Computation of X-ray and Neutron Scattering Patterns to Benchmark Atomistic Simulations against Experiments
Abstract
1. Introduction
2. Results
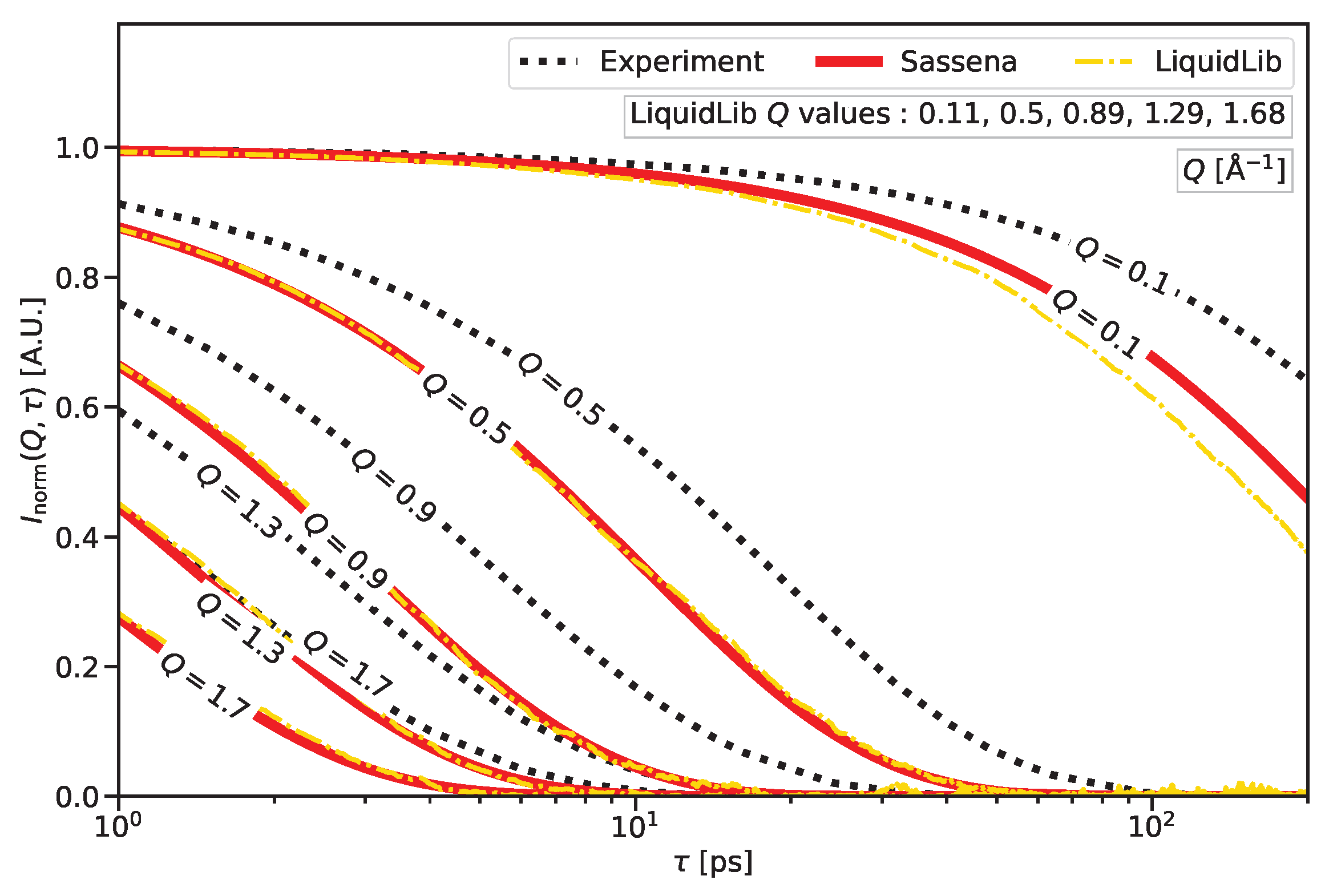
2.1. Atomistic Dynamics as Seen by the Incoherent Intermediate Scattering Function
2.2. Structure as Seen by Diffraction
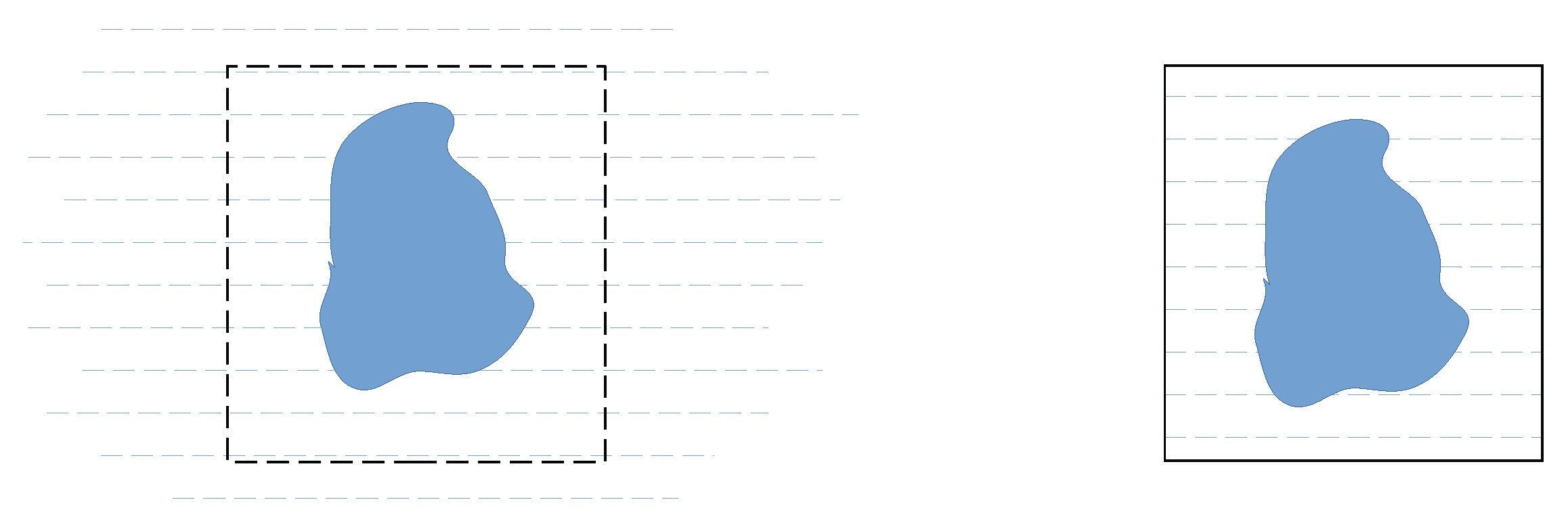
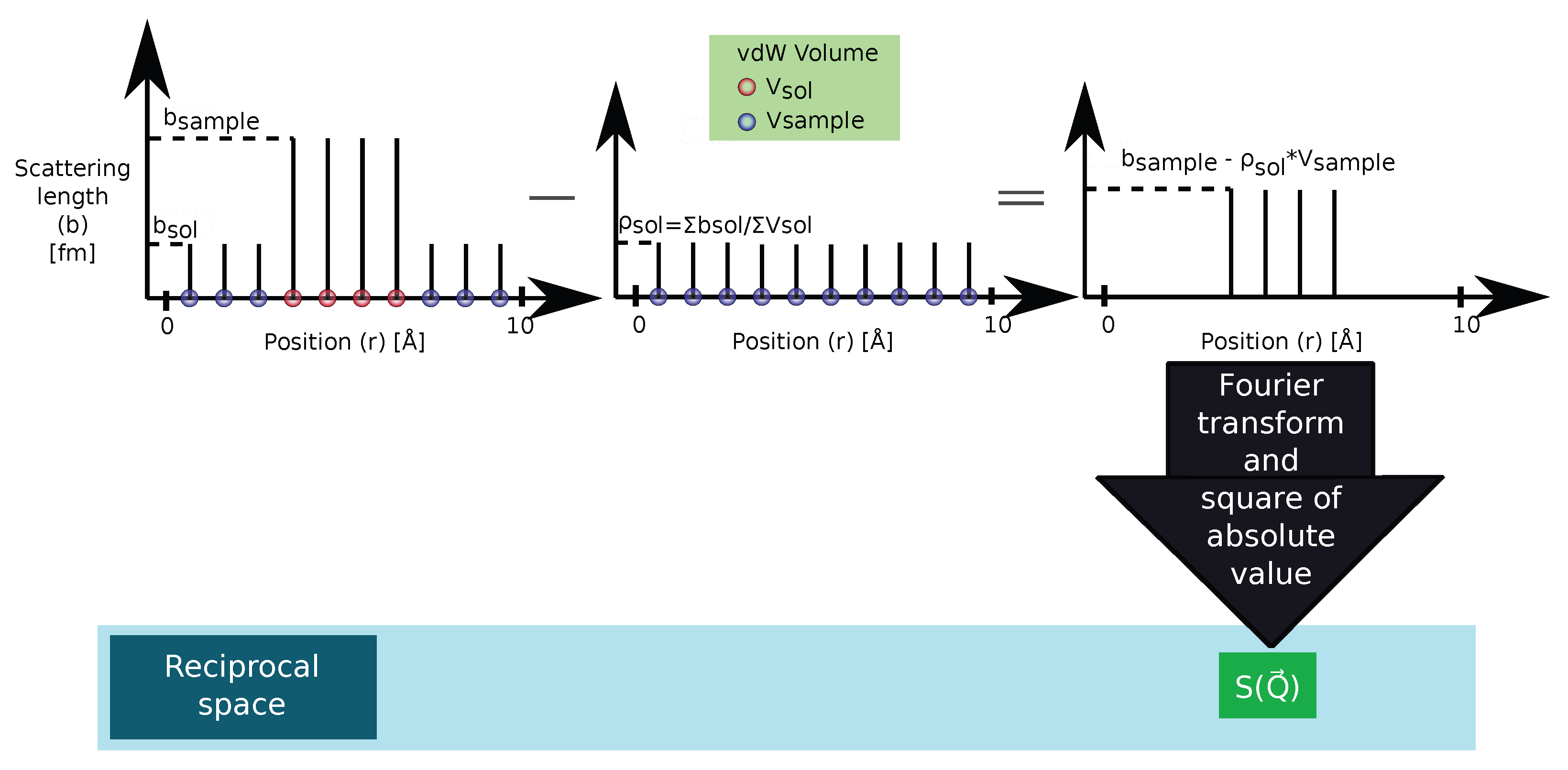
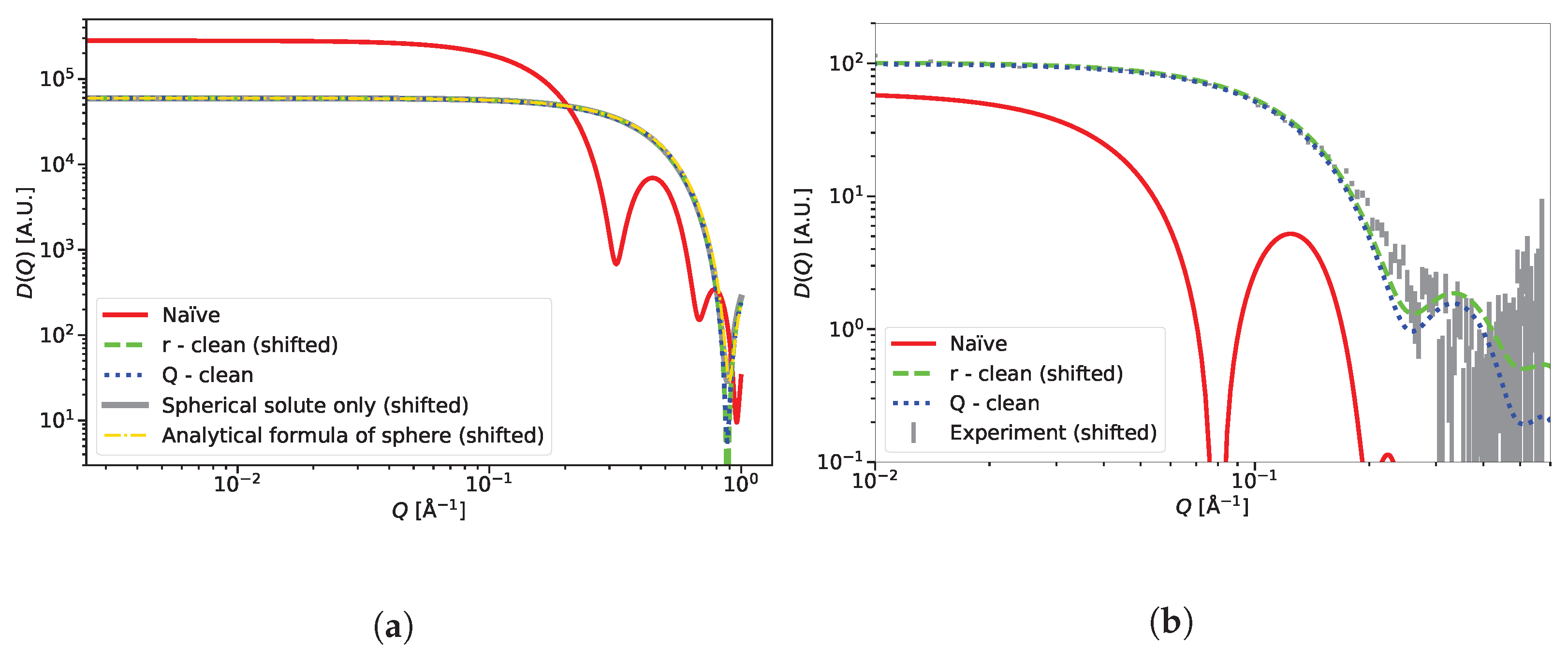
2.2.1. r-Clean: Subtracting the Average Scattering Length Density of the Solvent from the Scattering Lengths to Remove the Contrast between the Simulation Box and Surrounding Vacuum
2.2.2. Q-Clean: Subtracting the Scattering Amplitude of the Simulation Box in Reciprocal Space
2.2.3. Calculation of Example Diffractograms
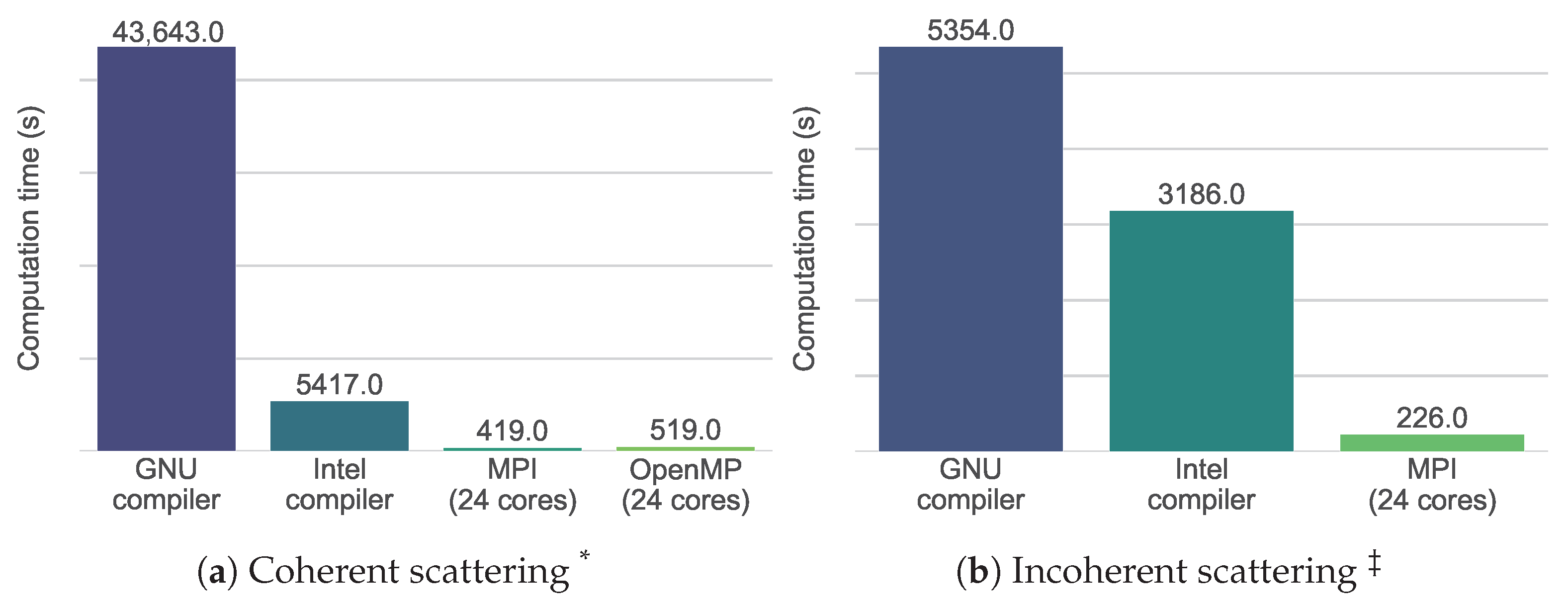
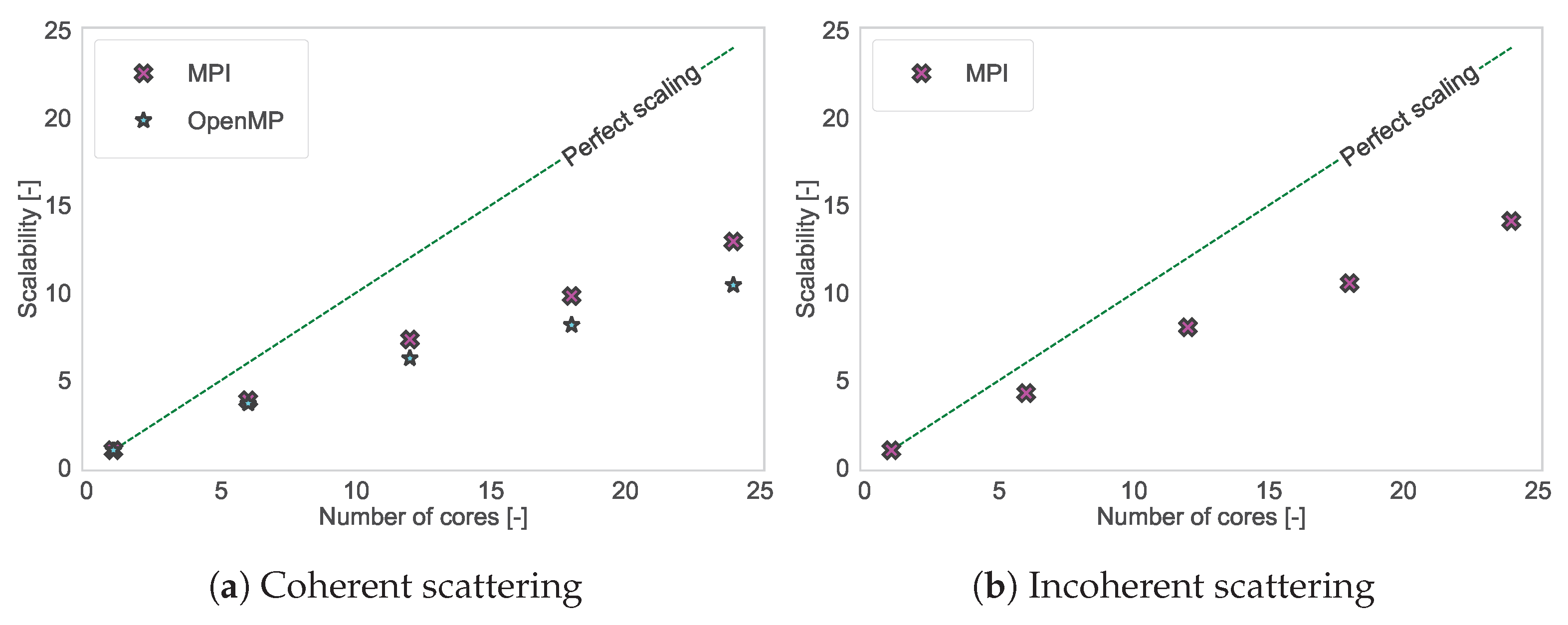
2.3. Increasing the Computation Speed
2.3.1. Single-Core Performance: Efficient Vectorization
2.3.2. Multi-Core Performance: Introduction of OpenMP
3. Discussion
3.1. Dynamics—Atomic
3.2. Structure—Atomic and Nanoscopic
3.3. Increasing the Computation Speed
4. Materials and Methods
4.1. Simulations
4.1.1. Water
4.1.2. Spherical Solute in Solvent
4.1.3. Lysozyme in Water
4.2. Calculation of Scattering Curves from Simulations
4.2.1. Probing the Dynamics
Water
4.2.2. Probing the Structure
Water
Spherical Solute in Solvent
Lysozyme in Water
4.3. Experiments
4.3.1. Water Dynamics
4.3.2. Water Structure
4.3.3. Spherical Solute in Solvent
4.3.4. Lysozyme in Water Structure
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
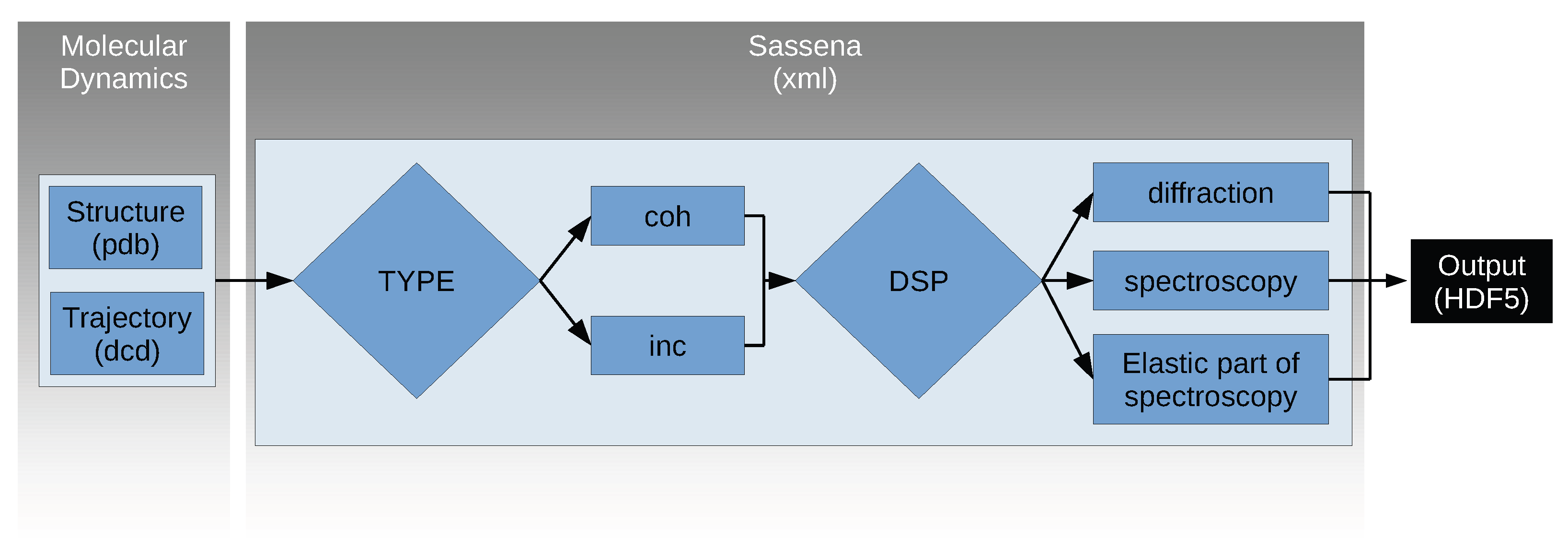
Appendix A. Algorithm of Sassena
Appendix B. Molecular Dynamics (MD) Simulation of Water
| Parameter | Value | Unit |
|---|---|---|
| bond length ((OH)) | 0.9572 | Å |
