided as a food source. Stage 6 larvae, approximately 2–3 cm in size, were selected for the purpose of the experiment. For each isolate and the controls, replicates of 10 and 5 larvae were used, respectively. The larvae were distributed in Petri dishes lined with wood shavings before injecting them with bacteria the next day. On day 1, the selected isolates for the experiment were sub-cultured onto CLED agar plates (Oxoid, Basingstoke Hampshire, UK) from cryo-beads. On day 2, three to five colonies were picked up and resuspended in 10 mL of liquid broth then incubated overnight at 37 °C. On day 3, a 1:100 dilution (1 mL bacterial suspension + 9 mL liquid broth) was grown for 3–4 h. The bacterial suspension was then centrifuged, the supernatant discarded, and the pelleted bacteria was washed three times with sterile 1× phosphate-buffered saline (PBS) to remove excess salts (Invitrogen, Agawam, MA, USA). The bacterial pellet was then resuspended in PBS to an absorbance of 0.2 (OD600), which was repeated for each isolate. The absorbance was measured using a spectrophotometer (Eppendorf BioPhotometer Spectrophotometer UV/VIS, St. Louis, MO, USA), and the colony-forming unit (CFU) counts were determined for each sample. For the test larvae, 10 μL of each sample (undiluted 0.2 OD600 suspension) was injected into the larvae. For Control 1 larvae, PBS only was injected, and for Control 2 larvae, they were kept untouched. For Control 3 larvae, Kp 99 non-hypermucovirulent (negative for any capsular serotype) bacterial suspension was injected. All larvae were injected through the haemocoel via the rear left pro-leg using an insulin syringe. Post injection, the larvae were incubated at 37 °C and their mortality was assessed every 24 h for 5 days. Over the course of 5 days, the larvae were examined with forceps by flipping them onto their backs and checking if there are signs of motility. Live larvae will quickly flip back, whereas sick larvae will be slow, the exterior will be hardened and darkened, and they will usually die within the coming 1–2 days. Dead larvae will have a hard, dry, and very dark exterior and will shrink in size. The results were observed and noted, and survival graphs were generated using GraphPad Prism 10.0.3 software. Observation data are presented in Supplementary Table S2.
4.8. Serum Resistance Assay
The susceptibility of bacteria to human serum was investigated using the previously described method [79] with some modifications. On day 1, the selected isolates for the experiment were sub-cultured onto agar plates from cryo-beads as well as K. pneumoniae ATCC 1705 that was used as a positive susceptible control in this experiment. On day 2, 3–5 colonies were picked up and resuspended in 10 mL of liquid broth then incubated overnight at 37 °C. On day 3, the bacterial suspension was diluted to 2 × 106 CFU/mL. Then, 25 µL of bacterial suspension and 75 µL of human serum (Sigma-Aldrich, St. Louis, MO, USA) were dispensed in microtitration trays and mixed then incubated at 37 °C. To assess viability, the samples were streaked onto CLED agar plates immediately at time zero, after 1 h, after 2 h, and finally after 3 h. Each isolate was tested 3 times. The streaked agar plates were incubated overnight. On day 4, CFU counts of each sample and the control at 4 different time points were recorded, and graphs were made using Graphpad Prism 10.0.3. (Supplementary Table S3 and Figures S6–S10).
5. Conclusions
In summary, to our knowledge, this is the first study in Oman to depict the molecular and genomic characteristics of hypervirulent K. pneumoniae isolates collected from all around the country, where analysis of WGS of the isolates showed a distinct pattern of k-loci depending on the particular STs, particularly ST-231 and ST-395, which are the two most prominent STs. Resistance to serum bactericidal activity and G. mellonella lethality by hypervirulent capsular serotypes K1 and K2 K. pneumoniae strains was demonstrated. The hvKp strains in this study are diverse in the phylogenetic origin, but the likelihood of transmission and future outbreaks is key to detect possible spread of hvKp isolates in healthcare settings via active surveillance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25031944/s1. Reference [80] is cited in the Supplementary Materials.
