Identification of Candidate Male-Reproduction-Related Genes from the Testis and Androgenic Gland of Macrobrachium nipponense, Regulated by PDHE1, through Transcriptome Profiling Analysis
Abstract
1. Introduction
2. Results
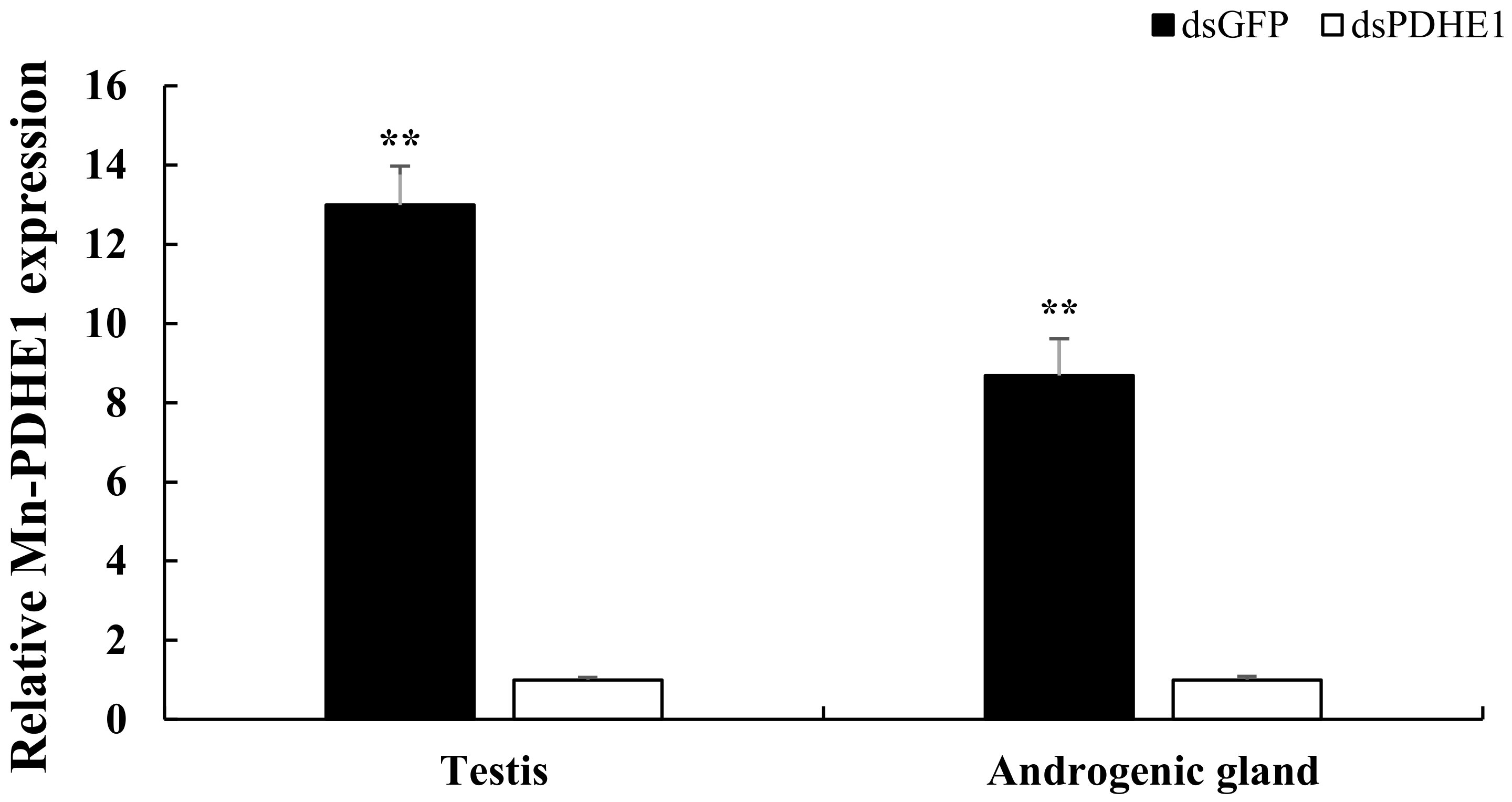
2.1. Measurement of the Efficiency of dsPDHE1
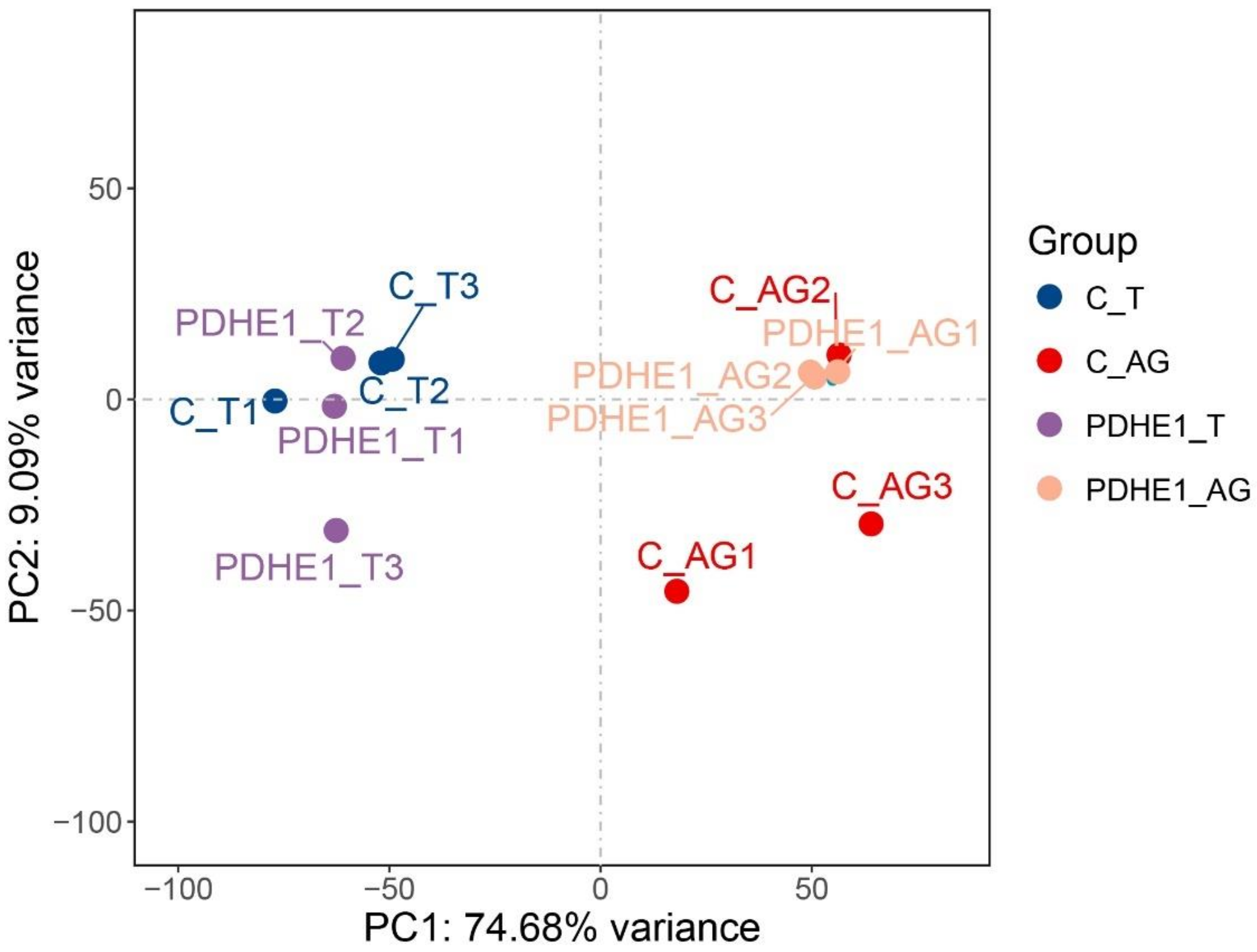
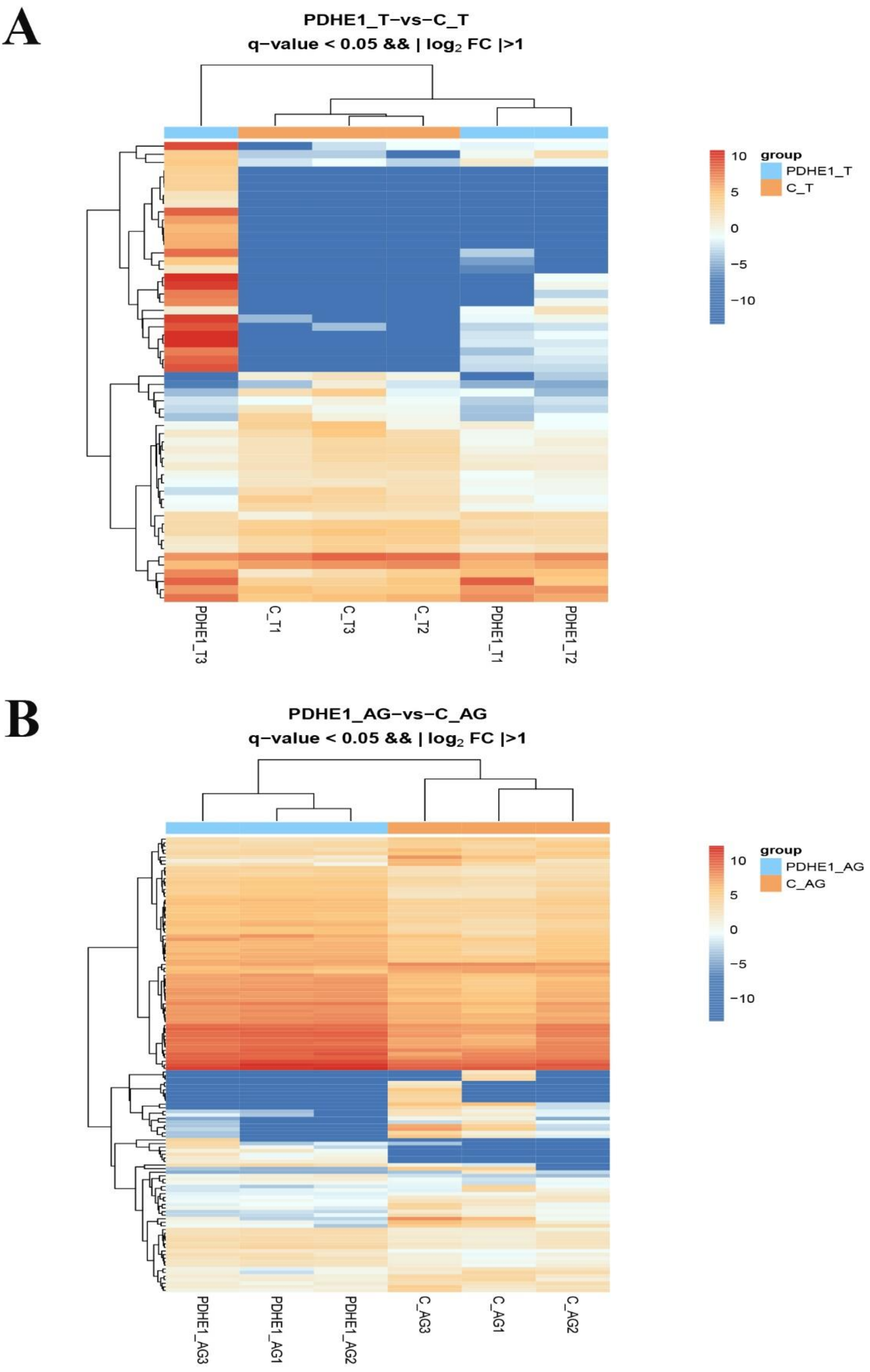
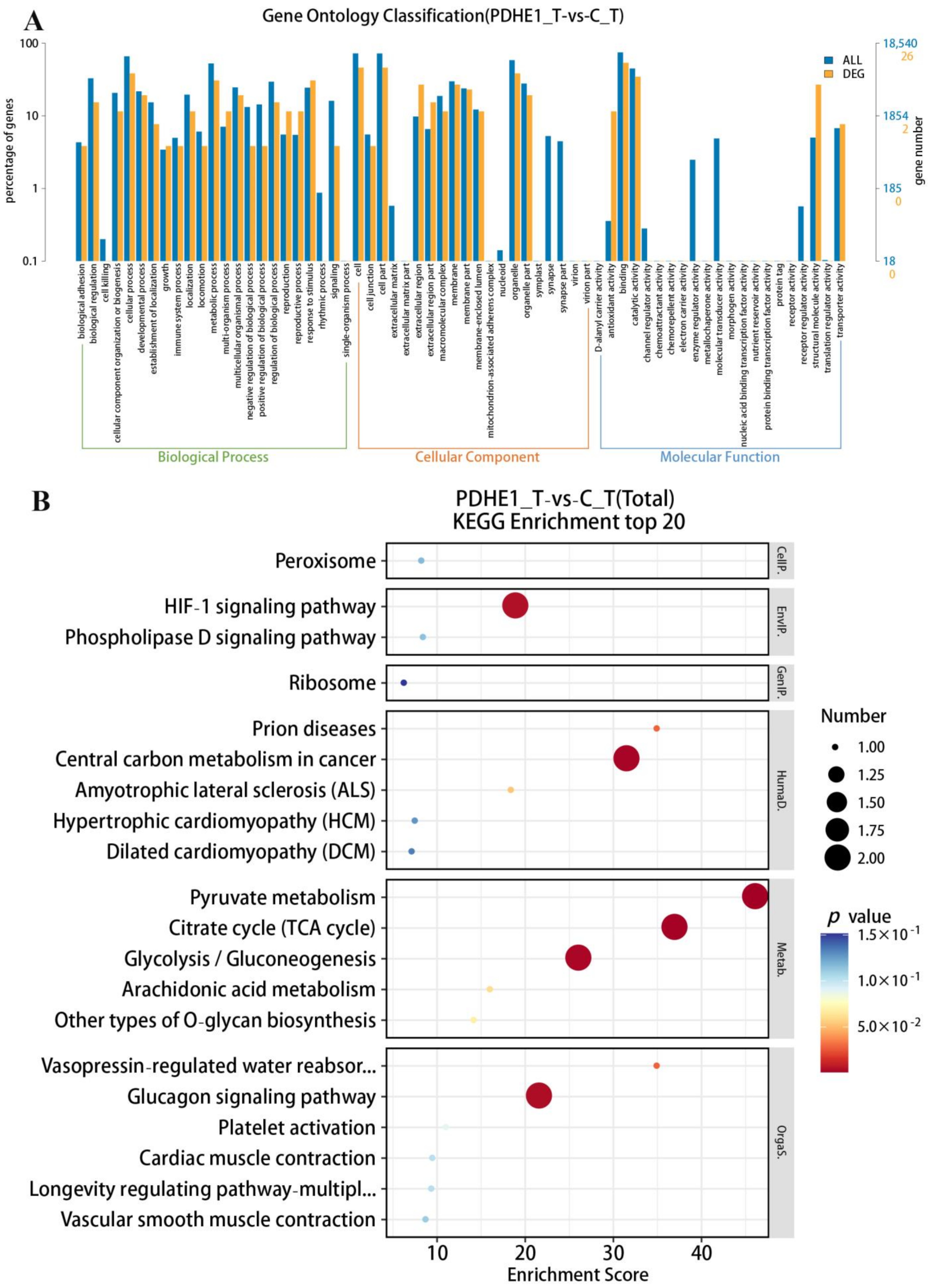
2.2. Transcriptome Profiling Analysis in Testis
2.3. Transcriptome Profiling Analysis in Androgenic Gland
2.4. Selection of Male-Reproduction-Related Genes
2.5. qPCR Analysis of DEGs
3. Materials and Methods
3.1. Sample Preparation
3.2. Synthesize of dsPDHE1 and dsGFP
3.3. RNAi Analysis
3.4. qPCR Analysis
3.5. Transcriptome Profiling Analysis
3.6. Statistical Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, H.T.; Jiang, S.F.; Xiong, Y.W. Current Status and Prospects of Farming the Giant River Prawn (Macrobrachium rosenbergii) and the oriental River Prawn (Macrobrachium nipponense) in china. Aquac. Res. 2012, 43, 993–998. [Google Scholar]
- Zhang, X.L.; Cui, L.F.; Li, S.M.; Liu, X.Z.; Han, X.; Jiang, K.Y.; Bureau of Fisheries, Ministry of Agriculture, P.R.C. Fisheries economic statistics. In China Fishery Yearbook; Beijing China Agricultural Press: Beijing, China, 2020; Volume 24. [Google Scholar]
- Jin, S.B.; Zhang, Y.; Guan, H.H.; Fu, H.T.; Jiang, S.F.; Xiong, Y.W.; Qiao, H.; Zhang, W.Y.; Gong, Y.S.; Wu, Y. Histological observation of gonadal development during post-larva in oriental river prawn, Macrobrachium nipponense. Chin. J. Fish. 2016, 29, 11–16. [Google Scholar]
- Metón, I.; Fernández, F.; Baanante, I.V. Short- and long-term effects of refeeding on key enzyme activities in glycolysis–gluconeogenesis in the liver of gilthead seabream (Sparus aurata). Aquaculture 2003, 225, 99–107. [Google Scholar] [CrossRef]
- Nordlie, R.C.; Foster, J.D.; Lange, A.J. Regulation OF glucose production BY the liver. Annu. Rev. Nutr. 1999, 19, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J. Biol. Chem. 2001, 276, 38329–38336. [Google Scholar] [CrossRef]
- Perham, R.N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: A paradigm in the design of a multifunctional protein. Biochemistry 1991, 30, 8501–8512. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.S.; Roche, T.E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990, 4, 3224–3233. [Google Scholar] [CrossRef] [PubMed]
- Linn, T.C.; Pettit, F.H.; Reed, L.J. Alpha-keto acid dehydrogenase complexes. X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc. Natl. Acad. Sci. USA 1969, 62, 234–241. [Google Scholar] [CrossRef]
- Linn, T.C.; Pettit, F.H.; Hucho, F.; Reed, L.J. Alpha-keto acid dehydrogenase complexes. XI. Comparative studies of regulatory properties of the pyruvate dehydrogenase complexes from kidney, heart, and liver mitochondria. Proc. Natl. Acad. Sci. USA 1969, 64, 227–234. [Google Scholar] [CrossRef]
- Patel, M.S.; Nemeria, N.S.; Furey, W.; Jordan, F. The pyruvate dehydrogenase complexes: Structure-based function and regulation. J. Biol. Chem. 2014, 289, 16615–16623. [Google Scholar] [CrossRef]
