Mitochondrial Homeostasis Regulating Mitochondrial Number and Morphology Is a Distinguishing Feature of Skeletal Muscle Fiber Types in Marine Teleosts
Abstract
1. Introduction
2. Results
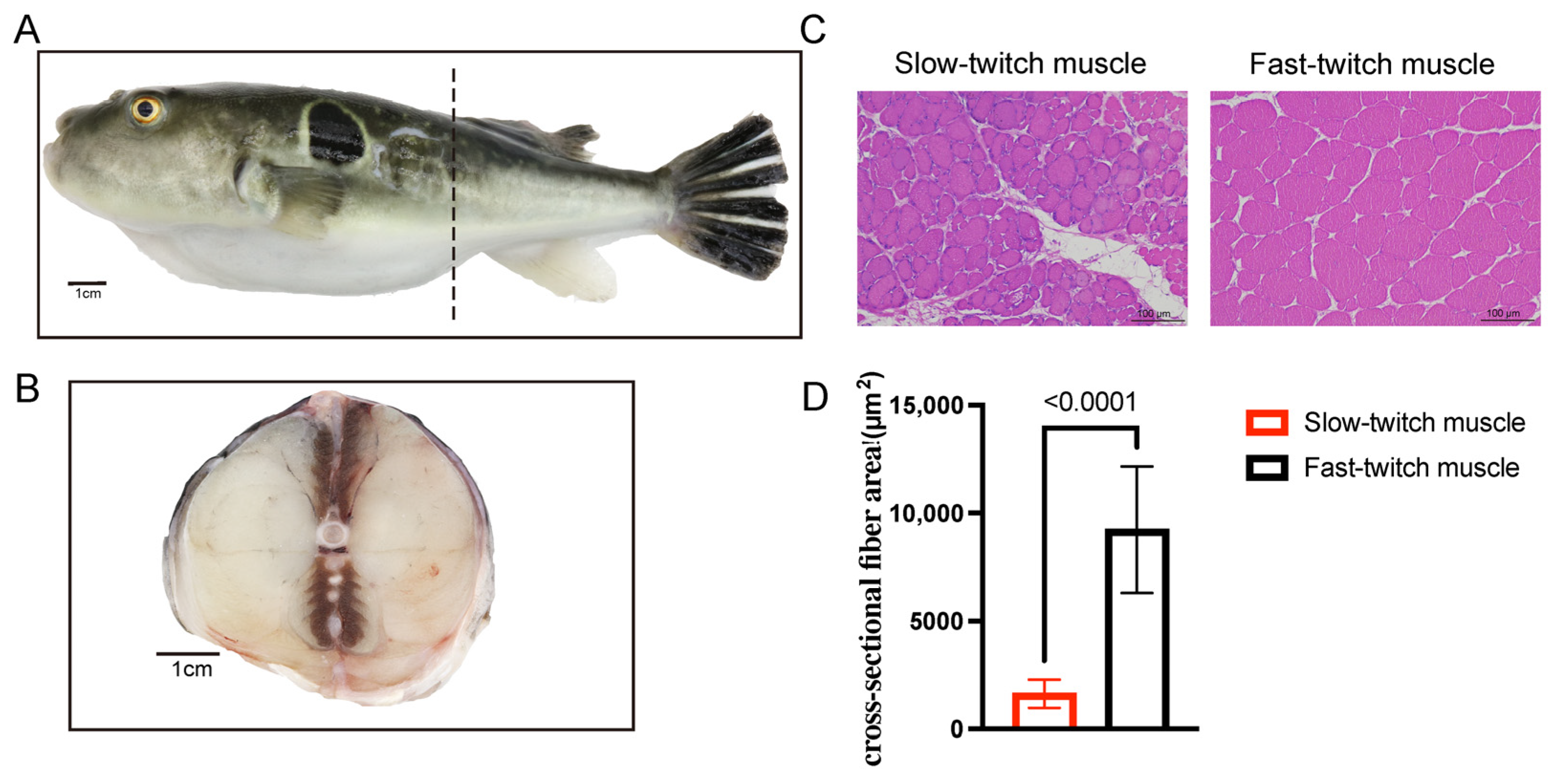
2.1. Histological Characteristics of SM and FM of T. rubripes
2.2. SM Contain More Mitochondria Than FM in T. rubripes
2.3. Gene Signatures of Energy Metabolism Are Different between SM and FM
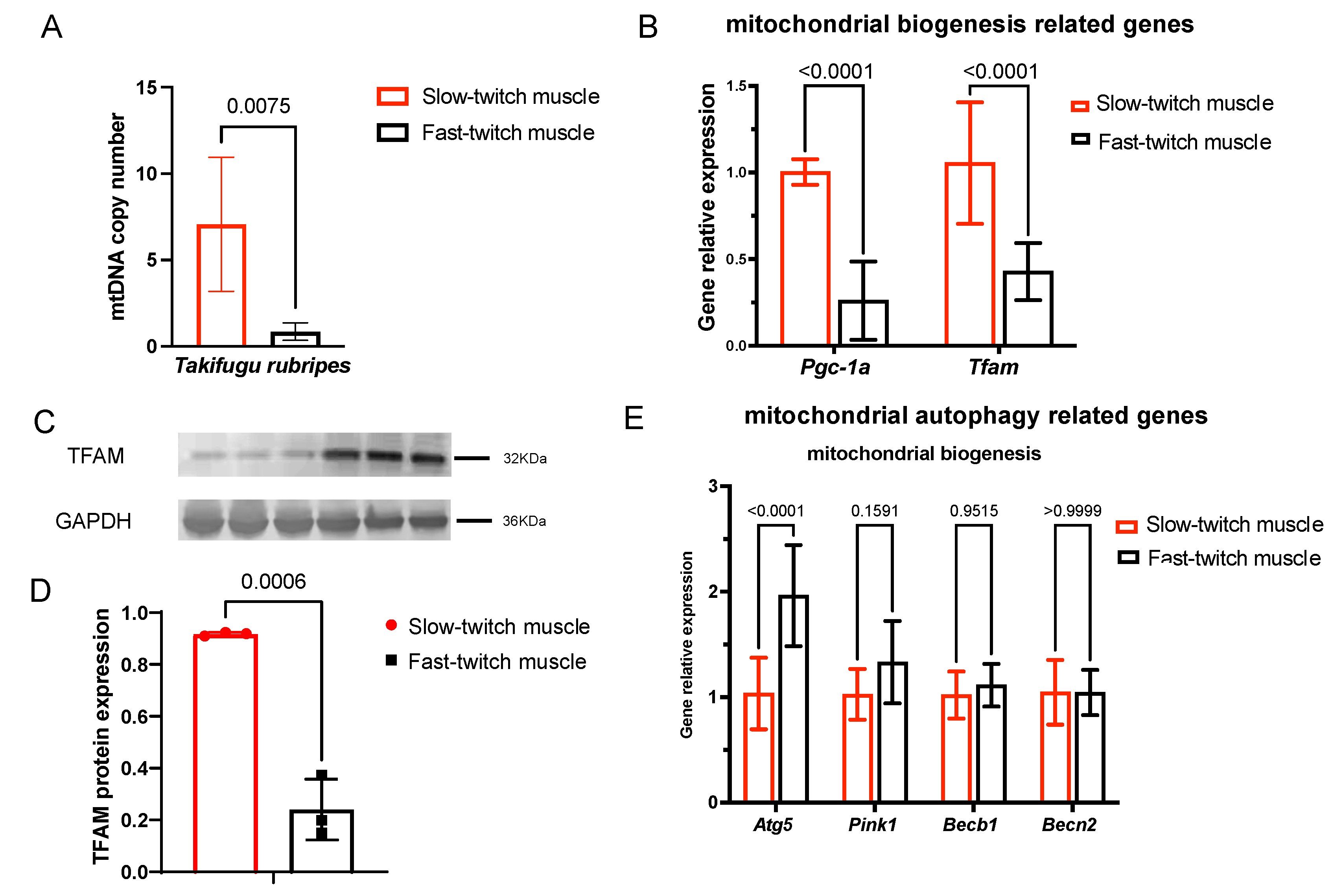
2.4. Slow-Twitch Muscle Performs Higher Mitochondrial Biogenesis in T. rubripes Than Fast-Twitch Muscle
2.5. Mitochondrial Autophagy in SM and FM of T. rubripes Shows No Difference
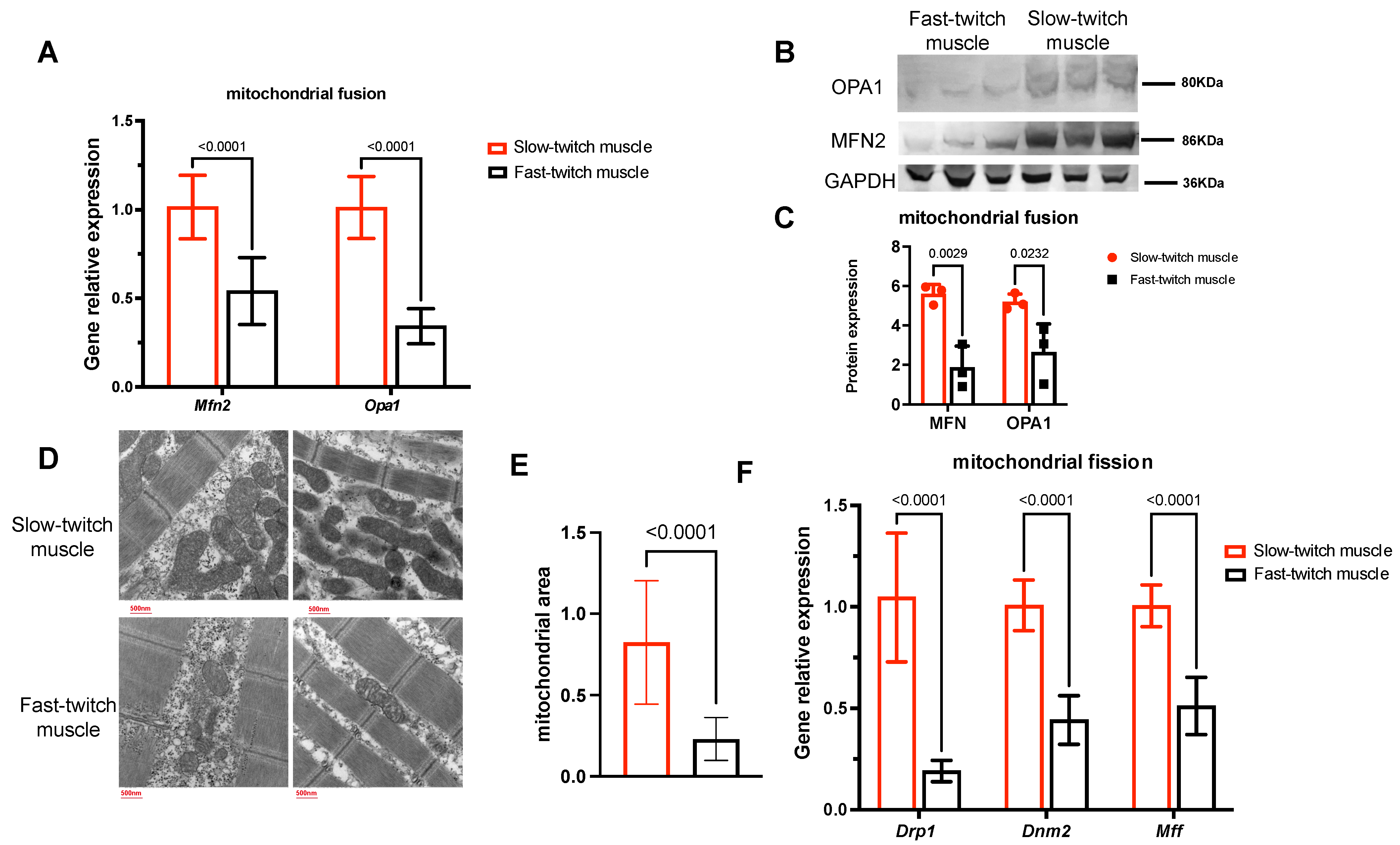
2.6. Slow-Twitch Muscles Have Higher Mitochondrial Fusion and Fission Status Compared to Fast-Twitch Muscle
3. Discussion
4. Materials and Methods
4.1. Fish Samples and Tissue Collection
4.2. Histological Analysis
4.3. Hsp60 Immunofluorescence Staining
4.4. Transmission Electron Microscopy
4.5. Western Blot Analysis
4.6. mRNA Expression Analysis
4.7. Mitochondrial DNA Copy Number Analysis
4.8. Gene Expression Heatmap of Energy Metabolism-Associated Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Moyes, C.D. Controlling muscle mitochondrial content. J. Exp. Biol. 2003, 206 Pt 24, 4385–4391. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Fu, T.; Kelly, D.P.; Vega, R.B. Skeletal muscle mitochondrial remodeling in exercise and diseases. Cell Res. 2018, 28, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Varuzhanyan, G.; Pham, A.H.; Chan, D.C. Mitochondrial Dynamics is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization. Cell Metab. 