Author Contributions
Conceptualization, Z.A.-J. and R.F.; methodology, B.A.-B.; software, B.A.-B. and Z.A.-S.; validation, M.R. and Z.A.-J.; formal analysis, B.A.-B. and Z.A.-S.; investigation, B.A.-B.; resources, A.A.-R., M.A.-M. and A.A.-J.; data curation, Z.A.-J. and Z.A.-S.; writing—original draft preparation, B.A.-B.; writing—review and editing, Z.A.-J.; visualization, M.R.; supervision, Z.A.-J.; project administration, Z.A.-J.; funding acquisition, Z.A.-J. All authors have read and agreed to the published version of the manuscript.
Funding
Funding was provided by Sultan Qaboos University internal grant (Project code: IG/MED/MICR/21/01).
Institutional Review Board Statement
The study was commenced after obtaining approval by the Medical Ethics Research committee (MREC#1896), College of Medicine and Health Sciences, Sultan Qaboos University, Muscat, Oman.
Informed Consent Statement
Not applicable.
Data Availability Statement
All supporting data can be found in the Supplementary Files. All whole-genome sequencing data are deposited in Genbank at accession number PRJNA999478. The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Acknowledgments
We would like to express our sincere thanks to the sequencing company provided by MicrobesNG (Birmingham, UK) (https://microbesng.com, accessed on 6 June 2021) for performing WGS and bioinformatics analysis. We would like to thank supervising staff, College of Agriculture, Department of Food Microbiology for providing instrumental support to conduct some experiments for cultivating the bacterial isolates.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae–Clinical and Molecular Perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, R.; Chen, Y.; Zhang, X.; Liu, L.; Luo, M.; Chen, J.; Chen, K.; Zeng, T.; Liu, B.; et al. Clinical and Molecular Characteristics and Antibacterial Strategies of Klebsiella pneumoniae in Pyogenic Infection. Microbiol. Spectr. 2023, 11, e0064023. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, B.; Wang, Y.; Jing, S.; Ning, W.; Liu, C.; Chen, C. A Wide Clinical Spectrum of Pulmonary Affection in Subjects with Community-Acquired Klebsiella pneumoniae Liver Abscess (CA-KPLA). J. Infect. Chemother. 2023, 29, 48–54. [Google Scholar] [CrossRef]
- Bagley, S.T. Habitat Association of Klebsiella Species. Infect. Control 1985, 6, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Rock, C.; Thom, K.A.; Masnick, M.; Johnson, J.K.; Harris, A.D.; Morgan, D.J. Frequency of Klebsiella pneumoniae Carbapenemase (KPC)–Producing and Non-KPC-Producing Klebsiella Species Contamination of Healthcare Workers and the Environment. Infect. Control Hosp. Epidemiol. 2014, 35, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Jeon, J.H.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell. Infect. Microbiol. 2017, 7, 483. [Google Scholar] [CrossRef]
- Larsen, J.; Enright, M.C.; Godoy, D.; Spratt, B.G.; Larsen, A.R.; Skov, R.L. Multilocus Sequence Typing Scheme for Staphylococcus aureus: Revision of the Gmk Locus. J. Clin. Microbiol. 2012, 50, 2538–2539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive Web: User-Friendly Capsule and Lipopolysaccharide Serotype Prediction for Klebsiella Genomes. J. Clin. Microbiol. 2018, 56, e00197-18. [Google Scholar] [CrossRef]
- Han, M.; Liu, C.; Xie, H.; Zheng, J.; Zhang, Y.; Li, C.; Shen, H.; Cao, X. Genomic and Clinical Characteristics of Carbapenem-Resistant Enterobacter Cloacae Complex Isolates Collected in a Chinese Tertiary Hospital during 2013–2021. Front. Microbiol. 2023, 14, 1127948. [Google Scholar] [CrossRef]
