1. Introduction
Metal complexes with different ligands can be used in medicine as antitumor and antibacterial drugs. A systematic study of their interactions with nucleic acids for the detection of the molecular mechanism of their biological activity is of the utmost importance in developing new drugs. Indeed, the DNA molecule is the main target of their therapeutic action.
DNA’s interaction with different coordination compounds of metals is among the most current research since the discovery and clinical development of the well-known anticancer drug cisplatin [
1,
2]. However, many aspects of the biological activity of platinum coordination compounds have not yet been identified [
3]. Indeed, the mechanism of action of cisplatin is still not yet fully elucidated [
4]. This demonstrates the relevance of studying the interaction of any complexes of metals with DNA in vitro to understand the molecular basis of their biological activity associated with their effect on DNA in cells.
DNA’s interaction with metal complexes depends on the nature of the metal, as well as on the type and location of ligands in the coordination sphere of ions. The coordination of metal to DNA bases may be accompanied by electrostatic interactions and the non-covalent binding of ligands from the coordination sphere of the central ion with DNA groups. Different ligands provide additional opportunities for the compound to specifically influence DNA’s properties. One of the most promising among the ligands used in coordination chemistry is phenanthroline. It is known that the phenanthroline ligands can enhance the antiproliferative activity of Pd(II) and Pt(II) complexes. Complexes of Pd(II) with phenanthroline have good pharmacokinetic and pharmacodynamic properties that make this class of compounds remarkable inhibitors, even of resistant cell growth [
5]. Simple 1,10-phenanthrolines as excellent N-donor bidentate chelating ligands have been widely used to construct such compounds because of their excellent coordinating ability. They have a large conjugated system and can easily show π-π interactions like stacking [
6,
7,
8] and other types of noncovalent contacts. Phenanthroline, both as a free molecule and as a ligand coordinated to metal centers, can bind with DNA base pairs. The ability of phenanthroline and its derivatives to intercalate into double-stranded DNA has been noted [
9,
10,
11].
The synthesis of different metal complexes with phenanthroline ligands and the study of their binding to the DNA molecule have been carried out [
11,
12,
13,
14,
15,
16,
17]. Some metal ions play an essential role in different biochemical functions. Biogenic ions Zn
2+, Cu
2+, and Mn
2+ are widely used for the synthesis of metal complexes for medicine due to their biocompatibility. Cadmium and its compounds show high toxicity [
18]. It is known that cadmium ions when connected with sulfhydryl groups of enzymes may destroy their functions [
19]. Cd(II) can replace Zn(II) in proteins [
20]. Highly toxic cisplatin shows the most striking results in the treatment of various tumors. Cadmium accumulates in cancer cells, so it can be used in some methods of antitumor therapy. The possible antitumor activity of different compounds is usually tested in vitro by studying their binding to DNA.
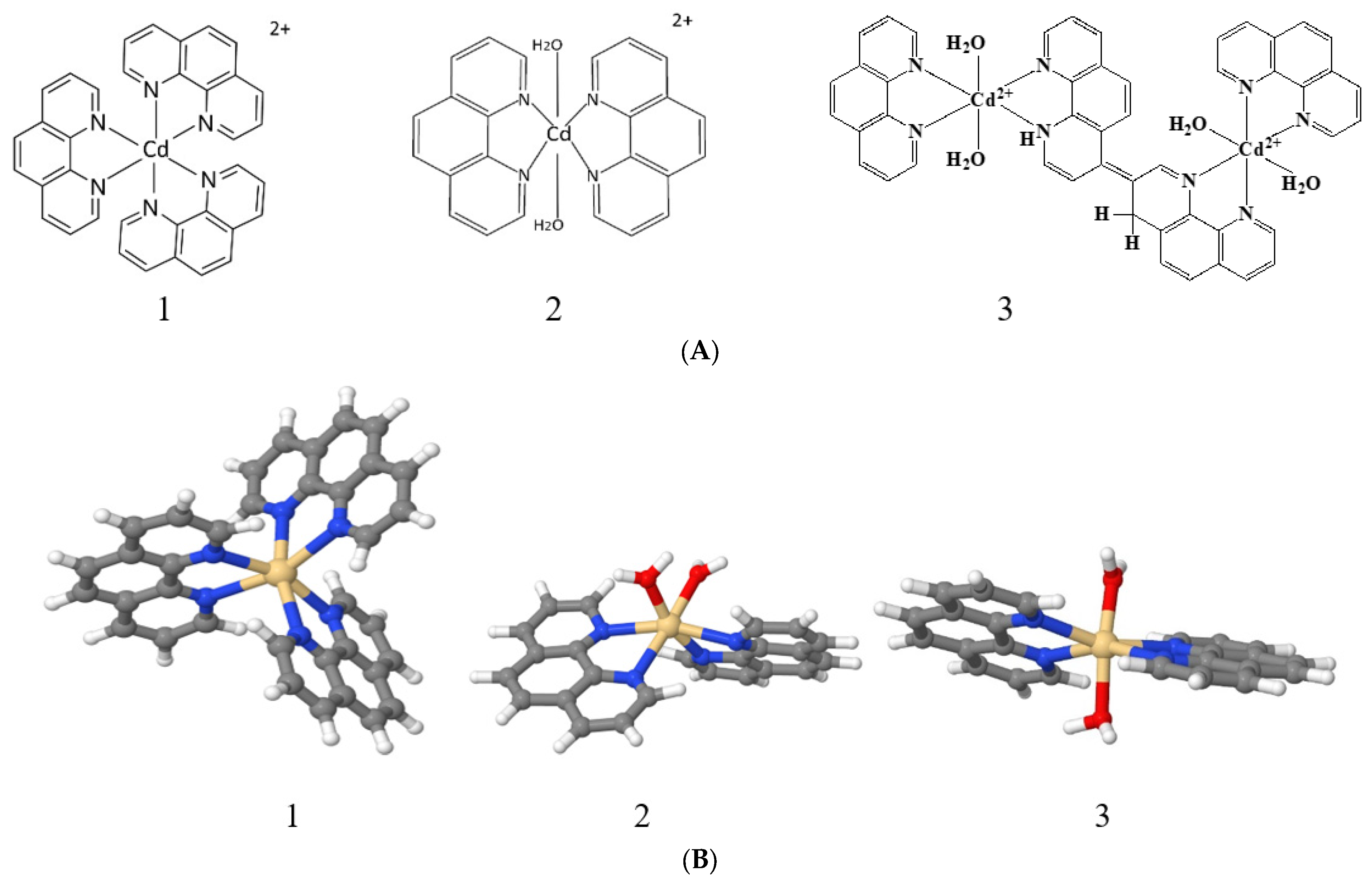
In our research, we examined DNA’s interactions with three cadmium compounds containing 1,10-phenanthroline: [Cd(phen)3](CH3CO2)2, [Cd(phen)2(H2O)2](CH3CO2)2, and [Cd2(phen)4(H2O)2](CH3CO2)4. The possibility of cadmium coordinating with DNA bases, the emergence of the stacking interaction of 1,10-phenanthroline ligands, and the eventuality of 1,10-phenanthroline intercalation between DNA bases during a complex formation were considered. To monitor the state of the secondary and tertiary structure of the DNA molecule during its interaction with cadmium compounds, methods of spectrophotometry, low-gradient viscometry, and dynamic light scattering were used. A preliminary computer simulation of DNA’s interactions with the compounds used was performed.