2015, 22, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Romanello, V.; Sandri, M. The connection between the dynamic remodeling of the mitochondrial network and the regulation of muscle mass. Cell. Mol. Life Sci. 2021, 78, 1305–1328. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, J.S.; Kumar, S.; Vijayan, M.; Bhatti, G.K.; Reddy, P.H. Therapeutic Strategies for Mitochondrial Dysfunction and Oxidative Stress in Age-Related Metabolic Disorders. Prog. Mol. Biol. Transl. Sci. 2017, 146, 13–46. [Google Scholar]
- Wilkins, H.M.; Weidling, I.; Koppel, S.; Wang, X.; von Schulze, A.; Swerdlow, R.H. Mitochondrial Function and Neurodegenerative Diseases. In The Molecular and Cellular Basis of Neurodegenerative Diseases; Academic Press: Cambridge, MA, USA, 2018; pp. 369–414. [Google Scholar]
- Romanello, V.; Sandri, M. Mitochondrial Quality Control and Muscle Mass Maintenance. Front. Physiol. 2015, 6, 422. [Google Scholar] [CrossRef]
- Kasahara, A.; Scorrano, L. Mitochondria: From cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014, 24, 761–770. [Google Scholar] [CrossRef]
- Otera, H.; Mihara, K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J. Biochem. 2011, 149, 241–251. [Google Scholar] [CrossRef]