- Carlsson, J.; Kujala, U.; Edlund, M.J.K. Pyruvate Dehydrogenase Activity in Streptococcus mutans. Infect. Immun. 1985, 49, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.B.; Hu, Y.N.; Fu, H.T.; Sun, S.M.; Jiang, S.F.; Xiong, Y.W.; Qiao, H.; Zhang, W.; Gong, Y.; Wu, Y. Analysis of Testis Metabolome and Transcriptome from the Oriental River Prawn (Macrobrachium nipponense) in Response to Different Temperatures and Illumination Times. Comp. Biochem. Phys. D 2020, 34, 100662. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.B.; Zhang, W.Y.; Xiong, Y.W.; Jiang, S.F.; Qiao, H.; Gong, Y.S.; Wu, Y.; Fu, H.T. Genetic regulation of male sexual development in the oriental river prawn Macrobrachium nipponense during reproductive vs. non-reproductive season. Aquacult. Int. 2022, 30, 2059–2079. [Google Scholar] [CrossRef]
- Jin, S.; Hu, Y.; Fu, H.; Jiang, S.; Xiong, Y.; Qiao, H.; Zhang, W.; Gong, Y.; Wu, Y. Identification and Characterization of the Pyruvate Dehydrogenase E1 Gene in the Oriental River Prawn, Macrobrachium nipponense. Front. Endocrinol. 2021, 12, 752501. [Google Scholar] [CrossRef] [PubMed]
- Sagi, A.; Cohen, D.; Wax, Y. Production of Macrobrachium rosenbergii in Momosex Population: Yield Characteristes Under Intensive Monoculture Conditions in Cages. Aquaculture 1986, 51, 265–275. [Google Scholar] [CrossRef]
- Sagi, A.; Cohen, D.; Milner, Y. Effect of Androgenic Gland Ablation on Morphotypic Differentiation and Sexual Characteristics of Male Freshwater Prawns, Macrobrachium rosenbergii. Gen. Comp. Endocr. 1990, 77, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.Y.; Li, J.L.; Qiu, G.F. Identification of putative regulatory region of insulin-like androgenic gland hormone gene (IAG) in the prawn Macrobrachium nipponense and proteins that interact with IAG by using yeast two-hybrid system. Gen. Comp. Endocr. 2016, 229, 112–118. [Google Scholar] [CrossRef]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.D.; Ventura, T.; Sagi, A. A sexual shift induced by silencing of a single insulin-like gene in crayfish: Ovarian upregulation and testicular degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 2009, 150, 1278–1286. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Sagi, A. Expression of an androgenic gland-specific insulin-like peptide during the course of prawn sexual and morpho-typic differentiation. ISRN Endocrinol. 2011, 2011, 476283. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing Sexual Differentiation: Full Functional Sex Reversal Achieved Through Silencing of a Single Insulin-Like Gene in the Prawn, Macrobrachium rosenbergii. Biol. Reprod. 2012, 86, 90. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.X. The Molecular Mechanism in Male Reproductive Tract of the Prawn, Macrobrachium rosenbergii; Zhejiang University: Hangzhou, China, 2006. [Google Scholar]
- Guo, Z.H. Study on Proliferation and Differentiation of Spermatogenic Cells from Macrobrachium nipponense In Vitro; Hebei University: Baoding, China, 2007. [Google Scholar]
