Identification of Yellow Seed Color Genes Using Bulked Segregant RNA Sequencing in Brassica juncea L.
Abstract
1. Introduction
2. Result
2.1. Critical Period of Seed Color Formation
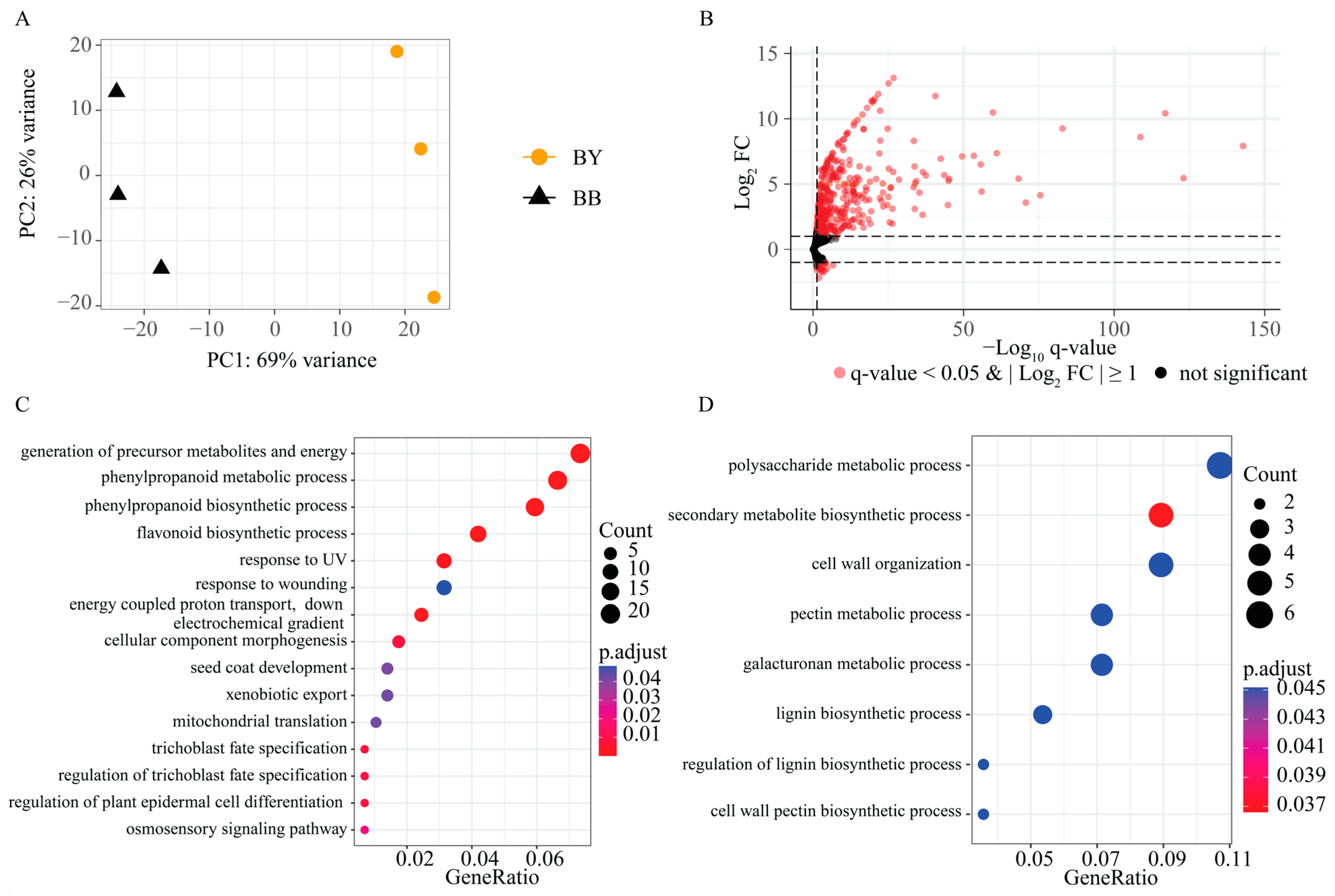
2.2. Alignment of BSR-Seq
2.3. Identification of DEGs between the BY and BB Bulks
2.4. QTL Mapping of Seed Color Using BSR-Seq
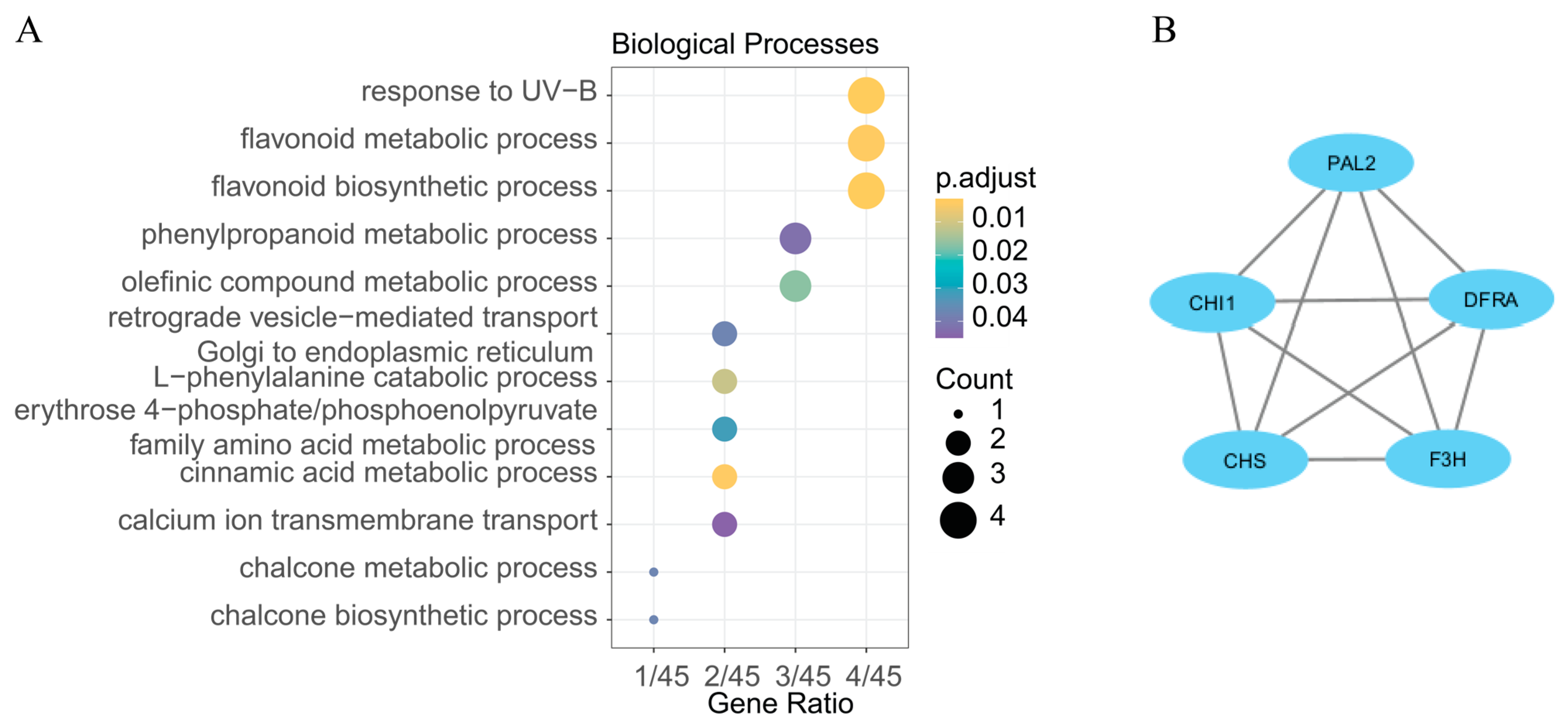
2.5. Overlapping Candidate Intervals and DEGs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Determination of Total Flavonoids, Anthocyanin, Total Phenol, and Melanin Content
4.3. Bulk Construction and Bulked Segregant RNA Sequencing
4.4. Differentially Expressed Gene (DEG) Analysis
4.5. Variant Detection and BSA (Bulk Segregant Analysis) Association Mapping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El-Esawi, M. Taxonomic relationships and biochemical genetic characterization of Brassica resources: Towards a recent platform for germplasm improvement and utilization. Annu. Res. Rev. Biol. 2015, 8, 1–11. [Google Scholar] [CrossRef]
- Wang, X.; Long, Y.; Wang, N.; Zou, J.; Ding, G.; Broadley, M.R.; White, P.J.; Yuan, P.; Zhang, Q.; Luo, Z.; et al. Breeding histories and selection criteria for oilseed rape in Europe and China identified by genome wide pedigree dissection. Sci. Rep. 2017, 7, 1916. [Google Scholar] [CrossRef]
- Nesi, N.; Delourme, R.; Brégeon, M.; Falentin, C.; Renard, M. Genetic and molecular approaches to improve nutritional value of Brassica napus L. Seed. Comptes Rendus Biol. 2008, 331, 763–771. [Google Scholar] [CrossRef]
- Zhang, X.K.; Yang, G.T.; Chen, L.; Yin, J.M.; Tang, Z.L.; Li, J.N. Physiological differences between yellow-seeded and black-seeded rapeseed (Brassica napus L.) with different testa characteristics during artificial ageing. Seed Sci. Technol. 2006, 34, 373–381. [Google Scholar] [CrossRef]
- Qu, C.; Zhu, M.; Hu, R.; Niu, Y.; Chen, S.; Zhao, H.; Li, C.; Wang, Z.; Yin, N.; Sun, F.; et al. Comparative genomic analyses reveal the genetic basis of the yellow-seed trait in Brassica napus. Nat. Commun. 2023, 14, 5194. [Google Scholar] [CrossRef]
- Rahman, M.H. Production of yellow-seeded Brassica napus through interspecific crosses. Plant Breed 2001, 120, 463–472. [Google Scholar] [CrossRef]
- Akhov, L.; Ashe, P.; Tan, Y.; Datla, R.; Selvaraj, G. Proanthocyanidin biosynthesis in the seed coat of yellow-seeded, canola quality Brassica napus YN01-429 is constrained at the committed step catalyzed by dihydroflavonol 4-reductase. Botany 2009, 87, 616–625. [Google Scholar] [CrossRef]
- Marles, M.S.; Gruber, M.Y. Histochemical characterisation of unextractable seed coat pigments and quantification of extractable lignin in the Brassicaceae. J. Sci. Food Agric. 2004, 84, 251–262. [Google Scholar] [CrossRef]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef]