- Crupi, A.N.; Nunnelee, J.S.; Taylor, D.J.; Thomas, A.; Vit, J.P.; Riera, C.E.; Gottlieb, R.A.; Goodridge, H.S. Oxidative muscles have better mitochondrial homeostasis than glycolytic muscles throughout life and maintain mitocnondrial function during aging. Aging-Us 2018, 10, 3327–3352. [Google Scholar] [CrossRef]
- Chen, H.C.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef]
- Cipolat, S.; De Brito, O.M.; Dal Zilio, B.; Scorrano, L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA 2004, 101, 15927–15932. [Google Scholar] [CrossRef]
- Pette, D.; Staron, R.S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 2000, 50, 500–509. [Google Scholar] [CrossRef]
- Wu, M.-P.; Chang, N.-C.; Chung, C.-L.; Chiu, W.-C.; Hsu, C.-C.; Chen, H.-M.; Sheu, J.-R.; Jayakumar, T.; Chou, D.-S.; Fong, T.-H. Analysis of Titin in Red and White Muscles: Crucial Role on Muscle Contractions Using a Fish Model. BioMed Res. Int. 2018, 2018, 5816875. [Google Scholar] [CrossRef]
- Aparicio, S.; Chapman, J.; Stupka, E.; Putnam, N.; Chia, J.-M.; Dehal, P.; Christoffels, A.; Rash, S.; Hoon, S.; Smit, A.; et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 2002, 297, 1301–1310. [Google Scholar] [CrossRef]
- Akolkar, D.B.; Kinoshita, S.; Yasmin, L.; Ono, Y.; Ikeda, D.; Yamaguchi, H.; Nakaya, M.; Erdogan, O.; Watabe, S. Fibre type-specific expression patterns of myosin heavy chain genes in adult torafugu Takifugu rubripes muscles. J. Exp. Biol. 2010, 213, 137–145. [Google Scholar] [CrossRef]
- Wang, H.; Cui, J.; Qiu, X.; Wang, X. Differences in DNA methylation between slow and fast muscle in Takifugu rubripes. Gene 2021, 801, 145853. [Google Scholar] [CrossRef]