- Qiu, G.F.; Du, N.S.; Lai, W. Studies on the Male Reproductive System of the Freshwater Prawn, Macrobrachium nipponense. J. Shanghai Ocean Uni. 1995, 4, 107–111. [Google Scholar]
- Qiao, H.; Fu, H.T.; Jin, S.B.; Wu, Y.; Jiang, S.F.; Gong, Y.S.; Xiong, Y.W. Constructing and Random Sequencing Analysis of Normalized cDNA Library of Testis Tissue from Oriental River Prawn (Macrobrachium nipponense). Comp. Biochem. Phys. D 2012, 7, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.B.; Fu, H.T.; Zhou, Q.; Sun, S.M.; Jiang, S.F.; Xiong, Y.W.; Gong, Y.S.; Qiao, H.; Zhang, W.Y. Transcriptome analysis of androgenic gland for discovery of novel genes from the oriental river prawn, Macrobrachium nipponense, using Illumina Hiseq 2000. PLoS ONE 2013, 8, e76840. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.B.; Fu, Y.; Hu, Y.N.; Fu, H.T.; Jiang, S.F.; Xiong, Y.W.; Qiao, H.; Zhang, W.Y.; Gong, Y.S.; Wu, Y. Transcriptome Profiling Analysis of the Testis After Eyestalk Ablation for Selection of the Candidate Genes Involved in the Male Sexual Development in Macrobrachium nipponense. Front. Genet. 2021, 12, 675928. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.B.; Fu, Y.; Hu, Y.N.; Fu, H.T.; Jiang, S.F.; Xiong, Y.W.; Qiao, H.; Zhang, W.Y.; Gong, Y.S.; Wu, Y. Identification of candidate genes from androgenic gland in Macrobrachium nipponense regulated by eyestalk ablation. Sci. Rep. 2021, 11, 19855. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Hu, Y.; Fu, H.; Jiang, S.; Xiong, Y.; Qiao, H.; Zhang, W.; Gong, Y.; Wu, Y. Identification and Characterization of the Succinate Dehydrogenase Complex Iron Sulfur Subunit B Gene in the Oriental River Prawn, Macrobrachium nipponense. Front. Genet. 2021, 12, 698318. [Google Scholar] [CrossRef]
- Zhang, W.; Xiong, Y.; Wang, P.; Chen, T.; Jiang, S.; Qiao, H.; Gong, Y.; Wu, Y.; Jin, S.; Fu, H. RNA interference analysis of potential functions of cyclin A in the reproductive development of male oriental river prawns (Macrobrachium nipponense). Front. Genet. 2022, 13, 1053826. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, P.; Xiong, Y.; Chen, T.; Jiang, S.; Qiao, H.; Gong, Y.; Wu, Y.; Jin, S.; Fu, H. RNA Interference Analysis of the Functions of Cyclin B in Male Reproductive Development of the Oriental River Prawn (Macrobrachium nipponense). Genes 2022, 13, 2079. [Google Scholar] [CrossRef]
- Jin, S.; Zhou, Z.; Zhang, W.; Xiong, Y.; Qiao, H.; Gong, Y.; Wu, Y.; Jiang, S.; Fu, H. RNAi Analysis of Potential Functions of Cyclin B3 in Reproduction of Male Oriental River Prawns (Macrobrachium nipponense). Animals 2023, 13, 1703. [Google Scholar] [CrossRef]
- Zhang, S.B.; Jiang, P.; Wang, Z.Q.; Long, S.R.; Liu, R.D.; Zhang, X.; Yang, W.; Ren, H.J.; Cui, J. Dsrna-Mediated Silencing of Nudix Hydrolase in Trichinella spiralis Inhibits the Larval Invasion and Survival in Mice. Exp. Parasitol. 2016, 162, 35–42. [Google Scholar] [CrossRef]