- Guo, X.; Chen, R.; Wang, Q.; Li, C.; Ge, H.; Qiao, J.; Li, Y. Global Prevalence, Characteristics, and Future Prospects of IncX3 Plasmids: A Review. Front. Microbiol. 2022, 13, 979558. [Google Scholar] [CrossRef]
- Bilal, H.; Zhang, G.; Rehman, T.; Han, J.; Khan, S.; Shafiq, M.; Yang, X.; Yan, Z.; Yang, X. First Report of Blandm-1 Bearing Incx3 Plasmid in Clinically Isolated St11 Klebsiella pneumoniae from Pakistan. Microorganisms 2021, 9, 951. [Google Scholar] [CrossRef]
- Hameed, M.F.; Chen, Y.; Wang, Y.; Shafiq, M.; Bilal, H.; Liu, L.; Ma, J.; Gu, P.; Ge, H. Epidemiological Characterization of Colistin and Carbapenem Resistant Enterobacteriaceae in a Tertiary: A Hospital from Anhui Province. Infect. Drug Resist. 2021, 14, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. PlasmidFinder and PMLST: In Silico Detection and Typing of Plasmid. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Nazir, A.; Zhao, Y.; Li, M.; Manzoor, R.; Tahir, R.A.; Zhang, X.; Qing, H.; Tong, Y. Structural Genomics of Repa, Repb1-Carrying Incfib Family Pa1705-Qnrs, P911021-Teta, and P1642-Teta, Multidrug-Resistant Plasmids from Klebsiella pneumoniae. Infect. Drug Resist. 2020, 13, 1889–1903. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population Genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, Y.; Li, G.; Liu, J.; Li, X.; Tian, L.; Sun, J.; Ou, H.Y.; Qu, H. Whole-Genome-Sequencing Characterization of Bloodstream Infection-Causing Hypervirulent Klebsiella pneumoniae of Capsular Serotype K2 and ST374. Virulence 2018, 9, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Lin, T.L.; Chen, Y.H.; Hsu, C.R.; Hsieh, P.F.; Wu, M.C.; Wang, J.T. Capsular Types of Klebsiella pneumoniae Revisited by Wzc Sequencing. PLoS ONE 2013, 8, e80670. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Fang, H.C.; Yang, H.C.; Lin, T.L.; Hsieh, P.F.; Tsai, F.C.; Keynan, Y.; Wang, J.T. Capsular Polysaccharide Synthesis Regions in Klebsiella pneumoniae Serotype K57 and a New Capsular Serotype. J. Clin. Microbiol. 2008, 46, 2231–2240. [Google Scholar] [CrossRef]
- Whitfield, C.; Roberts, I.S. Structure, Assembly and Regulation of Expression of Capsules in Escherichia coli. Mol. Microbiol. 1999, 31, 1307–1319. [Google Scholar] [CrossRef]
- Rahn, A.; Drummelsmith, J.; Whitfield, C. Conserved Organization in the Cps Gene Clusters for Expression of Escherichia coli Group 1 K Antigens: Relationship to the Colanic Acid Biosynthesis Locus and the Cps Genes from Klebsiella pneumoniae. J. Bacteriol. 1999, 181, 2307–2313. [Google Scholar] [CrossRef]
- Qian, C.; Zhang, S.; Xu, M.; Zeng, W.; Chen, L.; Zhao, Y.; Zhou, C.; Zhang, Y.; Cao, J.; Zhou, T. Genetic and Phenotypic Characterization of Multidrug-Resistant Klebsiella pneumoniae from Liver Abscess. Microbiol. Spectr. 2023, 11, e0224022. [Google Scholar] [CrossRef]
- Struve, C.; Roe, C.C.; Stegger, M.; Stahlhut, S.G.; Hansen, D.S.; Engelthaler, D.M.; Andersen, P.S.; Driebe, E.M.; Keim, P.; Krogfelt, K.A. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. mBio 2015, 6, e00630-15. [Google Scholar] [CrossRef]
- Cubero, M.; Grau, I.; Tubau, F.; Pallarés, R.; Dominguez, M.A.; Liñares, J.; Ardanuy, C. Hypervirulent Klebsiella pneumoniae Clones Causing Bacteraemia in Adults in a Teaching Hospital in Barcelona, Spain (2007–2013). Clin. Microbiol. Infect. 2016, 22, 154–160. [Google Scholar] [CrossRef]