- Gao, K.; Wang, Z.; Zhou, X.; Wang, H.; Kong, D.; Jiang, C.; Wang, X.; Jiang, Z.; Qiu, X. Comparative transcriptome analysis of fast twitch muscle and slow twitch muscle in Takifugu rubripes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 24, 79–88. [Google Scholar] [CrossRef]
- Asaduzzaman, M.D.; Akolkar, D.B.; Kinoshita, S.; Watabe, S. The expression of multiple myosin heavy chain genes during skeletal muscle development of torafugu Takifugu rubripes embryos and larvae. Gene 2013, 515, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Tan, X.; Sui, Y.; You, F. Muscle fibre type composition in the lateral muscle of olive flounder Paralichthys olivaceus. Acta Histochem. 2019, 121, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Martínez, I.I.; Gil Cano, F.; Zarzosa, G.R.; Vázquez, J.M.; Latorre, R.; Albors, O.L.; Arencibia, A.; Orenes, Y.M. Histochemical and Morphometric Aspects of the Lateral Musculature of Different Species of Teleost Marine Fish of the Percomorphi Order. Anat. Histol. Embryol. 2000, 29, 211–219. [Google Scholar] [CrossRef]
- Westerblad, H.; Bruton, J.D.; Katz, A. Skeletal muscle: Energy metabolism, fiber types, fatigue and adaptability. Exp. Cell Res. 2010, 316, 3093–3099. [Google Scholar] [CrossRef]
- Sahlin, K.; Tonkonogi, M.; Söderlund, K. Energy supply and muscle fatigue in humans. Acta Physiol. Scand. 1998, 162, 261–266. [Google Scholar] [CrossRef]