2. Results and Discussion
All compounds under study are acetates: [Cd(phen)3](CH3CO2)2, denoted as Cd(Phen)3; [Cd(phen)2(H2O)2](CH3CO2)2, denoted as Cd(Phen)2; and [Cd2(phen)4(H2O)2](CH3CO2)4, denoted as 2Cd(Phen)4.
When cadmium compounds Cd(Phen)3, Cd(phen)2, and 2Cd(Phen)4 are dissolved in water, complex ions with a charge of 2+ appear as a result of dissociation. The positive charge ensures the electrostatic attraction of cadmium ions to the negatively charged phosphate groups of DNA. The role of electrostatic interactions in the binding of cadmium compounds to DNA can be revealed by comparing the results of complex formation in solutions of low (0.005 M NaCl) and high (1 M NaCl) ionic strengths. An excess of counterions in 1 M NaCl provides the effective screening of the charge of the macromolecule.
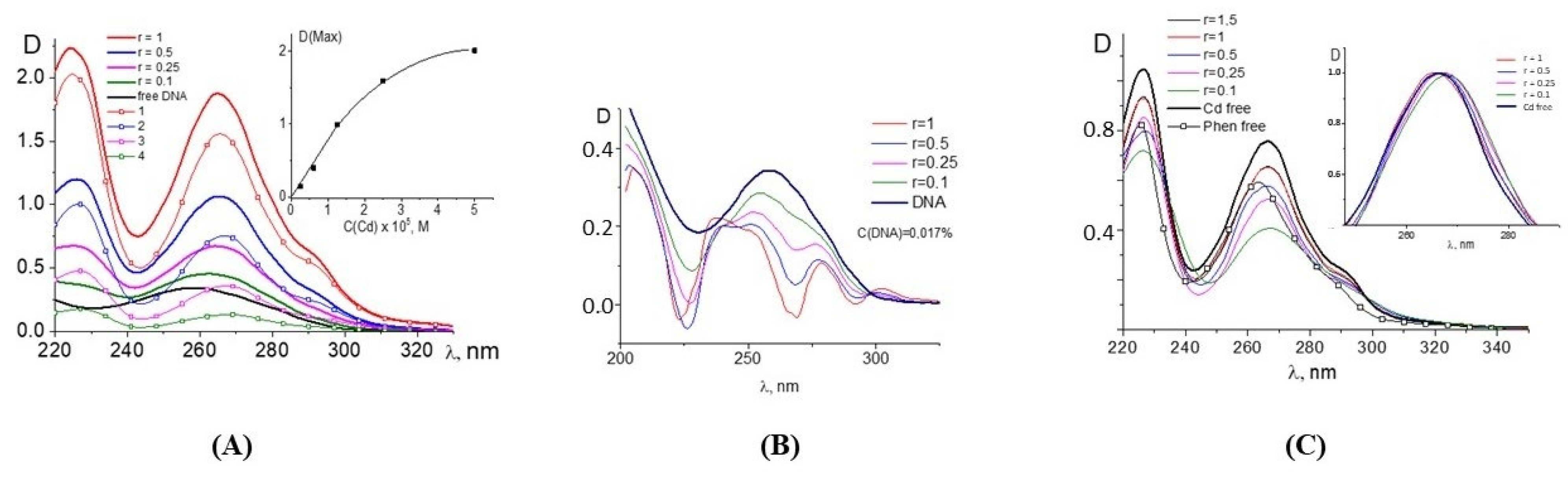
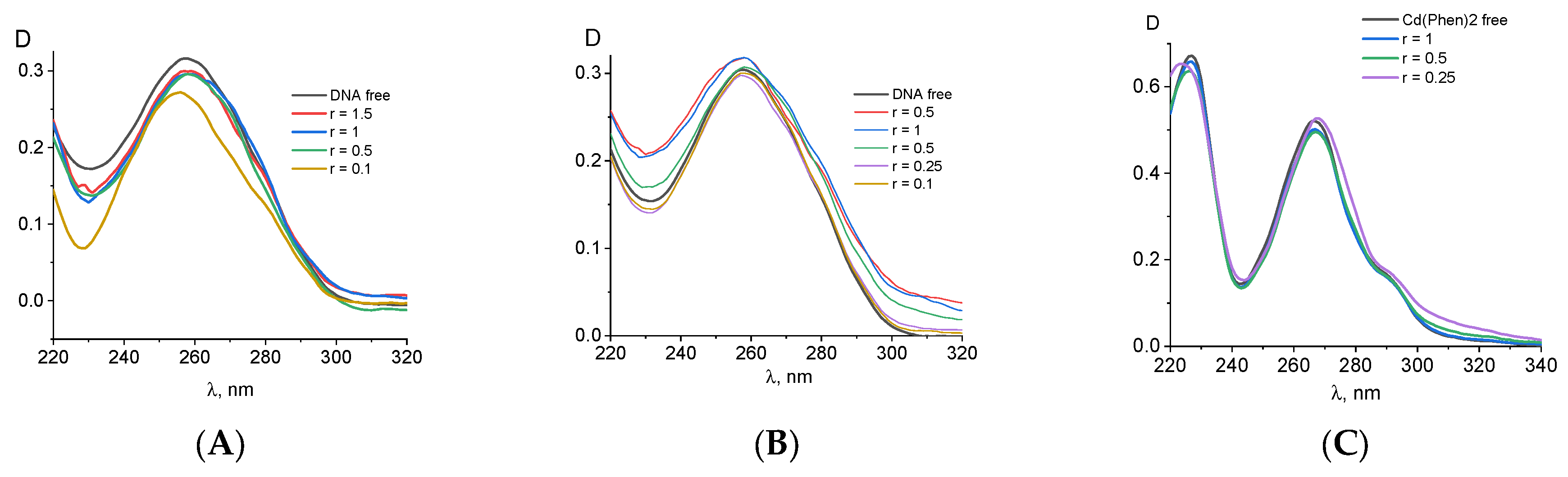
Figure 1 demonstrates the spectral properties of the components during a DNA interaction with Cd(Phen)3 in 0.005 M NaCl. Unfortunately, the absorption spectrum of 1,10-phenanthroline ligands, which are responsible for the spectral properties of the cadmium compound, lies in the same frequency range as the analyzed DNA absorption band. This circumstance makes it difficult to analyze the binding using spectrophotometry. One can see two well-resolved bands with a maximum at 226 nm and 266 nm in absorption spectra of Cd(Phen)3 and one unresolved band in the wavelength region of 280–310 nm. In the spectrum of free 1,10-phenanthroline (
Figure 1C), one can see two bands, shifted to the short-wavelength region relative to the absorption bands of Cd(Phen)3. The unresolved band in the region above 280 nm is practically not expressed.