- Yang, S.C.; Arasu, M.V.; Chun, J.H.; Jang, Y.S.; Lee, Y.H.; Kim, I.H.; Lee, K.T.; Hong, S.T.; Kim, S.J. Identification and determination of phenolic compounds in rapeseed meals (Brassica napus L.). J. Agric. Chem. Environ. 2015, 4, 14–23. [Google Scholar] [CrossRef]
- Marles, M.A.S.; Gruber, M.Y.; Scoles, G.J.; Muir, A.D. Pigmentation in the developing seed coat and seedling leaves of Brassica Carinata is controlled at the dihydroflavonol reductase locus. Phytochemistry 2003, 62, 663–672. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biolog. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Bradley, J.M.; Davies, K.M.; Deroles, S.C.; Bloor, S.J.; Lewis, D.H. The maize Lc regulatory gene up-regulates the flavonoid biosynthetic pathway of petunia. Plant J. 1998, 13, 381–392. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic Diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Glagoleva, A.Y.; Shoeva, O.Y.; Khlestkina, E.K. Melanin pigment in plants: Current knowledge and future perspectives. Front. Plant Sci. 2020, 11, 770. [Google Scholar] [CrossRef]
- Badani, A.G.; Snowdon, R.J.; Wittkop, B.; Lipsa, F.D.; Baetzel, R.; Horn, R.; De Haro, A.; Font, R.; Lühs, W.; Friedt, W. Colocalization of a partially dominant gene for yellow seed colour with a major QTL Influencing acid detergent fibre (ADF) content in different crosses of oilseed rape (Brassica napus). Genome 2006, 49, 1499–1509. [Google Scholar] [CrossRef]
- Liu, Z.W.; Fu, T.D.; Tu, J.X.; Chen, B.Y. Inheritance of seed colour and identification of RAPD and AFLP markers linked to the seed colour gene in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2005, 110, 303–310. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Chen, W.; Yi, B.; Wen, J.; Shen, J.X.; Ma, C.Z.; Chen, B.Y.; Tu, J.X.; Fu, T.D. Identification of two major QTL for yellow seed color in two crosses of resynthesized Brassica napus line No. 2127-17. Mol. Breeding 2011, 28, 335–342. [Google Scholar] [CrossRef]
- Stein, A.; Wittkop, B.; Liu, L.; Obermeier, C.; Friedt, W.; Snowdon, R.J. Dissection of a major QTL for seed colour and fibre content in Brassica napus reveals colocalization with candidate genes for phenylpropanoid biosynthesis and flavonoid deposition. Plant Breed 2013, 132, 382–389. [Google Scholar] [CrossRef]
- Chao, H.; Guo, L.; Zhao, W.; Li, H.; Li, M. A Major yellow-seed QTL on chromosome A09 significantly increases the oil content and reduces the fiber content of seed in Brassica napus. Theor. Appl. Genet. 2022, 135, 1293–1305. [Google Scholar] [CrossRef]