- Hu, Y.N.; Fu, H.T.; Qiao, H.; Sun, S.M.; Zhang, W.Y.; Jin, S.B.; Jiang, S.F.; Gong, Y.S.; Xiong, Y.W.; Wu, Y. Validation and evaluation of reference genes for Quantitative real-time PCR in Macrobrachium nipponense. Int. J. Mol. Sci. 2018, 19, 2258. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.l.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Itoh, M. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ferreira, J.A.; Zwinderman, A.H. On the Benjamini–Hochberg method. Ann. Statist. 2006, 34, 1827–1849. [Google Scholar] [CrossRef]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 2014, 71, 2577–2604. [Google Scholar] [CrossRef] [PubMed]
- Shahin, A.Y.; Ismail, A.M.; Shaaban, O.M. Supplementation of clomiphene citrate cycles with Cimicifuga racemosa or ethinyl oestradiol-a randomized trial. Reprod. BioMed. Online 2009, 19, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Lubert, S. Glycolysis. In Biochemistry, 4th ed.; W. H. Freeman and Company: New York, NY, USA, 1995; pp. 483–508. [Google Scholar]
- Yang, M.X.; Pan, M.Z.; Huang, D.; Liu, J.H.; Guo, Y.L.; Liu, Y.; Xiao, T.Y.; Mai, K.S.; Zhang, W.B. The uninhibited glucagon signaling pathway in Japanese flounder Paralichthys olivaceus: A link with the glucose intolerance? Aquaculture 2024, 579, 740172. [Google Scholar] [CrossRef]
- Westman, J.; Grinstein, S. Determinants of phagosomal pH during host-pathogen interactions. Front. Cell Dev. Biol. 2020, 8, 624958. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wang, S.; Zhang, C.; Hu, X.; Zhou, L.; Li, Y.; Xu, L. Selenium ameliorates cadmium-induced mouse leydig TM3 cell apoptosis via inhibiting the ROS/JNK/c-jun signaling pathway. Ecotoxicol. Environ. Saf. 2020, 192, 110266. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, S.; Cao, L.; Ren, X.; Li, Y.; Shao, J.; Xu, L. Lead acetate induces apoptosis in Leydig cells by activating PPARgamma/caspase-3/PARP pathway. Int. J. Environ. Health Res. 2021, 31, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Kraft, C.; Deplazes, A.; Sohrmann, M.; Peter, M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 2008, 10, 602–610. [Google Scholar] [CrossRef]
- Wyant, G.A.; Abu-Remaileh, M.; Frenkel, E.M.; Laqtom, N.N.; Dharamdasani, V.; Lewis, C.A.; Chan, S.H.; Heinze, I.; Ori, A.; Sabatini, D.M. Nufip1 is a ribosome receptor for starvation-induced ribophagy. Science 2018, 360, 751–758. [Google Scholar] [CrossRef]
- An, H.; Harper, J.W. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat. Cell Biol. 2018, 20, 135–143. [Google Scholar] [CrossRef]
- Inoue, H.; Ozaki, N.; Nagasawa, H. Purification and structural determination of a phosphorylated peptide with anti-calcification and chitin-binding activities in the exoskeleton of the crayfish, Procambarus clarkia. Biosci. Biotechnol. Biochem. 2001, 65, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Ohira, T.; Ozaki, N.; Nagasawa, H. Cloning and expression of a cDNA encoding a matrix peptide associated with calcification in the exoskeleton of the crayfish. Comp. Biochem. Physiol. B 2003, 136, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Ohira, T.; Ozaki, N.; Nagasawa, H. A novel calcium-binding peptide from the cuticle of the crayfish, Procambarus clarkia. Biochem. Biophys. Res. Commun. 2004, 318, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Ohira, T.; Nagasawa, H. Significance of the N- and C-terminal regions of CAP-1, a cuticle calcification-associated peptide from the exoskeleton of the crayfish, for calcification. Peptides 2007, 28, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, H. The molecular mechanism of calcification in aquatic organisms. Biosci. Biotechnol. Biochem. 2013, 77, 1991–1996. [Google Scholar] [CrossRef]