| bond stiffness ( OH)) | 450 | 1 kcal/mol/ |
| angle (HOH) | 104.52 | degree |
| angular stiffness HOH)) | 55 | kcal/mol/rad2 |
| charge | 0.415 | e |
| Lennard-Jones parameter (Oxygen) | 3.188 | Å |
| Lennard-Jones parameter (Oxygen) | 0.102 | kcal/mol |
| Lennard-Jones parameter (Hydrogen) | 0.0 | Å |
| Lennard-Jones parameter (Hydrogen) | 0.0 | kcal/mol |
Appendix C. Normalization of Scattering Curves
Appendix D. Quasielastic Neutron Scattering (QENS) Experiment on Water
| a [Å] | [ps] | [ps] | [kcal/mol] | L [Å] | [Å] |
|---|---|---|---|---|---|
| 0.98 | 1.25 | 0.0485 | 1.85 | 1.29 | 0.48 |
Appendix E. Molecular Dynamics (MD) Simulation of Lysozyme Immersed in Water
References
- Pokotilovski, Y.N. Constraints on New Interactions from Neutron Scattering Experiments. Phys. Atom. Nuclei 2006, 69, 924–931. [Google Scholar] [CrossRef]
- Ristig, M.L.; Gernoth, K.A. Particle Scattering, X-ray Diffraction, and Microstructure of Solids and Liquids; Springer: Berlin/Heidelberg, Germany, 2008; Volume 610. [Google Scholar] [CrossRef]
- Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N.; Teller, A.H.; Teller, E. Equation of State Calculations by Fast Computing Machines. J. Chem. Phys. 1953, 21, 1087–1092. [Google Scholar] [CrossRef]
- Xie, Y.; Zhou, J.; Jiang, S. Parallel Tempering Monte Carlo Simulations of Lysozyme Orientation on Charged Surfaces. J. Chem. Phys. 2010, 132, 065101. [Google Scholar] [CrossRef] [PubMed]
- Alder, B.J.; Wainwright, T.E. Phase Transition for a Hard Sphere System. J. Chem. Phys. 1957, 27, 1208–1209. [Google Scholar] [CrossRef]
- McCarthy, A.N.; Grigera, J.R. Effect of Pressure on the Conformation of Proteins. A Molecular Dynamics Simulation of Lysozyme. J. Mol. Graph. Model. 2006, 24, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Hinchliffe, A. Molecular Dynamics. In Molecular Modelling for Beginners, 2nd ed.; John Wiley & Sons Ltd.: Oxford, UK, 2008. [Google Scholar]