- Qu, C.; Fu, F.; Lu, K.; Zhang, K.; Wang, R.; Xu, X.; Wang, M.; Lu, J.; Wan, H.; Zhanglin, T.; et al. Differential accumulation of phenolic compounds and expression of related genes in black- and yellow-seeded Brassica napus. J. Exp. Bot. 2013, 64, 2885–2898. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Qian, L.; Zheng, M.; Chen, L.; Chen, H.; Yang, L.; You, L.; Yang, B.; Yan, M.; Gu, Y.; et al. Genomic insights into the origin, domestication and diversification of Brassica Juncea. Nat. Genet. 2021, 53, 1392–1402. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Ban, Y.; Yang, L.; Zhang, Y.; Li, H.; Xiao, E.; Xu, A.; Zhang, D. Fine mapping of the yellow seed locus in Brassica Juncea L. Genome 2012, 55, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, L.; Hong, M.; Zhang, Y.; Zu, F.; Wen, J.; Yi, B.; Ma, C.; Shen, J.; Tu, J.; et al. A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PLoS ONE 2012, 7, e44145. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, J.; Li, Z.; Li, X.; He, Z.; Zhang, L.; Sha, T.; Lyu, X.; Chen, S.; Gu, Y.; et al. Genomic signatures of vegetable and oilseed allopolyploid Brassica Juncea and genetic loci controlling the accumulation of glucosinolates. Plant. Biotech. J. 2021, 19, 2619–2628. [Google Scholar] [CrossRef]
- Wen, J.; Zhu, L.; Qi, L.; Ke, H.; Yi, B.; Shen, J.; Tu, J.; Ma, C.; Fu, T. Characterization of interploid hybrids from crosses between Brassica Juncea and B. Oleracea and the production of yellow-seeded B. napus. Theor. Appl. Genet. 2012, 125, 19–32. [Google Scholar] [CrossRef]
- Rashid, A.; Rakow, G.; Downey, R.K. Development of yellow seeded Brassica napus through interspecific crosses. Plant Breeding 1994, 112, 127–134. [Google Scholar] [CrossRef]
- Flores, P.C.; Yoon, J.S.; Kim, D.Y.; Seo, Y.W. Transcriptome analysis of MYB genes and patterns of anthocyanin accumulation during seed development in wheat. Evol. Bioinform. Online 2022, 18, 117693432210933. [Google Scholar] [CrossRef]
- Lu, N.; Jun, J.H.; Li, Y.; Dixon, R.A. An unconventional proanthocyanidin pathway in maize. Nat. Commun. 2023, 14, 4349. [Google Scholar] [CrossRef]
- Shen, S.; Tang, Y.; Zhang, C.; Yin, N.; Mao, Y.; Sun, F.; Chen, S.; Hu, R.; Liu, X.; Shang, G.; et al. Metabolite profiling and transcriptome analysis provide insight into seed coat color in Brassica juncea. Int. J. Mol. Sci. 2021, 22, 7215. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; Yang, W.; Xie, T.; Wang, Y.; Wang, X.; Jiang, J.; Wang, Y. Transcriptional profiling between yellow- and black-seeded Brassica napus reveals molecular modulations on flavonoid and fatty acid content. J. Int. Agric. 2022, 21, 2211–2226. [Google Scholar] [CrossRef]