- Boßelmann, F.; Romano, P.; Fabritius, H.; Raabe, D.; Epple, M. The composition of the exoskeleton of two crustacea: The American lobster Homarus americanus and the edible crab Cancer pagurus. Thermochim. Acta 2007, 463, 65–68. [Google Scholar] [CrossRef]
- Zhu, K.Y.; Merzendorfer, H.; Zhang, W.; Zhang, J.; Muthukrishnan, S. Biosynthesis, turnover, and functions of chitin in insects. Annu. Rev. Entomol. 2016, 61, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Westermann, P.; Heumann, W.; Bommer, U.A.; Bielka, H.; Nygard, O.; Hultin, T. Crosslinking of initiation factor eIF-2 to proteins of the small subunit of rat liver ribosomes. FEBS Lett. 1979, 97, 101–104. [Google Scholar] [CrossRef]
- Tolan, D.R.; Hershey, J.W.; Traut, R.T. Crosslinking of eukaryotic initiation factor eIF3 to the 40S ribosomal subunit from rabbit reticulocytes. Biochimie 1983, 65, 427–436. [Google Scholar] [CrossRef]
- Schafer, T.; Maco, B.; Petfalski, E.; Tollervey, D.; Bottcher, B.; Aebi, U.; Hurt, E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 2006, 441, 651–655. [Google Scholar] [CrossRef]
- Mitterer, V.; Murat, G.; Rety, S.; Blaud, M.; Delbos, L.; Stanborough, T.; Bergler, H.; Leulliot, N.; Kressler, D.; Pertschy, B. Sequential domain assembly of ribosomal protein S3 drives 40S subunit maturation. Nat. Commun. 2016, 7, 10336. [Google Scholar] [CrossRef]
- Jang, C.Y.; Lee, J.Y.; Kim, J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004, 560, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.Y.; Kim, H.D.; Kim, J. Ribosomal protein S3 interacts with TRADD to induce apoptosis through caspase dependent JNK activation. Biochem. Biophys. Res. Commun. 2012, 42, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Youn, H.; Lee, S.; Kim, E.; Kim, D.; Lee, S.; Lee, J.M.; Youn, B.H. RNF138-mediated ubiquitination of rpS3 is required for resistance of glioblastoma cells to radiation-induced apoptosis. Exp. Mol. Med. 2018, 50, e434. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, J. Reduction of invasion in human fibrosarcoma cells by ribosomal protein S3 in conjunction with Nm23-H1 and ERK. Biochim. Biophys. Acta 2006, 1763, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Yadavilli, S.; Mayo, L.D.; Higgins, M.; Lain, S.; Hegde, V.; Deutsch, W.A. Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair 2009, 8, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Hegde, V.; Yadavilli, S.; McLaughlin, L.D.; Deutsch, W.A. DNA repair efficiency in transgenic mice over expressing ribosomal protein S3. Mutat. Res. 2009, 666, 16–22. [Google Scholar] [CrossRef][Green Version]