- Fedorov, B.; Denesyuk, A. Sperm whale Myoglobin Structure in Solution Differs from its Structure in Crystal by a Shift of the ‘Hairpin’ Gh. FEBS Lett. 1978, 88, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Svergun, D.; Barberato, C.; Koch, M.H. CRYSOL—A Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Carlsson, F.; Malmsten, M.; Linse, P. Monte Carlo Simulations of Lysozyme Self-association in Aqueous Solution. J. Phys. Chem. B 2001, 105, 12189–12195. [Google Scholar] [CrossRef]
- Cardinaux, F.; Stradner, A.; Schurtenberger, P.; Sciortino, F.; Zaccarelli, E. Modeling Equilibrium Clusters in Lysozyme Solutions. Europhys. Lett. 2007, 77, 48004. [Google Scholar] [CrossRef]
- Chen, P.C.; Hub, J.S. Validating Solution Ensembles from Molecular Dynamics Simulation by Wide-Angle X-ray Scattering Data. Biophys. J. 2014, 107, 435–447. [Google Scholar] [CrossRef]
- Alvarez, F.; Arbe, A.; Colmenero, J. Unraveling the Coherent Dynamic Structure Factor of Liquid Water at the Mesoscale by Molecular Dynamics Simulations. J. Chem. Phys. 2021, 155, 244509. [Google Scholar] [CrossRef] [PubMed]
- Reich, V.; Majumdar, A.; Müller, M.; Busch, S. Comparison of Molecular Dynamics Simulations of Water with Neutron and X-ray Scattering Experiments. In Proceedings of the EPJ Web of Conferences, EDP Sciences, San Sebastian, Spain (Online), 23-27 May 2022; Volume 272, p. 01015. [Google Scholar] [CrossRef]
- Fedorov, B.; Ptitsyn, O.; Voronin, L. X-ray Diffuse Scattering of Globular Protein Solutions: Consideration of the Solvent Influence. FEBS Lett. 1972, 28, 188–190. [Google Scholar] [CrossRef] [PubMed]