- Jiang, J.; Zhu, S.; Yuan, Y.; Wang, Y.; Zeng, L.; Batley, J.; Wang, Y.P. Transcriptomic comparison between developing seeds of yellow- and black-seeded Brassica napus reveals that genes influence seed quality. BMC Plant Biol. 2019, 19, 203. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.Y.; Liu, L.Z.; Chai, Y.-R.; Chen, L.; Yang, T.; Jin, M.Y.; Ma, A.F.; Yan, X.Y.; Zhang, Z.S.; Li, J.N. Localization of QTLs for seed color using recombinant inbred lines of Brassica napus in different environments. Genome 2007, 50, 840–854. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xian, X.; Xu, X.; Qu, C.; Lu, K.; Li, J.; Liu, L. Genome-wide association mapping of seed coat color in Brassica napus. J. Agric. Food Chem. 2017, 65, 5229–5237. [Google Scholar] [CrossRef]
- Wang, F.; He, J.; Shi, J.; Zheng, T.; Xu, F.; Wu, G.; Liu, R.; Liu, S. embryonal control of yellow seed coat locus ECY1 is related to alanine and phenylalanine metabolism in the seed embryo of Brassica napus. G3 Genes Genom. Genet. 2016, 6, 1073–1081. [Google Scholar] [CrossRef]
- Lian, J.; Lu, X.; Yin, N.; Ma, L.; Lu, J.; Liu, X.; Li, J.; Lu, J.; Lei, B.; Wang, R.; et al. Silencing of BnTT1 family genes affects seed flavonoid biosynthesis and alters seed fatty acid composition in Brassica napus. Plant Sci. 2017, 254, 32–47. [Google Scholar] [CrossRef]
- Li, H.; Yu, K.; Zhang, Z.; Yu, Y.; Wan, J.; He, H.; Fan, C. Targeted mutagenesis of flavonoid biosynthesis pathway genes reveals functional divergence in seed coat colour, oil content and fatty acid composition in Brassica napus L. Plant Biotech. J. 2023, in press. [Google Scholar] [CrossRef]
- Zhai, Y.; Yu, K.; Cai, S.; Hu, L.; Amoo, O.; Xu, L.; Yang, Y.; Ma, B.; Jiao, Y.; Zhang, C.; et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotech. J. 2020, 18, 1153–1168. [Google Scholar] [CrossRef]
- Chai, Y.R.; Lei, B.; Huang, H.L.; Li, J.N.; Yin, J.M.; Tang, Z.L.; Wang, R.; Chen, L. TRANSPARENT TESTA12 genes from Brassica napus and parental species: Cloning, evolution, and differential involvement in yellow seed Trait. Mol. Genet. Genom. 2009, 281, 109–123. [Google Scholar] [CrossRef]
- Huang, Z.; Peng, G.; Liu, X.; Deora, A.; Falk, K.C.; Gossen, B.D.; McDonald, M.R.; Yu, F. Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front. Plant Sci. 2017, 8, 1448. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Li, Z.; Liu, Z.; Guo, Y.; Qiu, L.J. Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front. Plant Sci. 2017, 8, 919. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xie, J.; Zhang, H.; Guo, B.; Ning, S.; Chen, Y.; Lu, P.; Wu, Q.; Li, M.; Zhang, D.; et al. Mapping stripe rust resistance gene YrZH22 in Chinese wheat cultivar Zhoumai 22 by bulked segregant RNA-Seq (BSR-Seq) and comparative genomics analyses. Theor. Appl. Genet. 2017, 130, 2191–2201. [Google Scholar] [CrossRef] [PubMed]