- Kim, Y.; Kim, H.D.; Kim, J. Cytoplasmic ribosomal protein S3 (rpS3) plays a pivotal role in mitochondrial DNA damage surveillance. Biochim. Biophys. Acta 2013, 1833, 2943–2952. [Google Scholar] [CrossRef]
- Wan, F.; Anderson, D.E.; Barnitz, R.A.; Snow, A.; Bidere, N.; Zheng, L.; Hegde, V.; Lam, L.T.; Staudt, L.M.; Levens, D. Ribosomal protein S3: A KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 2007, 131, 927–939. [Google Scholar] [CrossRef]
- Yoon, I.S.; Chung, J.H.; Hahm, S.H.; Park, M.J.; Lee, Y.R.; Ko, S.I.; Kang, L.W.; Kim, T.S.; Kim, J.; Han, Y.S. Ribosomal protein S3 is phosphorylated by Cdk1/cdc2 during G2/M phase. BMB Rep. 2011, 44, 529–534. [Google Scholar] [CrossRef]
- Jang, C.Y.; Kim, H.D.; Zhang, X.; Chang, J.S.; Kim, J. Ribosomal protein S3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem. Biophys. Res. Commun. 2012, 429, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.C.; Zhang, W.Y.; Xiong, Y.W.; Chen, T.Y.; Jiang, S.F.; Qiao, H.; Gong, Y.S.; Wu, Y.; Jin, S.B.; Fu, H.T. RNA interference analysis of the potential functions of cyclin-dependent kinase 2 in sexual reproduction of male oriental river prawns (Macrobrachium nipponense). Aquacult. Int. 2023, 31, 2849–2866. [Google Scholar] [CrossRef]
- Sethi, M.K.; Buettner, F.F.R.; Krylov, V.B.; Takeuchi, H.; Nifantiev, N.E.; Haltiwanger, R.S.; Gerardy-Schahn, R.; Bakker, H. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J. Biol. Chem. 2010, 285, 1582–1586. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.V.; Sethi, M.K.; Leonardi, J.; Rana, N.A.; Buettner, F.F.R.; Haltiwanger, R.S.; Bakker, H.; Jafar-Nejad, H. Negative regulation of notch signaling by xylose. PLoS Genet. 2013, 9, e1003547. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Kankel, M.W.; Artavanis-Tsakonas, S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 2012, 13, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Cytochrome P-450 3A4: Regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Maurel, P. The CYP3 family. In Cytochrome P450 Metabolic and Toxicological Aspects; Ioannides, C., Ed.; CRC Press: Boca Raton, FL, USA, 1996; pp. 241–270. [Google Scholar]
- Chancellor, T.J.; Lee, J.; Thodeti, C.K.; Lele, T. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys. J. 2010, 99, 115–123. [Google Scholar] [CrossRef]
- Zhang, Q.; Bethmann, C.; Worth, N.F.; Davies, J.D.; Wasner, C.; Feuer, A.; Ragnauth, C.D.; Yi, Q.; Mellad, J.A.; Warren, D.T.; et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 2007, 16, 2816–2833. [Google Scholar] [CrossRef]
- Gros-Louis, F.; Dupré, N.; Dion, P.; Fox, M.A.; Laurent, S.; Verreault, S.; Sanes, J.R.; Bouchard, J.P.; Rouleau, G.A. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat. Genet. 2007, 39, 80–85. [Google Scholar] [CrossRef]