The linear dependence of the intensity of the middle band of Cd(Phen)3 on the concentration of the compound (
Figure 1A, inset) is disrupted at concentrations of more than 2 × 10
−5 M. This may indicate a hypochromic effect due to the formation of associates with the stacking structures of 1,10-phenanthroline ligands. At lower concentrations, the absence of associates of the compounds can be manifested. Note that the positive charge of the complex ions does not contribute to the formation of associates. Apparently, an increase in the concentration of the compound in solution promotes the formation of such associates when 1,10-phenanthroline ligands of different complex ions form dimer structures. The comparison of the absorption spectrum of the compound Cd(Phen)3, containing three 1,10-phenanthroline ligands, with the absorption spectrum of a 1,10-phenanthroline with the same concentration of chromophores shows a greater hypochromic effect for free 1,10-phenanthrolines than for 1,10-phenanthrolines in the coordination sphere of the compounds. Thus, in solutions of free 1,10-phenanthroline, the formation of dimers occurs more easily than in solutions of Cd(Phen)3.
Figure 1A shows the absorption spectra of DNA complexes with Cd(Phen)3 in 0.005 M NaCl when the DNA solution was added into Cd(Phen)3 solutions of different concentrations. C(DNA) in complexes was 0.0017%. The r value gives the ratio of the Cd concentration in the solution to the DNA base-pair molar concentration. We subtracted the absorbance of free Cd(Phen)3 and calculated the DNA absorption in the complexes (
Figure 1B) with the assumption that the spectral properties of the cadmium compound remained unchanged during the complex formation (which is incorrect). Indeed, the obtained DNA spectra demonstrate an unrealistic shape. Such spectra may be obtained due to a decrease in the absorption of Cd(Phen)3 in the complexes. The hypochromic effect occurs in two bands, and this can produce the observed result.
The results of another experiment with the same concentration of Cd(Phen)3 and different DNA concentrations also clearly demonstrate a hypochromic effect in the spectrum of Cd(Phen)3 upon binding (see calculated spectra of Cd(Phen)3 in complexes with DNA in
Figure 1C). Naturally, in such a calculation, we assume that the absorption of DNA in a complex coincides with that of free DNA. The hypochromic effect is most noticeable in solutions with high DNA concentrations (small r value) and with an excess of binding sites for the compounds on DNA. We believe that, in this case, the appearance of stacking interactions of chromophores in the complexes can be observed. This may be either the result of the intercalation of 1,10-phenanthroline between DNA bases or the formation of a complex when different 1,10-phenanthroline ligands form stacked associates. Note that free 1,10-phenanthroline has another spectral band (see
Figure 1C). A small shift in the absorption band of Cd(Phen)3 with a decreasing r value also indicates the binding and changes in Cd(Phen)3 spectra in the complexes.
The increase in the reduced viscosity of DNA solutions at a constant DNA concentration is observed with the growth of the Cd(Phen)3 concentration (
Figure 2A) at low ionic strength (0.005 M NaCl). This result may be explained by several reasons: the intercalation of the 1,10-phenanthroline ligands of the compound between DNA bases, an increase in the persistent length of DNA during the formation of its complexes with cadmium compounds, and the appearance of intermolecular DNA–DNA aggregates. The relation between the intrinsic viscosity of DNA [η] and the parameters of the macromolecule is given by the Flory formula:
where Ф is a Flory parameter, M is the DNA molecular mass,
L is the hydrodynamic length of the polymer chain, and
A is the length of the Kuhn segment (chain rigidity parameter).
For high molecular DNA samples, A = 2p, where p is the persistent length, <h
2>
1/2 and <h
02>
1/2 are the mean-square distances between the ends of the DNA chain in the real and in the ideal solutions, and α is the coefficient of linear swelling, α = <h
2>
1/2/<h
02>
1/2. The intrinsic viscosity [
η] of DNA is found by extrapolating the reduced viscosity to a zero DNA concentration at zero gradient:
The relative change in the reduced viscosity of solutions at the same DNA concentration may reflect the change in the DNA volume V ~ <h02>3/2 α3 = (2Lp)3/2 α3.
Since the compound Cd(Phen)3 has a positive charge, it is natural to assume its electrostatic attraction to the negatively charged DNA. However, electrostatic binding should cause a drop in the solution viscosity due to the shielding of the DNA charges. The existence of other types of binding must be regarded (this does not exclude the electrostatic interaction). One of the popular models of the binding of metal complexes with 1,10-phenanthroline ligands to DNA is intercalation or the insertion of 1,10-phenqnthroline between the planar bases of deoxyribonucleic acid. For ligands during intercalation, specific changes in their absorption spectra along with an increase in the viscosity of DNA solutions must be observed [
21,
22].
The intercalation of ligands stabilizes the double-stranded structure. DNA intercalators can be cationic or neutral. They cause DNA helix unwinding and helix elongation (increase in
L value). The unwinding of a DNA helix causes the base pairs to separate, creating an opening of about 0.34 nm (3.4 Å). This type of binding causes changes in the spectral properties of ligands, such as DNA-induced hypochromism and a shift of the absorption band. DNA lengthening and unwinding usually are determined from the change in the viscosity of a solution of linear DNA upon the addition of ligands. Hydrodynamic measurements are very sensitive to length changes in DNA. The significant increase in the viscosity of the DNA solution with complexes is obviously due to the increase in the overall DNA contour length
L [
23]. The intercalation binding obeys the rule of nearest-neighbor exclusion [
24]. Indeed, the intercalating ligand can be inserted in such a way that the neighboring DNA base pair does not participate in the intercalation. Hence, we can conclude that the increase in the intrinsic viscosity of DNA due to the elongation of the macromolecule during intercalation at the maximum possible binding without a change in the DNA rigidity according to the Flory formula should not exceed 80%. For the reduced viscosity of DNA solutions, this change may be different.