- Ninio, J.; Luzzati, V.; Yaniv, M. Comparative Small-Angle X-ray Scattering Studies on Unacylated, Acylated and Cross-linked Escherichia Coli Transfer RNAIVal. J. Mol. Biol. 1972, 71, 217–229. [Google Scholar] [CrossRef]
- Fedorov, B.; Ptitsyn, O.; Voronin, L. X-ray Diffuse Scattering by Proteins in Solution. Consideration of Solvent Influence. J. Appl. Crystallogr. 1974, 7, 181–186. [Google Scholar] [CrossRef]
- McGreevy, R.; Pusztai, L. Reverse Monte Carlo Simulation: A New Technique for the Determination of Disordered Structures. Mol. Simul. 1988, 1, 359–367. [Google Scholar] [CrossRef]
- Soper, A.K. Computer Simulation as a Tool for the Interpretation of Total Scattering Data from Glasses and Liquids. Mol. Simul. 2012, 38, 1171–1185. [Google Scholar] [CrossRef]
- Tucker, M.G.; Keen, D.A.; Dove, M.T.; Goodwin, A.L.; Hui, Q. RMCProfile: Reverse Monte Carlo for Polycrystalline Materials. J. Phys. Condens. Matter 2007, 19, 335218. [Google Scholar] [CrossRef] [PubMed]
- Ivanović, M.T.; Bruetzel, L.K.; Lipfert, J.; Hub, J.S. Temperature-Dependent Atomic Models of Detergent Micelles Refined against Small-Angle X-Ray Scattering Data. Angew. Chem. Int. Ed. 2018, 57, 5635–5639. [Google Scholar] [CrossRef]
- Chen, P.C.; Shevchuk, R.; Strnad, F.M.; Lorenz, C.; Karge, L.; Gilles, R.; Stadler, A.M.; Hennig, J.; Hub, J.S. Combined Small-Angle X-ray and Neutron Scattering Restraints in Molecular Dynamics Simulations. J. Chem. Theory Comput. 2019, 15, 4687–4698. [Google Scholar] [CrossRef]
- Youngs, T. Dissolve: Next Generation Software for the Interrogation of Total Scattering Data by Empirical Potential Generation. Mol. Phys. 2019, 117, 3464–3477. [Google Scholar] [CrossRef]
- Wolf, C.M.; Guio, L.; Scheiwiller, S.; Pakhnyuk, V.; Luscombe, C.; Pozzo, L.D. Strategies for the Development of Conjugated Polymer Molecular Dynamics Force Fields Validated with Neutron and X-ray Scattering. ACS Polym. Au 2021, 1, 134–152. [Google Scholar] [CrossRef]