- Quarrie, S.A.; Lazic-Jancic, V.; Kovacevic, D.; Steed, A.; Pekic, S. Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. J. Exp. Bot. 1999, 50, 1299–1306. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Pirie, A.; Mullins, M.G. Changes in anthocyanin and phenolics content of grapevine leaf and fruit tissues treated with sucrose, nitrate, and abscisic acid. Plant Physiol. 1976, 58, 468–472. [Google Scholar] [CrossRef]
- Stavenga, D.G.; Leertouwer, H.L.; Dudek, B.; Van Der Kooi, C.J. Coloration of flowers by flavonoids and consequences of pH dependent absorption. Front. Plant Sci. 2021, 11, 600124. [Google Scholar] [CrossRef]
- Mansfeld, B.N.; Grumet, R. QTLseqr: An R package for bulk segregant analysis with next-generation sequencing. Plant Genom. 2018, 11, 180006. [Google Scholar] [CrossRef]
- Magwene, P.M.; Willis, J.H.; Kelly, J.K. The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput. Biol. 2011, 7, e1002255. [Google Scholar] [CrossRef]



| Flavonoids (OD Unit 325 nm/g DW) | Anthocyanins (OD Unit 530 nm/g DW) | Melanin (OD Unit 290 nm/g DW) | Total Phenolics (mg/g DW) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wugong | Wuqi | Difference | Wugong | Wuqi | Difference | Wugong | Wuqi | Difference | Wugong | Wuqi | Difference | |
| 9 DAP | 121.47 | 127.85 | −6.38 | 238.20 | 69.17 | 169.03 ** | 26.68 | 17.60 | 9.07 ** | 230.20 | 120.60 | 109.60 ** |
| 16 DAP | 109.51 | 126.12 | −16.61 | 726.75 | 75.33 | 651.42 ** | 59.89 | 48.22 | 11.66 ** | 319.30 | 180.90 | 138.40 ** |
| 23 DAP | 523.20 | 337.39 | 185.81 ** | 2217.00 | 182.80 | 2034.20 ** | 109.66 | 87.50 | 22.15 ** | 793.30 | 255.60 | 537.70 ** |
| 30 DAP | 887.38 | 770.32 | 117.07 ** | 2337.33 | 247.83 | 2089.50 ** | 224.68 | 136.51 | 88.16 ** | 1070.40 | 544.00 | 526.40 ** |
| 38 DAP | 1926.05 | 1400.00 | 526.05 ** | 1256.34 | 305.43 | 950.91 ** | 397.66 | 195.95 | 201.71 ** | 1157.40 | 655.50 | 501.90 ** |
| 45 DAP | 2035.07 | 1778.57 | 256.50 ** | 1195.00 | 300.08 | 894.92 ** | 698.76 | 203.55 | 495.21 ** | 1229.70 | 868.00 | 601.70 ** |
| QTL | Chrom | Start | End | FPKM in BB | FPKM in BY | Gene ID | At_ID |
|---|---|---|---|---|---|---|---|
| qSCA09-3 | A09 | 8,929,403 | 8,931,985 | 914.09 | 33.40 | BjuOA09G16530 | AT1G62750 |
| qSCA09-3 | A09 | 9,495,191 | 9,500,364 | 14.39 | 1.43 | BjuOA09G17120 | AT1G62120 |
| qSCA09-3 | A09 | 11,771,003 | 11,772,142 | 182.41 | 19.20 | BjuOA09G20180 | AT2G03750 |
| qSCA09-3 | A09 | 11,913,894 | 11,916,103 | 224.83 | 36.06 | BjuOA09G20410 | AT5G42370 |
| qSCA09-3 | A09 | 13,787,173 | 13,788,148 | 54.53 | 125.96 | BjuOA09G22750 | AT1G26470 |
| qSCA09-3 | A09 | 14,094,583 | 14,098,369 | 51.18 | 7.05 | BjuOA09G23160 | AT5G46690 |
| qSCA09-3 | A09 | 14,113,829 | 14,115,475 | 307.67 | 145.43 | BjuOA09G23180 | AT5G46700 |
| qSCA09-3 | A09 | 14,279,969 | 14,280,275 | 283.29 | 103.14 | BjuOA09G23340 | AT5G46871 |
| qSCA09-3 | A09 | 14,415,615 | 14,416,772 | 98.94 | 288.38 | BjuOA09G23480 | AT5G47060 |