- Attali, R.; Warwar, N.; Israel, A.; Gurt, I.; McNally, E.; Puckelwartz, M.; Glick, B.; Nevo, Y.; Ben-Neriah, Z.; Melki, J. Mutation of SYNE-1, encoding an essential component of the nuclear lamina, is responsible for autosomal recessive arthrogryposis. Hum. Mol. Genet. 2009, 18, 3462–3469. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, R.; Thiruchenthooran, V.; Jayadas, T.T.P.; Eswaramohan, T.; Santhirasegaram, S.; Sivabalakrishnan, K.; Naguleswaran, A.; Uzest, M.; Cayrol, B.; Voisin, S.N.; et al. Transcriptomic, proteomic and ultrastructural studies on salinity-tolerant Aedes aegypti in the context of rising sea levels and arboviral disease epidemiology. BMC Genom. 2021, 22, 253. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Gui, W.Y.; Chen, B.; Chen, L. Transcriptome profiling of maternal stress-induced wing dimorphism in pea aphids. Ecol. Evol. 2019, 9, 11848–11862. [Google Scholar] [CrossRef]
- Saavedra-Rodriguez, K.; Campbell, C.L.; Lozano, S.; Penilla-Navarro, P.; Lopez-Solis, A.; Solis-Santoyo, F.; Rodriguez, A.D.; Perera, R.; Black IV, W.C. Permethrin resistance in Aedes aegypti: Genomic variants that confer knockdown resistance, recovery, and death. PLoS Genet. 2021, 17, e1009606. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.O. The cross-links in resilin identified as dityrosine and trityrosine. Biochim. Biophys. Acta 1964, 93, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Gosline, J.; Lillie, M.; Carrington, E.; Guerette, P.; Ortlepp, C.; Savage, K. Elastic proteins: Biological roles and mechanical properties. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002, 357, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Weis-Fogh, T. A rubber-like protein in insect cuticle. J. Exp. Biol. 1960, 37, 887–907. [Google Scholar] [CrossRef]
- Gorb, S.N. Serial elastic elements in the damselfly wing: Mobile vein joints contain resilin. Naturwissenschaften 1999, 86, 552–555. [Google Scholar] [CrossRef]
- Neville, A.C.R.; Rothschild, M. Fleas-insects which fly with their legs. Proc. R. Entomol. Soc. Lond. 1967, 32, 9–10. [Google Scholar]
- Rothschild, M.; Schlein, J. The jumping mechanism of Xenopsylla cheopis. I. Exoskeletal structures and musculature. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1975, 271, 457–490. [Google Scholar]
- Bennet-Clark, H. Resonators in insect sound production: How insects produce loud pure-tone songs. J. Exp. Biol. 1999, 202, 3347–3357. [Google Scholar] [CrossRef]
- Pontremoli, C.; Barbero, N.; Viscardi, G.; Visentin, S. Mucin–drugs interaction: The case of theophylline, prednisolone and cephalexin. Bioorg. Med. Chem. 2015, 23, 6581–6586. [Google Scholar] [CrossRef]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef]
- Longman, R.J.; Poulsom, R.; Corfield, A.P.; Warren, B.F.; Wright, N.A.; Thomas, M.G. Alterations in the composition of the supramucosal defense barrier in relation to disease severity of ulcerative colitis. J. Histochem. Cytochem. 2006, 54, 1335–1348. [Google Scholar] [CrossRef]


| Gene | Accession Number | Species | E-Value | Fold Change (dsPDHE1 vs. dsGFP) | |
|---|---|---|---|---|---|
| Testis | Androgenic Gland | ||||
| Calcification-associated peptide (Cap) | WGF13344.1 | Macrobrachium nipponense | 6.87 × 10−8 | 21.07 | −10.38 |
| Chitin binding-like protein (CBP) | ROT74831.1 | Penaeus vannamei | 3.79 × 10−5 | 3.25 | −2.53 |
| Ribosomal protein S3 (RPS3) | KAF6343595.1 | Pipistrellus kuhlii | 0.00011 | −3.12 | −2.78 |