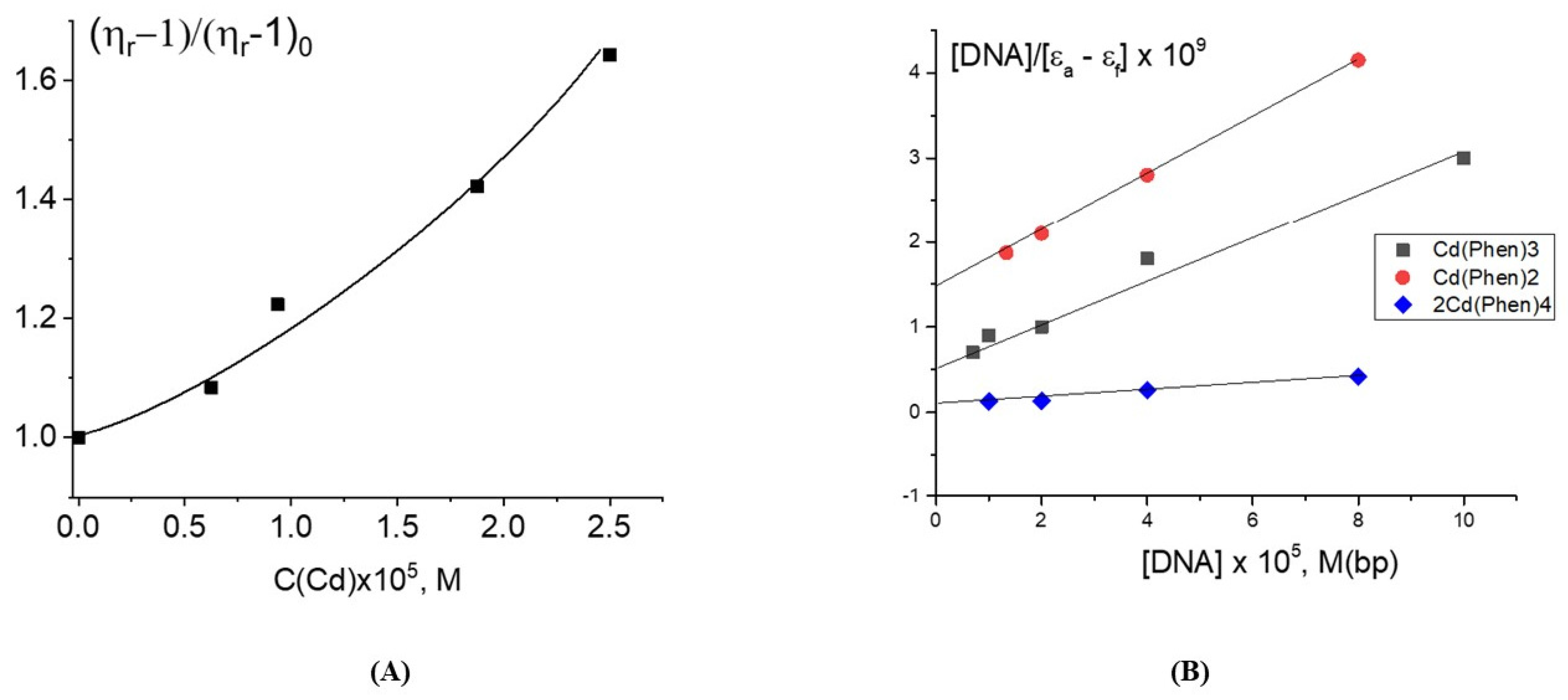
Figure 2A shows a large increase in the DNA solution viscosity with a growth of the Cd(Phen)3 concentration. The intercalation of the 1,10-phenanthroline ligands of the compound Cd(Phen)3 between the bases of the macromolecule is sterically hindered: it is inconvenient due to the specific orientation of 1,10-phenanthrolines in the coordination sphere of the central ion. This becomes possible only as a result of the release of 1,10-phenanthroline from the coordination sphere of the cadmium. At the same time, an increase in the persistent length of DNA during binding also cannot explain the data obtained. Indeed, the increase in viscosity is too high. Based on the data reviewed, we also cannot unambiguously confirm or refute the assumption about intercalation. But the formation of DNA–DNA aggregates can be confirmed experimentally. The viscosity of the DNA solution in this case increases, and its dependence on the Cd(Phen) concentration is non-linear.
Estimation of the binding constant of Cd(Phen)3 in 0.005 M NaCl using the Wolfe-Shimer plot (
Figure 2B) gave K
b = (5.0 ± 1.0) × 10
4 M
−1.
This value indicates the absence of coordination bonds between cadmium and DNA. Possibly as a result of the electrostatic attraction of positively charged complex ions to negatively charged DNA phosphates, a complex formation occurs, which can be classified as external binding. However, the results of the experiment discussed above indicate a change in the spectral properties of the 1,10-phenanthroline ligands of the compound upon the binding. Compensation of the charge of a compound upon binding to DNA can provoke the association of hydrophobic 1,10-phenanthroline ligands, attracting complex ions to each other, including those bound to distant DNA sites or even bound to another DNA molecule. As a result, Cd(Phen)3-Cd(Phen)3 associates with dimers of 1,10-phenanthroline, and the intermolecular DNA–DNA aggregates can be formed. Indeed, for DNA connected to externally bound complex ions containing hydrophobic ligands, the polymer–solvent affinity changes significantly, which promotes the formation of intermolecular contacts.