| qSCA09-3 | A09 | 14,667,647 | 14,669,807 | 67.84 | 165.38 | BjuOA09G23850 | AT5G47650 |
| qSCA09-3 | A09 | 15,097,228 | 15,100,123 | 13.20 | 0.00 | BjuOA09G24380 | AT1G62200 |
| qSCA09-3 | A09 | 16,516,247 | 16,518,639 | 28.12 | 1.67 | BjuOA09G26270 | AT4G04710 |
| qSCA09-3 | A09 | 16,848,640 | 16,849,093 | 52.31 | 110.67 | BjuOA09G26840 | AT1G64590 |
| qSCA09-3 | A09 | 16,951,516 | 16,952,504 | 44.29 | 108.10 | BjuOA09G27000 | AT4G12580 |
| qSCA09-3 | A09 | 12,155,595 | 12,157,288 | 5727.33 | 3.97 | BjuOA09G20700 | AT5G42800 |
| qSCA09-3 | A09 | 12,694,439 | 12,698,035 | 44.80 | 1.13 | BjuOA09G21470 | AT1G51540 |
| qSCA09-3 | A09 | 13,580,386 | 13,583,345 | 36.50 | 87.50 | BjuOA09G22510 | AT1G52360 |
| qSCA09-7 | A09 | 38,731,200 | 38,733,688 | 158.05 | 0.26 | BjuOA09G35390 | At1G32150 |
| qSCA09-7 | A09 | 38,797,903 | 38,800,210 | 42.41 | 10.28 | BjuOA09G35460 | AT1G32080 |
| qSCA09-7 | A09 | 38,972,655 | 38,973,252 | 40.24 | 90.11 | BjuOA09G35740 | AT2G37420 |
| qSCA09-7 | A09 | 39,070,816 | 39,072,813 | 1244.90 | 551.60 | BjuOA09G35800 | AT1G31830 |
| qSCA09-7 | A09 | 39,216,208 | 39,218,452 | 54.76 | 117.49 | BjuOA09G35960 | AT1G31660 |
| qSCA09-7 | A09 | 40,466,891 | 40,467,856 | 14.78 | 51.11 | BjuOA09G37390 | AT5G26770 |
| qSCA09-7 | A09 | 40,486,136 | 40,487,627 | 58.47 | 139.95 | BjuOA09G37430 | AT1G73490 |
| qSCA09-7 | A09 | 40,724,953 | 40,725,511 | 33.64 | 82.22 | BjuOA09G37660 | AT1G33390 |
| qSCA09-7 | A09 | 42,041,441 | 42,044,539 | 60.86 | 0.00 | BjuOA09G39430 | AT1G28020 |
| qSCA09-7 | A09 | 42,288,652 | 42,290,857 | 18.15 | 0.00 | BjuOA09G39810 | AT1G27500 |
| qSCA09-7 | A09 | 42,429,480 | 42,430,621 | 10.62 | 50.20 | BjuOA09G40150 | AT1G63660 |
| qSCA09-7 | A09 | 42,537,814 | 42,540,009 | 27.12 | 93.94 | BjuOA09G40310 | AT5G18880 |
| qSCA09-7 | A09 | 42,765,028 | 42,767,784 | 249.60 | 74.33 | BjuOA09G40600 | AT1G25390 |
| qSCA09-7 | A09 | 42,796,085 | 42,797,338 | 52.17 | 2.46 | BjuOA09G40630 | AT3G20620 |
| qSCA09-7 | A09 | 42,873,467 | 42,875,000 | 29.26 | 1.41 | BjuOA09G40750 | AT1G14800 |
| qSCA09-7 | A09 | 42,894,662 | 42,895,078 | 18.93 | 0.85 | BjuOA09G40790 | AT1G21380 |
| qSCA09-7 | A09 | 42,895,126 | 42,896,198 | 12.29 | 0.28 | BjuOA09G40800 | AT1G14800 |
| qSCA09-7 | A09 | 42,915,568 | 42,917,540 | 78.09 | 32.19 | BjuOA09G40860 | AT1G25520 |
| qSCA09-7 | A09 | 43,276,364 | 43,278,604 | 34.41 | 3.97 | BjuOA09G41440 | AT1G26680 |
| qSCA09-7 | A09 | 43,368,606 | 43,369,235 | 26.12 | 1.14 | BjuOA09G41640 | AT5G13930 |
| qSCA09-7 | A09 | 43,401,376 | 43,402,160 | 69.82 | 12.92 | BjuOA09G41660 | AT1G26920 |
| qSCA09-7 | A09 | 44,111,932 | 44,114,261 | 184.46 | 646.93 | BjuOA09G42790 | AT1G23200 |
| qSCA09-7 | A09 | 44,403,018 | 44,404,800 | 16.43 | 0.86 | BjuOA09G43350 | AT1G22620 |