- Saha, B. Green Computing. Int. J. Comput. Trends Technol. (IJCTT) 2014, 14, 46–50. [Google Scholar] [CrossRef]
- Shalf, J. The Future of Computing Beyond Moore’s Law. Philos. Trans. R Soc. Math. Phys. Eng. Sci. 2020, 378, 20190061. [Google Scholar] [CrossRef] [PubMed]
- Grama, A. Introduction to Parallel Computing; Pearson Education: London, UK, 2003; pp. 2–9. [Google Scholar]
- Thompson, A.P.; Aktulga, H.M.; Berger, R.; Bolintineanu, D.S.; Brown, W.M.; Crozier, P.S.; in’t Veld, P.J.; Kohlmeyer, A.; Moore, S.G.; Nguyen, T.D.; et al. LAMMPS-A Flexible Simulation Tool for Particle-Based Materials Modeling at the Atomic, Meso, and Continuum Scales. Comput. Phys. Commun. 2022, 271, 108171. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations Through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Pabit, S.A.; Meisburger, S.P.; Pollack, L.; Case, D.A. Accurate Small and Wide Angle X-ray Scattering Profiles from Atomic Models of Proteins and Nucleic Acids. J. Chem. Phys. 2014, 141, 22D508. [Google Scholar] [CrossRef] [PubMed]
- Gotz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- McCulley, C.H.; Walker, A.R. Dimer Interface Destabilization of Photodissociative Dronpa Driven by Asymmetric Monomer Dynamics. J. Phys. Chem. B 2023, 127, 9248–9257. [Google Scholar] [CrossRef] [PubMed]
- Róg, T.; Murzyn, K.; Hinsen, K.; Kneller, G.R. nMoldyn: A Program Package for a Neutron Scattering Oriented Analysis of Molecular Dynamics Simulations. J. Comput. Chem. 2003, 24, 657–667. [Google Scholar] [CrossRef]
- Konrad Hinsen. nMOLDYN3—Version 3.0.12. Available online: https://github.com/khinsen/nMOLDYN3/ (accessed on 27 December 2023).