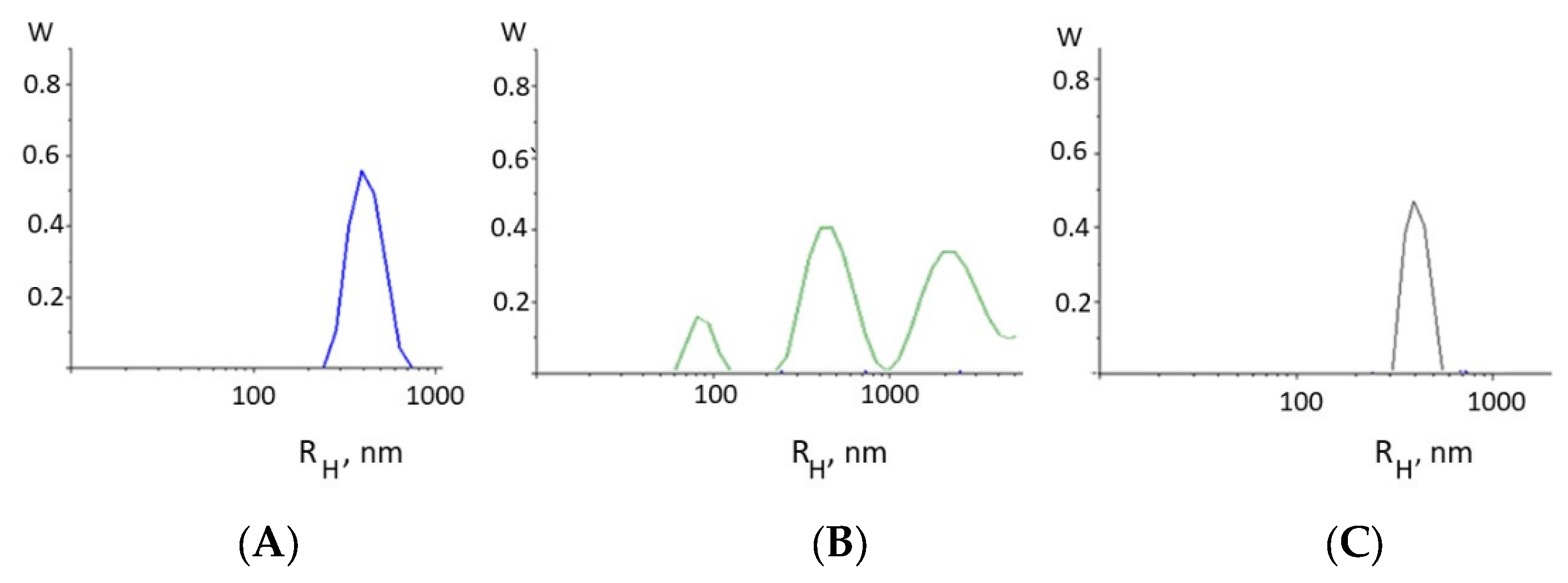
The formation of intermolecular DNA–DNA bonds is evidenced, for example, from the results of the dynamic light scattering method (
Figure 3). Indeed, the appearance of intermolecular DNA–DNA associates in the solution as a result of DNA complexation with Cd(Phen)3 molecules follows from the type of the particle size distribution function obtained from DLS measurements (
Figure 3B). For DNA (
Figure 3A), the results demonstrate the presence in the solution of particles with a hydrodynamic radius of 350 ± 50 nm, which corresponds to the mode responsible for the translational motion of the macromolecule as a whole.
We can estimate the gyration radius of the DNA used with a molecular mass of 11 × 106 g/mol as <Rg2>1/2 = 310 nm. Indeed, the hydrodynamic (contour) length of DNA is L = 5700 nm. The root-mean-square end-to-end distance of the chain in an ideal solution is <h02> = LA, and the radius of gyration is <Rg2> = (1/6) <h02>. However, in 0.005 M NaCl, the size of the macromolecule will increase as a result of polyelectrolyte swelling, but due to the high rigidity of DNA, the linear swelling coefficient in this case is small. In principle, the value of the gyration radius obtained using the DLS method quite adequately reflects the actual size of a molecular coil under this condition.
For a DNA complex with Cd(Phen)3, this mode shifts and corresponds to particles with a hydrodynamic radius of 450 ± 50 nm. This agrees well with the viscometry result. The small mode in this case may be attributed to an association of free cadmium compounds in a solution. The data also indicate the emergence of large particles like intermolecular DNA aggregates in the solution.
The tendency to form aggregates in DNA solutions with Cd(Phen)3 is also evidenced from the results of examining systems in 1 M NaCl, with the excess salt. DNA solutions with a high enough concentration of Cd(Phen)3 in this case are unstable. The turbidity in the solutions indicates a phase separation. No turbidity was observed at low concentrations of the compound in DNA solutions or in solutions without DNA.
The analysis of all experimental data obtained for DNA complexes with Cd(Phen)3 indicates that complexes were formed with the participation of the 1,10-phenanthroline ligands of the compound, which remain part of the cadmium coordination sphere (the spectrum of free 1,10-phenanthroline differs significantly). The hypochromic effect in the absorption spectrum of the compound during its binding to DNA shows the formation of stack-like associates of 1,10-phenanthrolines, although this is sterically inconvenient for Cd(Phen)3. This can occur outside the DNA helix between ligands belonging to two neighboring complex ions bound to the macromolecule. The insertion of 1,10-phenanthroline ligands between heterocyclic DNA bases (intercalation model) is possible but unlikely. The value of the binding constant indicates the absence of a coordination bond between cadmium and DNA during the interaction.
The data obtained for DNA complexes with the compound Cd(Phen)2 with two 1,10-phenanthroline ligands are presented in
Figure 4. Two positions in the coordination sphere of the cadmium ion are occupied by water molecules, which can be replaced by incoming DNA groups. The absorption spectra of DNA complexes with Cd(Phen)2 in 0.005 M NaCl are similar to those observed for DNA-Cd(Phen)3 complexes. The result of their processing also shows the hypochromic effect in spectra of a cadmium compound after its binding to DNA. This is evident both from the drop in absorption for the calculated spectra of the compound in complexes and from the form of the calculated DNA spectra in complexes (
Figure 4A,B). It follows that, upon binding, the spectral properties of phenanthroline ligands change almost identically for both compounds. At the same time, spectral changes in complexes are greater for Cd(Phen)2 compared to Cd(Phen)3 (
Figure 4C). Note that in the absorption spectra of flat ligands that form stacking structures upon external binding to DNA, a hypochromic effect with no shift was observed [
25]. This is typical for complexes of the compound Cd(Phen)3 with DNA.