| qSCA09-7 | A09 | 44,436,051 | 44,436,732 | 29.02 | 69.92 | BjuOA09G43370 | AT1G22590 |
| qSCA09-7 | A09 | 44,589,631 | 44,591,503 | 81.63 | 18.31 | BjuOA09G43580 | AT1G22280 |
| qSCA09-7 | A09 | 45,670,071 | 45,671,807 | 2727.30 | 475.40 | BjuOA09G45170 | AT3G51240 |
| qSCA09-7 | A09 | 45,707,274 | 45,708,740 | 520.50 | 1235.15 | BjuOA09G45260 | AT3G51300 |
| qSCA09-7 | A09 | 46,016,674 | 46,017,162 | 20.52 | 1.07 | BjuOA09G45830 | AT1G79990 |
| qSCA09-7 | A09 | 46,019,207 | 46,019,883 | 200.76 | 46.18 | BjuOA09G45840 | AT5G38980 |
| qSCA09-7 | A09 | 46,109,849 | 46,111,124 | 38.86 | 7.12 | BjuOA09G45990 | AT3G51930 |
| qSCA09-7 | A09 | 46,507,649 | 46,510,728 | 24.76 | 65.24 | BjuOA09G46830 | AT1G72880 |
| qSCA09-7 | A09 | 46,669,636 | 46,671,877 | 90.67 | 193.86 | BjuOA09G47060 | AT2G23790 |
| qSCA09-7 | A09 | 46,798,481 | 46,801,460 | 2511.67 | 870.15 | BjuOA09G47290 | AT3G53260 |
| qSCA09-7 | A09 | 47,714,123 | 47,726,747 | 37.96 | 0.28 | BjuOA09G49050 | AT3G55120 |
| SNP between BB and BB Bulks | SNP between “AU213” and “T84-66” (V2) | |||||||
|---|---|---|---|---|---|---|---|---|
| CHROM | Position | Ref | Alt | Annotation | AU213 Position | Base in AU213 | Base in T84-66 | Gene ID |
| A09 | 46,798,684 | T | C | 3_prime_UTR_variant | 46,798,684 | T | C | BjuOA09G47290 |
| A09 | 46,799,098 | G | A | synonymous_variant | 46,799,098 | G | A | BjuOA09G47290 |
| A09 | 46,799,171 | T | C | missense_variant | 46,799,171 | T | C | BjuOA09G47290 |
| A09 | 46,799,228 | G | C | missense_variant | 46,799,228 | G | C | BjuOA09G47290 |
| A09 | 46,799,260 | G | A | synonymous_variant | 46,799,260 | G | A | BjuOA09G47290 |
| A09 | 46,799,300 | T | C | missense_variant | 46,799,300 | T | C | BjuOA09G47290 |
| A09 | 46,799,477 | A | T | missense_variant | 46,799,477 | A | T | BjuOA09G47290 |
| A09 | 46,799,528 | A | C | missense_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,799,543 | A | G | missense_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,799,606 | A | G | missense_variant | 46,799,606 | A | G | BjuOA09G47290 |
| A09 | 46,799,690 | G | C | missense_variant | 46,799,690 | G | C | BjuOA09G47290 |
| A09 | 46,799,861 | T | C | missense_variant | 46,799,861 | T | C | BjuOA09G47290 |
| A09 | 46,799,897 | C | T | missense_variant | 46,799,897 | C | T | BjuOA09G47290 |
| A09 | 46,799,987 | C | A | missense_variant | 46,799,987 | C | A | BjuOA09G47290 |
| A09 | 46,800,041 | G | T | missense_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,800,056 | C | T | stop_gained | - | - | - | BjuOA09G47290 |
| A09 | 46,800,095 | G | C | missense_variant | 46,800,095 | G | C | BjuOA09G47290 |
| A09 | 46,800,143 | C | T | missense_variant | 46,800,143 | C | T | BjuOA09G47290 |