- Zheng, R.; Zhang, Q.; Guo, Y.; Feng, Y.; Liu, L.; Zhang, A.; Zhao, Y.; Yang, X.; Xia, X. Outbreak of Plasmid-Mediated NDM-1-Producing Klebsiella pneumoniae ST105 among Neonatal Patients in Yunnan, China. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Liu, J.W.; Su, L.H.; Chien, C.C.; Li, C.C.; Yang, K.D. Hypermucoviscosity Associated with Klebsiella pneumoniae-Mediated Invasive Syndrome: A Prospective Cross-Sectional Study in Taiwan. Int. J. Infect. Dis. 2010, 14, e688–e692. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.Y.; Fung, C.P.; Liu, Y.M.; Wu, K.M.; Chen, Y.T.; Li, L.H.; Liu, T.T.; Kirby, R.; Tsai, S.F. Genetic Diversity of Capsular Polysaccharide Biosynthesis in Klebsiella pneumoniae Clinical Isolates. Microbiology 2009, 155, 4170–4183. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Siek, K.E.; Johnson, S.J.; Nolan, L.K. DNA Sequence of a ColV Plasmid and Prevalence of Selected Plasmid-Encoded Virulence Genes among Avian Escherichia Coli Strains. J. Bacteriol. 2006, 188, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Vill, L.; Dobiasova, H.; Fortini, D.; Feudi, C.; Carattoli, A. Plasmid Content of a Clinically Relevant Klebsiella pneumoniae Clone from the Czech Republic Producing CTX-M-15 and QnrB1. Antimicrob. Agents Chemother. 2013, 57, 1073–1076. [Google Scholar] [CrossRef] [PubMed]
- Khajanchi, B.K.; Hasan, N.A.; Choi, S.Y.; Han, J.; Zhao, S.; Colwell, R.R.; Cerniglia, C.E.; Foley, S.L. Comparative Genomic Analysis and Characterization of Incompatibility Group FIB Plasmid Encoded Virulence Factors of Salmonella Enterica Isolated from Food Sources. BMC Genom. 2017, 18, 570. [Google Scholar] [CrossRef]
- Pedersen, T.; Tellevik, M.G.; Kommedal, Ø.; Lindemann, P.C.; Moyo, S.J.; Janice, J.; Blomberg, B.; Samuelsen, Ø.; Langeland, N. Horizontal Plasmid Transfer among Klebsiella pneumoniae Isolates Is the Key Factor for Dissemination of Extended-Spectrum β-Lactamases among Children in Tanzania. mSphere 2020, 5, e00428-20. [Google Scholar] [CrossRef] [PubMed]
- Lutgring, J.D.; Zhu, W.; De Man, T.J.B.; Avillan, J.J.; Anderson, K.F.; Lonsway, D.R.; Rowe, L.A.; Batra, D.; Rasheed, J.K.; Limbago, B.M. Phenotypic and Genotypic Characterization of Enterobacteriaceae Producing Oxacillinase-48-like Carbapenemases, United States. Emerg. Infect. Dis. 2018, 24, 700–709. [Google Scholar] [CrossRef]
- Ragupathi, N.K.D.; Bakthavatchalam, Y.D.; Mathur, P.; Pragasam, A.K.; Walia, K.; Ohri, V.C.; Veeraraghavan, B. Plasmid Profiles among Some ESKAPE Pathogens in a Tertiary Care Centre in South India. Indian. J. Med. Res. 2019, 149, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.A.D.; Brisse, S. Multilocus Sequence Typing of Klebsiella pneumoniae Nosocomial Isolates. J. Clin. Microbiol. 2005, 43, 4178–4182. [Google Scholar] [CrossRef]
- Choi, M.; Hegerle, N.; Nkeze, J.; Sen, S.; Jamindar, S.; Nasrin, S.; Sen, S.; Permala-Booth, J.; Sinclair, J.; Tapia, M.D.; et al. The Diversity of Lipopolysaccharide (O) and Capsular Polysaccharide (K) Antigens of Invasive Klebsiella pneumoniae in a Multi-Country Collection. Front. Microbiol. 2020, 11, 1249. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Han, R.; Yin, D.; Jiang, B.; Ding, L.; Guo, Y.; Wu, S.; Wang, C.; Zhang, H.; Hu, F. A Nationwide Genomic Study of Clinical Klebsiella pneumoniae Carrying Bla OXA-232 and RmtF in China. Microbiol. Spectr. 2023, 11, e0386322. [Google Scholar] [CrossRef]