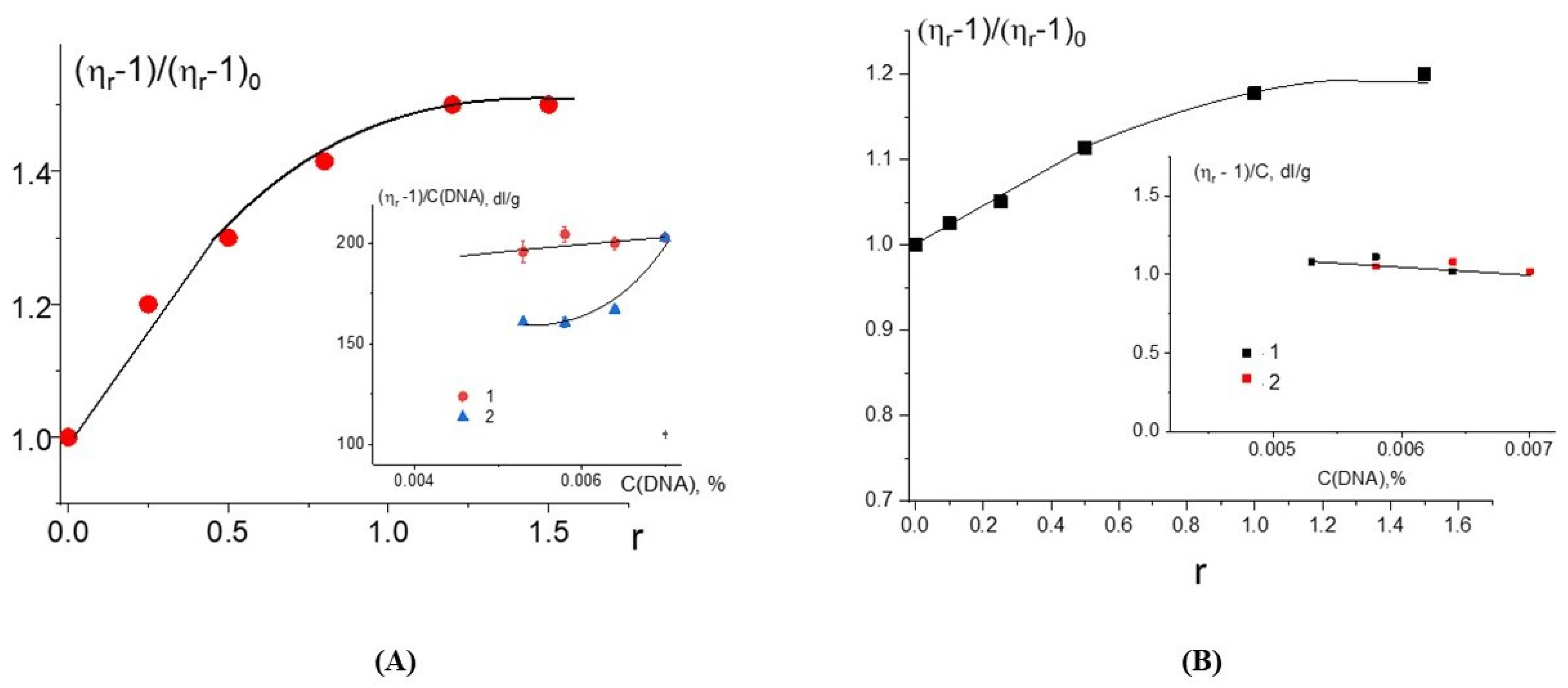
The growth of the relative change in the reduced viscosity of DNA solutions with the increasing r value at a constant DNA concentration for DNA complexes with Cd(Phen)2 (
Figure 5A) may be associated primarily with intercalation. But we cannot exclude another reason (increase in the persistent length of DNA, for example) for the growth of the viscosity. At Cd(Phen)2 concentrations more than 5 × 10
−5 M (at r > 1), the increase in viscosity stops. It is possible that the saturation of one of the types of binding is observed. This to some extent argues against the formation of DNA aggregates in solution, since in this case, it is impossible for the viscosity to remain unchanged with an increasing concentration of the compound that provokes the formation of such aggregates. This conclusion is also supported by the data of the DLS method. We cannot exclude the emergence of intermolecular aggregates, but
Figure 3C did not show the presence of large particles in the DNA solution at the concentration used. Apparently, when binding, the Cd(Phen)2 compound is not located on the surface of the DNA but is located in its grooves.
The assessment of the binding constant for this compound to DNA (
Figure 2B) gives K
b = (3 ± 1) × 10
5 M
−1, which is almost an order of magnitude greater than for Cd(Phen)3. The 1,10-phenanthroline ligands of the compound Cd(Phen)2 in complexes with DNA are in the cadmium-bound state (the absorption spectra of free 1,10-phenanthroline and 1,10-phenanthroline in complexes are shifted to shorter wavelengths). The simulation shows that Cd(Phen)2 has two structures: with cis- and trans-orientations of 1,10-phenanthrolines. The orientation of 1,10-phenanthroline in the coordination sphere of cadmium must be convenient for the intercalation if the complex ion is in a DNA groove due to the electrostatic attraction and good steric conditions.
Whether a coordination of a cadmium ion to DNA is observed for Cd(Phen)2 can be checked via the experiment with different dilutions of a pre-prepared complex (stock solution) of DNA with Cd(Phen)2 in 0.005 M NaCl. This stock solution was kept for 8 h before the study after preparing (mixing together of solutions with DNA and Cd(Phen)2) for the reliable formation of the coordination bonds. This solution was diluted to test the type of concentration dependence either with a NaCl solution (when C(Cd)/C(DNA) = constant) or with a solution of Cd(Phen)2 (in this case, C(Cd) = const). If strong nonequilibrium binding occurs in complexes (coordination bonds), both dilutions will lead to the same dependence of the reduced viscosity on the DNA concentration [
16]. For the case when the equilibrium between the fraction of free and the fraction of bonded compounds changes with the decrease in DNA concentration, these two methods of dilution will lead to different concentration dependencies observed in our experiment (see insert in
Figure 5A). Thus, the compound Cd(Phen)2 does not form a coordination bond with DNA. The intercalation model with the location of a compound in the minor or major groove of DNA between two negatively charged phosphates that neutralize the charge of the cadmium may be offered. It should be emphasized once again that, despite the great similarity in the spectral changes of Cd(Phen)2 and Cd(Phen)3 in complexes with DNA, the difference in binding constants suggests that a stronger binding is observed for Cd(Phen)2. Indeed, in addition to the electrostatic binding, the intercalation of 1,10-phenanthroline occurs.