| A09 | 46,800,194 | T | C | missense_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,800,197 | C | T | missense_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,800,242 | T | C | missense_variant | 46,800,242 | T | C | BjuOA09G47290 |
| A09 | - | - | - | - | 46,800,326 | T | C | BjuOA09G47290 |
| A09 | 46,800,374 | A | G | missense_variant | 46,800,374 | A | G | BjuOA09G47290 |
| A09 | 46,800,401 | G | A | missense_variant | 46,800,401 | G | A | BjuOA09G47290 |
| A09 | - | - | - | - | 46,800,511 | A | G | BjuOA09G47290 |
| A09 | - | - | - | - | 46,800,762 | C | G | BjuOA09G47290 |
| A09 | 46,801,226 | A | G | synonymous_variant | 46,801,226 | A | G | BjuOA09G47290 |
| A09 | 46,801,270 | C | T | missense_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,801,286 | G | A | 5_prime_UTR_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,801,307 | G | A | 5_prime_UTR_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,801,337 | A | T | 5_prime_UTR_variant | - | - | - | BjuOA09G47290 |
| A09 | 46,801,386 | G | A | 5_prime_UTR_variant | 46,801,386 | G | A | BjuOA09G47290 |
| A09 | 46,801,436 | A | G | 5_prime_UTR_variant | 46,801,436 | A | G | BjuOA09G47290 |
| A09 | 45,670,196 | C | G | synonymous_var | - | - | - | BjuOA09G45170 |
| A09 | - | - | - | - | 45,670,220 | A | G | BjuOA09G45170 |
| A09 | 45,670,292 | T | C | synonymous_var | - | - | - | BjuOA09G45170 |
| A09 | 45,670,968 | A | G | synonymous_varian | - | - | - | BjuOA09G45170 |
| A09 | 45,671,218 | T | G | missense_variant | - | - | - | BjuOA09G45170 |
| A09 | 45,671,381 | G | T | synonymous | - | - | - | BjuOA09G45170 |
| A09 | 45,671,597 | G | C | synonymous_var | - | - | - | BjuOA09G45170 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Lu, H.; Liu, X.; Liu, L.; Zhang, W.; Huang, Z.; Li, K.; Xu, A. Identification of Yellow Seed Color Genes Using Bulked Segregant RNA Sequencing in Brassica juncea L. Int. J. Mol. Sci. 2024, 25, 1573. https://doi.org/10.3390/ijms25031573
Wang Y, Lu H, Liu X, Liu L, Zhang W, Huang Z, Li K, Xu A. Identification of Yellow Seed Color Genes Using Bulked Segregant RNA Sequencing in Brassica juncea L. International Journal of Molecular Sciences. 2024; 25(3):1573. https://doi.org/10.3390/ijms25031573
Chicago/Turabian StyleWang, Yang, Hong Lu, Xiang Liu, Lu Liu, Wenying Zhang, Zhen Huang, Keqi Li, and Aixia Xu. 2024. "Identification of Yellow Seed Color Genes Using Bulked Segregant RNA Sequencing in Brassica juncea L." International Journal of Molecular Sciences 25, no. 3: 1573. https://doi.org/10.3390/ijms25031573
APA StyleWang, Y., Lu, H., Liu, X., Liu, L., Zhang, W., Huang, Z., Li, K., & Xu, A. (2024). Identification of Yellow Seed Color Genes Using Bulked Segregant RNA Sequencing in Brassica juncea L. International Journal of Molecular Sciences, 25(3), 1573. https://doi.org/10.3390/ijms25031573