- Goret, G.; Aoun, B.; Pellegrini, E. MDANSE: An Interactive Analysis Environment for Molecular Dynamics Simulations. J. Chem. Inf. Model. 2017, 57, 1–5. [Google Scholar] [CrossRef]
- UKRI STFC ISIS Neutron and Muon Facility. MDANSE—Version 1.5.2. Available online: https://github.com/ISISNeutronMuon/MDANSE/ (accessed on 27 December 2023).
- Walter, N.P.; Jaiswal, A.; Cai, Z.; Zhang, Y. LiquidLib: A Comprehensive Toolbox for Analyzing Classical and Ab Initio Molecular Dynamics Simulations of Liquids and Liquid-like Matter with Applications to Neutron Scattering Experiments. Comput. Phys. Commun. 2018, 228, 209–218. [Google Scholar] [CrossRef]
- Z-laboratory. LiquidLib—Version 1.0. Available online: https://github.com/Z-Laboratory/LiquidLib/tree/master (accessed on 8 December 2023).
- Lindner, B.; Smith, J.C. Sassena—X-ray and Neutron Scattering Calculated from Molecular Dynamics Trajectories Using Massively Parallel Computers. Comput. Phys. Commun. 2012, 183, 1491–1501. [Google Scholar] [CrossRef]
- Lindner, B. Towards a Unification of Supercomputing, Molecular Dynamics Simulation and Experimental Neutron and X-ray Scattering Techniques. Ph.D. Thesis, University of Tennessee, Knoxville, TN, USA, 2012. [Google Scholar]
- Lindner, B. Sassena—Version 1.4.2. 2017. Available online: https://github.com/camm/sassena (accessed on 1 March 2023).
- Majumdar, A.; Lindner, B. Sassena—Version 1.4.3. 2023. Available online: https://codebase.helmholtz.cloud/DAPHNE4NFDI/sassena (accessed on 27 December 2023). [CrossRef]
- Lion, D.; Chiu, A.; Stumm, M.; Yuan, D. Investigating Managed Language Runtime Performance: Why JavaScript and Python Are 8x and 29x Slower Than C++, Yet Java and Go Can Be Faster? In Proceedings of the 2022 USENIX Annual Technical Conference (USENIX ATC 22), San Diego, CA, USA, 11–13 July 2022; USENIX Association: Carlsbad, CA, USA, 2022; pp. 835–852. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Theoretical and Computational Biophysics Group-University of Illinois at Urbana-Champaign. VMD—Version 1.9.4a48. Available online: https://www.ks.uiuc.edu/Research/vmd/ (accessed on 27 December 2023).
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [PubMed]
- MDAnalysis. MDAnalysis—Version 2.7.0. Available online: https://github.com/MDAnalysis/mdanalysis (accessed on 27 December 2023).
- Brehm, M.; Thomas, M.; Gehrke, S.; Kirchner, B. TRAVIS—A Free Analyzer for Trajectories from Molecular Simulation. J. Chem. Phys. 2020, 152, 164105. [Google Scholar] [CrossRef] [PubMed]
- Brehm, M. TRAVIS. Available online: https://www.travis-analyzer.de/ (accessed on 27 December 2023).
- Skånberg, R.; Hotz, I.; Ynnerman, A.; Linares, M. VIAMD: A Software for Visual Interactive Analysis of Molecular Dynamics. J. Chem. Inf. Model. 2023, 63, 7382–7391. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.E.; Barber, M.N. Scaling Theory for Finite-Size Effects in the Critical Region. Phys. Rev. Lett. 1972, 28, 1516–1519. [Google Scholar] [CrossRef]
- Brisard, S.; Levitz, P. Small-Angle Scattering of Dense, Polydisperse Granular Porous Media: Computation Free of Size Effects. Phys. Rev. E 2013, 87, 013305. [Google Scholar] [CrossRef]
- Olds, D.P.; Duxbury, P.M. Efficient Algorithms for Calculating Small-angle Scattering from Large Model Structures. J. Appl. Crystallogr. 2014, 47, 1077–1086. [Google Scholar] [CrossRef]
- Dohn, A.O.; Markmann, V.; Nimmrich, A.; Haldrup, K.; Møller, K.B.; Nielsen, M.M. Eliminating Finite-size Effects on the Calculation of X-ray Scattering from Molecular Dynamics Simulations. J. Chem. Phys. 2023, 159, 124115. [Google Scholar] [CrossRef]
- Teixeira, J.; Bellissent-Funel, M.C.; Chen, S.H.; Dianoux, A.J. Experimental Determination of the Nature of Diffusive Motions of Water Molecules at Low Temperatures. Phys. Rev. A 1985, 31, 1913. [Google Scholar] [CrossRef]
- Sears, V.F. Neutron Scattering Lengths and Cross Sections. Neutron News 1992, 3, 26–37. [Google Scholar] [CrossRef]
- Brown, P.; Fox, A.; Maslen, E.; O’Keefe, M.; Willis, B. Intensity of Diffracted Intensities. Int. Tables Crystallogr. C 2006, 554, 554–595. [Google Scholar] [CrossRef]
- Arbe, A.; Nilsen, G.J.; Stewart, J.R.; Alvarez, F.; Sakai, V.G.; Colmenero, J. Coherent Structural Relaxation of Water from Meso-to Intermolecular Scales Measured Using Neutron Spectroscopy with Polarization Analysis. Phys. Rev. Res. 2020, 2, 022015. [Google Scholar] [CrossRef]
- Qvist, J.; Schober, H.; Halle, B. Structural Dynamics of Supercooled Water from Quasielastic Neutron Scattering and Molecular Simulations. J. Chem. Phys. 2011, 134, 144508. [Google Scholar] [CrossRef]
- Piskulich, Z.A.; Thompson, W.H. Examining the Role of Different Molecular Interactions on Activation Energies and Activation Volumes in Liquid Water. J. Chem. Theory Comput. 2021, 17, 2659–2671. [Google Scholar] [CrossRef]
- Sivia, D.S. Elementary Scattering Theory: For X-ray and Neutron Users; Oxford University Press: Oxford, UK, 2011. [Google Scholar]
- Bondi, A. Van Der Waals Volumes and Radii. J. Phys. Chem. 1964, 68, 441–451. [Google Scholar] [CrossRef]
- Lide, D.R. CRC Handbook of Chemistry and Physics; CRC Press: Boca Raton, FL, USA, 2004; Volume 85. [Google Scholar]
- Trewhella, J.; Vachette, P.; Bierma, J.; Blanchet, C.; Brookes, E.; Chakravarthy, S.; Chatzimagas, L.; Cleveland, T.E.; Cowieson, N.; Crossett, B.; et al. A Round-Robin Approach Provides a Detailed Assessment of Biomolecular Small-Angle Scattering Data Reproducibility and Yields Consensus Curves for Benchmarking. Acta Crystallogr. Sect. Struct. Biol. 2022, 78, 1315–1336. [Google Scholar] [CrossRef]
- Parhami, B. SIMD Machines: Do They Have a Significant Future? ACM SIGARCH Comput. Archit. News 1995, 23, 19–22. [Google Scholar] [CrossRef]
- Hofmann, J.; Treibig, J.; Hager, G.; Wellein, G. Comparing the Performance of Different x86 SIMD Instruction Sets for a Medical Imaging Application on Modern Multi-and Manycore chips. In Proceedings of the 2014 Workshop on Programming Models for SIMD/Vector Processing, Orlando, FL, USA, 16 February 2014; pp. 57–64. [Google Scholar] [CrossRef]
- Gottschlag, M.; Brantsch, P.; Bellosa, F. Automatic Core Specialization for AVX-512 Applications. In Proceedings of the 13th ACM International Systems and Storage Conference, Haifa, Israel, 13–15 October 2020; pp. 25–35. [Google Scholar] [CrossRef]
- Intel Corporation. Intel Advisor 2020—Initial Release. 2019. Available online: https://www.intel.com/content/dam/develop/external/us/en/documents/intel-advisor-2020-release-notes-562228.pdf (accessed on 20 December 2021).