| Uncharacterized protein LOC113829596 (UP) | XP_027238594.1 | Penaeus vannamei | 4.28 × 10−6 | 6.89 | 6.37 |
| Glucoside xylosyltransferase 2 | XP_045611801.1 | Procambarus clarkii | 8.33 × 10−5 | −21.36 | |
| Skeletal muscle actin 6 | QPC96639.1 | Macrobrachium nipponense | 3.61 × 10−5 | −19.23 | |
| Polycystic kidney disease protein 1 | XP_047488675.1 | Penaeus chinensis | 4.12 × 10−5 | −13.26 | |
| Cytochrome P450 3A30 | XP_037798812.1 | Penaeus monodon | 4.11 × 10−5 | −12.37 | |
| Nesprin-1 | XP_045608233.1 | Procambarus clarkii | 5.18 × 10−5 | −10.39 | |
| Histidine-rich protein DDB_G0274557 | XP_047473847.1 | Penaeus chinensis | 6.77 × 10−9 | −22.65 | |
| Cuticle protein AM1199 | XP_042888222.1 | Penaeus japonicus | 8.87 × 10−9 | −22.47 | |
| Cuticle protein 21 | XP_045617846.1 | Procambarus clarkii | 2.58 × 10−5 | −21.50 | |
| Pro-resilin-like | XP_047479276.1 | Penaeus chinensis | 4.35 × 10−5 | −19.85 | |
| Mucin-19-like | XP_037798457.1 | Penaeus monodon | 0.00012 | −17.23 | |
| Gene | Primer Sequence |
|---|---|
| dsPDHE1-F | TAATACGACTCACTATAGGGGTGCTCTTAGCACTGGAGGC |
| dsPDHE1-R | TAATACGACTCACTATAGGGCCAAGTAGTGGAAGGCAGGA |
| PDHE1-F | TGACCTTAACGGCAACGAGG |
| PDHE1-R | TGACCTTAACGGCAACGAGG |
| CAP-F | TGGAAGCTCGCCGTAGTTTT |
| CAP-R | CTTGACGAAGTGCTGGTTGC |
| CBP-F | CAAGGGCGTCTTCGAGTTCT |
| CBP-R | AAGTTGCCAGGTTCGGGAAT |
| RPS3-F | ATCATGGAGTCTGGCGCAAA |
| RPS3-R | ATACCCAGCACACCTTGACG |
| UP-F | TGTTGGGCGGGAGTTGAAAT |
| UP-R | GTCCTCCTCACCTTGCATCC |
| EIF-F | CATGGATGTACCTGTGGTGAAAC |
| EIF-R | CATGGATGTACCTGTGGTGAAAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, S.; Xiong, Y.; Zhang, W.; Qiao, H.; Wu, Y.; Jiang, S.; Fu, H. Identification of Candidate Male-Reproduction-Related Genes from the Testis and Androgenic Gland of Macrobrachium nipponense, Regulated by PDHE1, through Transcriptome Profiling Analysis. Int. J. Mol. Sci. 2024, 25, 1940. https://doi.org/10.3390/ijms25031940
Jin S, Xiong Y, Zhang W, Qiao H, Wu Y, Jiang S, Fu H. Identification of Candidate Male-Reproduction-Related Genes from the Testis and Androgenic Gland of Macrobrachium nipponense, Regulated by PDHE1, through Transcriptome Profiling Analysis. International Journal of Molecular Sciences. 2024; 25(3):1940. https://doi.org/10.3390/ijms25031940
Chicago/Turabian StyleJin, Shubo, Yiwei Xiong, Wenyi Zhang, Hui Qiao, Yan Wu, Sufei Jiang, and Hongtuo Fu. 2024. "Identification of Candidate Male-Reproduction-Related Genes from the Testis and Androgenic Gland of Macrobrachium nipponense, Regulated by PDHE1, through Transcriptome Profiling Analysis" International Journal of Molecular Sciences 25, no. 3: 1940. https://doi.org/10.3390/ijms25031940
APA StyleJin, S., Xiong, Y., Zhang, W., Qiao, H., Wu, Y., Jiang, S., & Fu, H. (2024). Identification of Candidate Male-Reproduction-Related Genes from the Testis and Androgenic Gland of Macrobrachium nipponense, Regulated by PDHE1, through Transcriptome Profiling Analysis. International Journal of Molecular Sciences, 25(3), 1940. https://doi.org/10.3390/ijms25031940