The third compound was the binuclear cadmium complex 2Cd(Phen)4. The internal coordination sphere of both cadmium ions in this compound is the same as in Cd(Phen)2. The processing of the absorption spectra of DNA complexes with the binuclear compound (
Figure 6) indicates a great similarity with spectral changes observed for DNA-Cd(Phen)2 complexes. The Wolfe-Shimer plot for the binuclear compound gives a binding constant of K
b = (5 ± 1) × 10
5 M
−1. The experiment with two different methods of dilution of the initial DNA-2Cd(Phen)4 complex for the construction of the concentration dependence shows similar results typical for strong nonequilibrium binding (see insert in
Figure 5B). The binding is stronger than observed for the Cd(Phen)2 interaction with DNA. We can assume the coordination binding of a compound to DNA with the intercalation of 1,10-phenanthroline. This is possible when a complex ion penetrates to the major groove of DNA. Then, the cadmium can coordinate with N7 guanine, and 1,10-phenanthroline can intercalate. DNA intercalation can involve the insertion of one intercalating moiety (mono-intercalator) and two intercalating moieties (bis-intercalator). From a comparison of the dependence of the viscosity of DNA solutions on r for compounds Cd(Phen)2 and 2Cd(Phen)4, it follows that only one of the two complex ions of the 2Cd(Phen)4 compound interacts with DNA. In addition, the presence of two independent types of binding (the formation of a coordination bond and the intercalation of 1,10-phenanthroline) can be assumed. The increase in viscosity of DNA solutions with 2Cd(Phen)4 is less than with Cd(Phen)2, but the saturation of binding is observed at the same r value.
The absorption spectra of DNA complexes with three cadmium compounds in 1 M NaCl show approximately the same results. In 1 M NaCl, the interaction of compounds with DNA is not so strong.
Figure 7 shows some results of processing these spectra. The estimation of the binding constants for three compounds in 1 M NaCl gives values around 10
4 M
−1. This result indicates that the electrostatic interactions in DNA binding with compounds in 0.005 M NaCl are very important. In 1 M NaCl, the turbidity and precipitation at a high concentration of cadmium compounds in DNA solutions appeared over time. The solutions of DNA with 2Cd(Phen)4 show relative stability. We can conclude that for DNA complexes with cadmium compounds containing hydrophobic ligands, this solvent with a high salt concentration is thermodynamically poor. These results confirm the influence of hydrophobic ligands on intermolecular interactions in DNA solutions with the cadmium compounds used in 0.005 M NaCl.
Simulation Results
Unlike the experiment results, the simulation shows that Cd(Phen)3 in water (system I) and in 1 M NaCl solution (system II) did not form associates under the conditions used. In a salt solution, they can approach each other at a distance of more than 4 Å (between their hydrogen atoms).
Cd(Phen)2 in the cis-conformation in water (system III) also did not form associates; however, in a 1 M NaCl (system IV), the formation of two dimers among nine complex ions of Cd(Phen)2 was observed. At first, Cd(Phen)2 molecules stick together via 1,10-phenanthroline residue (the residues are parallel). This is an unstable state, which then transforms into a stable state with two 1,10-phenathroline residues opposite each other (
Figure 8A). In this state, there are no water molecules between the Cd(Phen)2. When the salt ions were removed from system IV (system V), both dimers disintegrated. In the manually created system with a higher initial start concentration of Cd(Phen)2 in the form of dimers (system VI) in water for 30 ns, all dimers disintegrated too. So, the simulations show the impossibility of the existence of cis-Cd(Phen)2 dimers in unsalted solutions.
A total of 6 out of 18 dimers disintegrated in 100 ns in the manually created system with Cd(Phen)2 initially in the form of dimers in a solution of 0.15M NaCl (system VII); that is, at such a salt concentration, the existence of both dimers and monomers is possible. In 0.5 M NaCl (system VIII), 2 out of 18 dimers disintegrated in 50 ns, which means that in this salt concentration, Cd(Phen)2 exists predominantly in the form of dimers.
Cd(Phen)2 in the trans-conformation in water (system IX), as well as in the cis-conformation, does not form the dimers. In a 1 M NaCl solution (system X), the stable dimers are not formed, although unstable forms are possible (with an average lifetime no longer than 1 ns) when the trans-Cd(Phen)2 isomers cling together via the 1,10-phenanthroline residues with a parallel orientation (
Figure 8B).
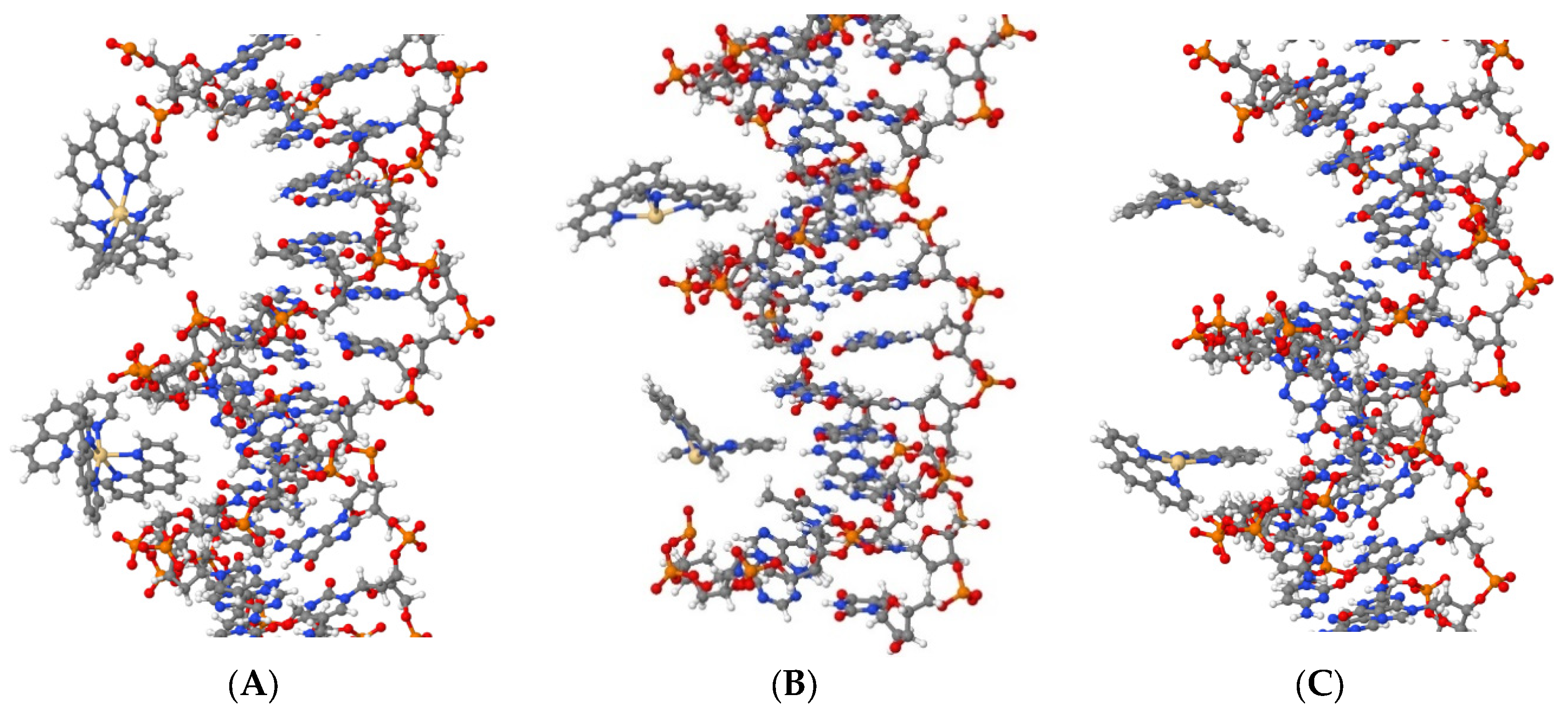
The simulation shows that all three compounds, Cd(Phen)3 (system DI), cis-Cd(Phen)2 (system DII), and trans-Cd(Phen)2 (systems DIII), are attracted to DNA phosphates due to their positive charge. In addition, they can penetrate (fully or partially) into the major and minor grooves of DNA (see
Figure 9). Cd(Phen)3 predominantly enters the major groove, and the penetration into the minor group is partial, i.e., by one 1,10-phenanthroline residue. Cd(Phen)3 cannot enter the minor groove fully due to its large size. Cd(Phen)2 (in cis- and trans-conformations) enters predominantly into the minor groove of DNA. It can be noted that they are attracted to phosphates. Note that the simulation used cannot visualize the intercalation of the 1,10-phenanthroline ligand. At this stage, we can say that the task regarding the DNA interaction with the considered compounds requires a more detailed analysis, which we will carry out in further work.