- GCC Team. GCC, The GNU Compiler Collection—Version 8.3.0. 2019. Available online: https://gcc.gnu.org/ (accessed on 20 December 2021).
- Intel Corporation. Intel® oneAPI DPC++/C++ Compiler—Version 2021.8.0. 2003. Available online: https://www.intel.com/content/www/us/en/developer/tools/oneapi/dpc-compiler.html#gs.4pn6zz (accessed on 25 March 2023).
- Intel Corporation. Intel® oneAPI Math Kernel Library. 2023. Available online: https://www.intel.com/content/www/us/en/developer/tools/oneapi/onemkl.html (accessed on 25 March 2023).
- Gabriel, E.; Fagg, G.E.; Bosilca, G.; Angskun, T.; Dongarra, J.J.; Squyres, J.M.; Sahay, V.; Kambadur, P.; Barrett, B.; Lumsdaine, A.; et al. Open MPI: Goals, Concept, and Design of a Next Generation MPI Implementation. In Proceedings of the 11th European PVM/MPI Users’ Group Meeting, Budapest, Hungary, 19–22 September 2004; pp. 97–104. [Google Scholar] [CrossRef]
- Kormanyos, C. Real-time C++: Efficient Object-Oriented and Template Microcontroller Programming; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar] [CrossRef]
- Dagum, L.; Menon, R. OpenMP: An Industry Standard API for Shared-Memory Programming. IEEE Comput. Sci. Eng. 1998, 5, 46–55. [Google Scholar] [CrossRef]
- Amdahl, G.M. Validity of the Single Processor Approach to Achieving Large Scale Computing Capabilities. In Proceedings of the Spring Joint Computer Conference, Atlantic City, NJ, USA, 18–20 April 1967; pp. 483–485. [Google Scholar] [CrossRef]
- Gustafson, J.L. Reevaluating Amdahl’s Law. Commun. ACM 1988, 31, 532–533. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Price, D.J.; Brooks, C.L., III. A Modified TIP3P Water Potential for Simulation with Ewald Summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef] [PubMed]
- Woolf, L. Tracer Diffusion of Tritiated Water (THO) in Ordinary Water (H2O) Under Pressure. J. Chem. Soc. Faraday Trans. Phys. Chem. Condens. Phases 1975, 71, 784–796. [Google Scholar] [CrossRef]
- Frenkel, D.; Vos, R.; de Kruif, C.D.; Vrij, A. Structure Factors of Polydisperse Systems of Hard Spheres: A Comparison of Monte Carlo Simulations and Percus–Yevick theory. J. Chem. Phys. 1986, 84, 4625–4630. [Google Scholar] [CrossRef]
- Scherrer, P. Bestimmung der Größe und der inneren Struktur von Kolloidteilchen mittels Röntgenstrahlen. Nachrichten Von Ges. Wiss. GöTtingen Math.-Phys. Kl. 1918, 1918, 98–100. [Google Scholar]
- Balaguer, M.; Solís, C.; Serra, J.M. Study of the Transport Properties of the Mixed Ionic Electronic Conductor Ce1-xTbxO2-δ+ Co (x = 0.1, 0.2) and Evaluation as Oxygen-Transport Membrane. Chem. Mater. 2011, 23, 2333–2343. [Google Scholar] [CrossRef]
- Salacuse, J.J.; Denton, A.R.; Egelstaff, P.A. Finite-Size Effects in Molecular Dynamics Simulations: Static Structure Factor and Compressibility. I. Theoretical Method. Phys. Rev. E 1996, 53, 2382–2389. [Google Scholar] [CrossRef]
- Patterson, A.L. The Scherrer Formula for X-Ray Particle Size Determination. Phys. Rev. 1939, 56, 978–982. [Google Scholar] [CrossRef]
- Cao, T.; Ji, X.; Wu, J.; Zhang, S.; Yang, X. Correction of Diffusion Calculations When Using Two Types of Non-Rectangular Simulation Boxes in Molecular Simulations. J. Mol. Model. 2019, 25, 1–10. [Google Scholar] [CrossRef]
- Debye, P. Zerstreuung von Röntgenstrahlen. Ann. Der Phys. 1915, 351, 809–823. [Google Scholar] [CrossRef]
- Guinier, A. X-ray Diffraction in Crystals, Imperfect Crystals, and Amorphous Bodies; Courier Corporation: Chelmsford, MA, USA, 1994. [Google Scholar]
- Farrow, C.L.; Billinge, S.J. Relationship Between the Atomic Pair Distribution Function and Small-Angle Scattering: Implications for Modeling of Nanoparticles. Acta Crystallogr. Sect. Found. Crystallogr. 2009, 65, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Marcin Wojdyr. Debyer. 2015. Available online: https://github.com/wojdyr/debyer (accessed on 27 December 2023).