- Mancini, S.; Poirel, L.; Tritten, M.L.; Lienhard, R.; Bassi, C.; Nordmann, P. Emergence of an MDR Klebsiella pneumoniae ST231 Producing OXA-232 and RmtF in Switzerland. J. Antimicrob. Chemother. 2017, 73, 821–823. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Kalpoe, J.; Poirel, L.; Nordmann, P. European Dissemination of a Single OXA-48-Producing Klebsiella pneumoniae Clone. Clin. Microbiol. Infect. 2011, 17, E24–E26. [Google Scholar] [CrossRef]
- Gijón, D.; Tedim, A.P.; Valverde, A.; Rodríguez, I.; Morosini, M.-I.; Coque, T.M.; Manrique, M.; Pareja, E.; Tobes, R.; Ruiz-Garbajosa, P.; et al. Early OXA-48-Producing Enterobacterales Isolates Recovered in a Spanish Hospital Reveal a Complex Introduction Dominated by Sequence Type 11 (ST11) and ST405 Klebsiella pneumoniae Clones. mSphere 2020, 5, e00080-20. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like Carbapenemases: The Phantom Menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, X.; Torres, V.V.L.; Liu, H.; Rocker, A.; Zhang, Y.; Wang, J.; Chen, L.; Bi, W.; Lin, J.; et al. An Outbreak of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae in an Intensive Care Unit of a Major Teaching Hospital in Wenzhou, China. Front. Public. Health 2019, 7, 229. [Google Scholar] [CrossRef]
- Maiden, M.C.J.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef] [PubMed]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; Aljallal, A.; Radwan, H.H.; Shibl, A.M. Characterization of Carbapenemases, ESBLs, and Plasmid-Mediated Quinolone Determinants in Carbapenem-Insensitive Escherichia Coli and Klebsiella pneumoniae in Riyadh Hospitals. J. Infect. Public Health 2018, 11, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Alsharapy, S.A.; Gharout-Sait, A.; Muggeo, A.; Guillard, T.; Cholley, P.; Brasme, L.; Bertrand, X.; Moghram, G.S.; Touati, A.; De Champs, C. Characterization of Carbapenem-Resistant Enterobacteriaceae Clinical Isolates in Al Thawra University Hospital, Sana’a, Yemen. Microb. Drug Resist. 2020, 26, 211–217. [Google Scholar] [CrossRef]
- López-Camacho, E.; Paño-Pardo, J.R.; Ruiz-Carrascoso, G.; Wesselink, J.J.; Lusa-Bernal, S.; Ramos-Ruiz, R.; Ovalle, S.; Gómez-Gil, R.; Pérez-Blanco, V.; Pérez-Vázquez, M.; et al. Population Structure of OXA-48-Producing Klebsiella pneumoniae ST405 Isolates during a Hospital Outbreak Characterised by Genomic Typing. J. Glob. Antimicrob. Resist. 2018, 15, 48–54. [Google Scholar] [CrossRef]
- Kutlu, H.H.; Dolapçı, İ.; Avcı, M.; Tekeli, A. The Emergence of Klebsiella pneumoniae Sequence Type 395 Non-Susceptible to Carbapenems and Colistin from Turkey. Indian. J. Med. Microbiol. 2023, 46, 100419. [Google Scholar] [CrossRef]
- Sandfort, M.; Hans, J.B.; Fischer, M.A.; Reichert, F.; Cremanns, M.; Eisfeld, J.; Pfeifer, Y.; Heck, A.; Eckmanns, T.; Werner, G.; et al. Increase in NDM-1 and NDM-1/OXA-48-Producing Klebsiella pneumoniae in Germany Associated with the War in Ukraine, 2022. Eurosurveillance 2022, 27, 2200926. [Google Scholar] [CrossRef]
- Shon, A.S.; Bajwa, R.P.S.; Russo, T.A. Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae: A New and Dangerous Breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef]
- Tichaczek-Goska, D.; Witkowska, D.; Cisowska, A.; Jankowski, S.; Hendrich, A.B. The Bactericidal Activity of Normal Human Serum against Enterobacteriaceae Rods with Lipopolysaccharides Possessing O-Antigens Composed of Mannan. Adv. Clin. Exp. Med. 2012, 21, 289–299. [Google Scholar] [PubMed]