4. Conclusions
Based on all data obtained, it can be argued that cadmium compounds Cd(Phen)3, Cd(Phen)2, and 2Cd(Phen)4 interact with DNA. There are many similarities in their interactions with DNA. This is especially true for the spectral changes recorded during the formation of complexes. This indicates the similarity in the interactions of the phenanthroline ligands of the compounds with DNA. The hypochromic effect in the absorption spectra of the compounds in the complexes with DNA points to stacking interactions involving 1,10 phenanthroline. This may be both a consequence of intercalation and the result of the formation of phenanthroline dimers upon external binding of compounds to DNA. An external binding is preferable for compound Cd(Phen)3. The location of hydrophobic ligands on the outside of the DNA helix provokes the formation of intermolecular DNA-DNA linkages and the appearance of aggregates in the solution. This is confirmed by DLS data. Without excluding the possibility of external binding of compounds Cd(Phen)2 and 2Cd(Phen)4 to DNA, we still believe that, for these compounds, the main type of binding, in addition to electrostatic, is intercalation. In this case, phenanthroline remains in the coordination sphere of the complex ion.
The possibility of the formation of the dimers of the Cd(Phen)2 compound with the stacking of 1,10-phenanthrolines during the external binding was confirmed by computer simulations for systems containing salt. Therefore, the stacking interaction of phenanthroline ligands is very likely after the compensation of the charge of the complex ions during their binding to DNA phosphates. Thus, the external binding of Cd(Phen)2 to DNA is also possible. However, the termination of the growth of the viscosity of DNA solutions at r > 1 may indicate the filling of possible binding sites on DNA for Cd(Phen)2. This is different from the external binding of Cd(Phen)3 to DNA, which induces the emergence of aggregates. Finally, simulations showed that compound Cd(Phen)3 does not penetrate DNA grooves, unlike compound Cd(Phen)2, which is a necessary step for the Intercalation.
We can assume the formation of a coordination bond between cadmium and DNA only for the complexes of DNA with 2Cd(Phen)4. Indeed, the difference in the binding of compounds to DNA, which appears in viscometry studies, especially in experiments with different dilutions of complexes, indicates that the strong binding with the formation of the coordination bonds is realized only for the binuclear compound. In this case, most likely, the intercalation of 1,10 phenanthroline also occurs. We believe that for the mononuclear compound Cd(Phen)2, the intercalation model is suitable but without coordination of the cadmium ion to DNA. Note that the binding constant can be determined only for equilibrium binding. This definition is incorrect for complexes with coordination bonds. However, most likely, at least two types of binding are realized for a binuclear complex, without considering the electrostatic interaction of cadmium ions with DNA phosphate groups. When a binuclear cadmium compound binds to DNA, only one complex ion participates in the complex formation. It may be that the coordination of the Cd ion to DNA in a major groove is accompanied by the simultaneous insertion of 1,10-phenanthroline between the bases. It is also possible that, on the contrary, coordination binding occurs in the major groove of DNA, and the intercalation of 1,10-phenanthroline also occurs from the minor or major grooves.
At high salt concentrations, the DNA interaction with cadmium compounds gets weaker. Under these conditions, a tendency toward DNA aggregation appears. In DNA solutions the hydrophobic 1,10-phenanthrolines from the coordination sphere of cadmium provoke the formation of intermolecular DNA-DNA bonds.
Computer simulation data confirm the formation of DNA complexes with compounds Cd(Phen)3 and Cd(Phen)2 with the penetration of complex ions into the major (for Cd(Phen)3) and minor (for Cd(Phen)2) grooves of DNA. Further research in this area is required.
The main goal of the research was to determine the molecular mechanism of the DNA interaction with cadmium compounds. A heterocyclic 1,10-phenanthroline exerts in vitro antimicrobial activity against a broad spectrum of bacteria. Metal coordination compounds also exhibit biological activity. The combination of the properties of the components of metal coordination compounds makes it possible to enhance the therapeutic effect. As shown in our study, the location of the ligands in the coordination sphere of the ion determines the interaction of the cadmium compound with DNA. The antimicrobial activity of phenanthroline can be significantly modulated by modifying the structure of the compounds. The cadmium compounds with two 1,10-phenanthroline ligands in the coordination sphere of an ion provide a greater opportunity to form biologically significant complexes via intercalation. At the same time, phenanthroline can manifest itself as an intercalator, and simultaneously, cadmium can coordinate with DNA bases only in binuclear cadmium compounds.