- Jin, H.; Jespersen, D.; Mehrotra, P.; Biswas, R.; Huang, L.; Chapman, B. High Performance Computing using MPI and OpenMP on Multi-Core Parallel Systems. Parallel Comput. 2011, 37, 562–575. [Google Scholar] [CrossRef]
- Rabenseifner, R.; Hager, G.; Jost, G. Hybrid MPI/OpenMP Parallel Programming on Clusters of Multi-Core SMP Nodes. In Proceedings of the 2009 17th Euromicro International Conference on Parallel, Distributed and Network-Based Processing, Weimar, Germany, 18–20 February 2009; pp. 427–436. [Google Scholar] [CrossRef]
- van Rossum, G. Python Tutorial, Technical Report CS-R9526; Technical Report; Centrum voor Wiskunde en Informatica (CWI): Amsterdam, The Netherlands, 1995. [Google Scholar]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef]
- Soper, A.K. The Radial Distribution Functions of Water as Derived from Radiation Total Scattering Experiments: Is There Anything We Can Say for Sure? Int. Sch. Res. Not. 2013, 2013, 279463. [Google Scholar] [CrossRef]
- Guinier, A.; Fournet, G.; Walker, C.B.; Yudowitch, K.L. Small-Angle Scattering of X-rays; Wiley: New York, NY, USA, 1955. [Google Scholar]
- Kikhney, A.G.; Borges, C.R.; Molodenskiy, D.S.; Jeffries, C.M.; Svergun, D.I. SASBDB: Towards an Automatically Curated and Validated Repository for Biological Scattering data. Protein Sci. 2020, 29, 66–75. [Google Scholar] [CrossRef]
- Aslan, N.; Horstmann, C.; Metz, O.; Kotlyar, O.; Dornheim, M.; Pistidda, C.; Busch, S.; Lohstroh, W.; Müller, M.; Pranzas, K. High-Pressure Cell for In Situ Neutron Studies of Hydrogen Storage Materials. J. Neutron Res. 2019, 21, 125–135. [Google Scholar] [CrossRef]
- Sandia Corporation. LAMMPS—Version 22 December 2022. Available online: https://www.lammps.org/ (accessed on 1 March 2023).
- Sandia Corporation. A Flexible Version of TIP3P. 2003–2023. Available online: https://docs.lammps.org/Howto_tip3p.html (accessed on 27 December 2023).
- Sears, V. Theory of Cold Neutron Scattering by Homonuclear Diatomic Liquids: II. Hindered Rotation. Can. J. Phys. 1966, 44, 1299–1311. [Google Scholar] [CrossRef]
- Bée, M. General Aspects of Neutron Scattering. In Quasielastic Neutron Scattering; Chapter General Aspects of Neutron Scattering; IOP Publishing Ltd.: Bristol, UK, 1988. [Google Scholar]
- Egelstaff, P. An Introduction to the Liquid State; Academic: London, UK, 1967. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; Van Der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- GROMACS. GROMACS—Version 2020.1-Ubuntu-2020.1-1. Available online: http://www.gromacs.org (accessed on 27 December 2023).
- Carter, D.; He, J.; Ruble, J.; Wright, B. The Structure of The Orthorohmbic Form of Hen Egg-White Lysozyme at 1.5 Angstroms Resolution. 1997. Available online: https://www.wwpdb.org/pdb?id=pdb_00001aki (accessed on 27 December 2023).
- Lemkul, J.A. GROMACS Tutorial: Lysozyme in Water. 2018. Available online: http://www.mdtutorials.com/gmx/lysozyme/index.html (accessed on 27 December 2023).
- Jorgensen, W.L.; Tirado-Rives, J. The OPLS [Optimized Potentials for Liquid Simulations] Potential Functions for Proteins, Energy Minimizations for Crystals of Cyclic Peptides and Crambin. J. Am. Chem. Soc. 1988, 110, 1657–1666. [Google Scholar] [CrossRef]
- Damm, W.; Frontera, A.; Tirado-Rives, J.; Jorgensen, W.L. OPLS All-Atom Force Field for Carbohydrates. J. Comput. Chem. 1997, 18, 1955–1970. [Google Scholar] [CrossRef]
- Armstrong, J.A.; Bresme, F. Water Polarization Induced by Thermal Gradients: The Extended Simple Point Charge Model (SPC/E). J. Chem. Phys. 2013, 139, 014504. [Google Scholar] [CrossRef]




| Name of Sample | Mole Fraction of H2O | avg. [fm] | SLD [ ] |
|---|---|---|---|
| D2O | |||
| HDO | |||
| null | |||
| H2O |
| Parameter | Coherent | Incoherent | ||
|---|---|---|---|---|
| Base Configuration * | Parameter Variations | Base Configuration ‡ | Parameter Variations | |
| Number of atoms | 10,125 | 5184, 1029 | 10,125 | 5184, 1029 |
| Length of trajectory | 10,001 | 5001, 1001 | 101 | 51, 11 |
| Number of orientations | 100 | 50, 10 | 100 | 50, 10 |
| Number of Q lengths | 100 | 50, 10 | 100 | 50, 10 |
| Name | Specification |
|---|---|
| Number of sockets | 2 |
| Number of cores per socket | 12 |
| Number of threads per core | 2 |
| CPU model | Intel(R) Xeon(R) Silver 4116 CPU @ 2.10 GHz (Skylake-SP) |
| CPU architecture | x86_64 |
| CPU frequency (minimum) | 800.121 MHz |
| L1d cache (i+d) | 32 kB + 32 kB |
| L2 cache | 1024 kB |
| L3 cache | 16,896 kB |
| Instruction set (vectorization) | SSE2, AVX2 and AVX-512 |
| Memory (per socket) | ∼96 GB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumdar, A.; Müller, M.; Busch, S. Computation of X-ray and Neutron Scattering Patterns to Benchmark Atomistic Simulations against Experiments. Int. J. Mol. Sci. 2024, 25, 1547. https://doi.org/10.3390/ijms25031547
Majumdar A, Müller M, Busch S. Computation of X-ray and Neutron Scattering Patterns to Benchmark Atomistic Simulations against Experiments. International Journal of Molecular Sciences. 2024; 25(3):1547. https://doi.org/10.3390/ijms25031547
Chicago/Turabian StyleMajumdar, Arnab, Martin Müller, and Sebastian Busch. 2024. "Computation of X-ray and Neutron Scattering Patterns to Benchmark Atomistic Simulations against Experiments" International Journal of Molecular Sciences 25, no. 3: 1547. https://doi.org/10.3390/ijms25031547
APA StyleMajumdar, A., Müller, M., & Busch, S. (2024). Computation of X-ray and Neutron Scattering Patterns to Benchmark Atomistic Simulations against Experiments. International Journal of Molecular Sciences, 25(3), 1547. https://doi.org/10.3390/ijms25031547