- Benge, G.R. Bactericidal Activity of Human Serum against Strains of Klebsiella from Different Sources. J. Med. Microbiol. 1988, 27, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Yeh, K.M.; Chiu, S.K.; Lin, C.L.; Huang, L.Y.; Tsai, Y.K.; Chang, J.C.; Lin, J.C.; Chang, F.Y.; Siu, L.K. Surface Antigens Contribute Differently to the Pathophysiological Features in Serotype K1 and K2 Klebsiella pneumoniae Strains Isolated from Liver Abscesses. Gut Pathog. 2016, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Tomas, J.M.; Benedi, V.J.; Ciurana, B.; Jofre, J. Role of Capsule and O Antigen in Resistance of Klebsiella pneumoniae to Serum Bactericidal Activity. Infect. Immun. 1986, 54, 85–89. [Google Scholar] [CrossRef] [PubMed]
- DeLeo, F.R.; Kobayashi, S.D.; Porter, A.R.; Freedman, B.; Dorward, D.W.; Chen, L.; Kreiswirth, B.N. Survival of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 258 in Human Blood. Antimicrob. Agents Chemother. 2017, 61, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Kumabe, A.; Kenzaka, T. String Test of Hypervirulent Klebsiella Pneumonia. QJM Int. J. Med. 2014, 107, 1053. [Google Scholar] [CrossRef] [PubMed]
- Ruangpan, L. Chapter 3. Minimal Inhibitory Concentration (MIC) Test and Determination of Antimicrobial Resistant Bacteria. In Laboratory Manual of Standardized Methods for Antimicrobial Sensitivity Tests for Bacteria Isolated from Aquatic Animals and Environment; Aquaculture Department, Southeast Asian Fisheries Development Center: Bangkok, Thailand, 2004. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing A CLSI Supplement for Global Application; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-Metagenomes from Highly Chimeric Reads. In Research in Computational Molecular Biology; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin, Germany, 2013; Volume 7821. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An Integrated Platform for Visualization and Analysis of High-Throughput Sequence-Based Experimental Data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A Genomic Surveillance Framework and Genotyping Tool for Klebsiella pneumoniae and Its Related Species Complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.Y. ICEberg 2.0: An Updated Database of Bacterial Integrative and Conjugative Elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0. Molecular Biology and Evolution. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 15 January 2021).
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL): An Online Tool for Phylogenetic Tree Display and Annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Seemann, T. Snippy: Rapid Haploid Variant Calling and Core SNP Phylogeny. 2015. Available online: https://github.com/tseemann/snippy (accessed on 25 January 2021).
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ménard, G.; Rouillon, A.; Cattoir, V.; Donnio, P.Y. Galleria Mellonella as a Suitable Model of Bacterial Infection: Past, Present and Future. Front. Cell. Infect. Microbiol. 2021, 11, 782733. [Google Scholar] [CrossRef] [PubMed]
- Bugla-Płoskońska, G.; Kiersnowski, A.; Futoma-Kołoch, B.; Doroszkiewicz, W. Killing of Gram-Negative Bacteria with Normal Human Serum and Normal Bovine Serum: Use of Lysozyme and Complement Proteins in the Death of Salmonella Strains O48. Microb. Ecol. 2009, 58, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, X.; Luo, M.; Liu, P.; Su, K.; Qing, Y.; Chen, S.; Qiu, J.; Li, Y. Molecular Characterisations of Integrons in Clinical Isolates of Klebsiella Pneumoniae in a Chinese Tertiary Hospital. Microb. Pathog. 2017, 104, 164–170. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).