- Dorn, G.W., II; Vega, R.B.; Kelly, D.P. Mitochondrial biogenesis and dynamics in the developing and diseased heart. Genes Dev. 2015, 29, 1981–1991. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef]
- Liang, C.; Lee, J.-S.; Inn, K.-S.; Gack, M.U.; Li, Q.; Roberts, E.A.; Vergne, I.; Deretic, V.; Feng, P.; Akazawa, C.; et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008, 10, 776–787. [Google Scholar] [CrossRef]
- Leduc-Gaudet, J.-P.; Hussain, S.N.A.; Barreiro, E.; Gouspillou, G. Mitochondrial Dynamics and Mitophagy in Skeletal Muscle Health and Aging. Int. J. Mol. Sci. 2021, 22, 8179. [Google Scholar] [CrossRef]
- Geeves, M.A.; Holmes, K.C. The Molecular Mechanism of Muscle Contraction. Adv. Protein Chem. 2005, 71, 161–193. [Google Scholar] [PubMed]
- Romanello, V.; Sandri, M. Implications of mitochondrial fusion and fission in skeletal muscle mass and health. Semin. Cell Dev. Biol. 2023, 143, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.E.; White, K.; Davey, T.; Philips, J.; Ogden, R.T.; Lawless, C.; Warren, C.; Hall, M.G.; Ng, Y.S.; Falkous, G.; et al. Quantitative 3D Mapping of the Human Skeletal Muscle Mitochondrial Network. Cell Rep. 2019, 26, 996–1009.e4. [Google Scholar] [CrossRef]
- Eisner, V.; Picard, M.; Hajnóczky, G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol. 2018, 20, 755–765. [Google Scholar] [CrossRef]
- Hood, D.A.; Memme, J.M.; Oliveira, A.N.; Triolo, M. Maintenance of Skeletal Muscle Mitochondria in Health, Exercise, and Aging. Annu. Rev. Physiol. 2019, 81, 19–41. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kinoshita, T.; Pandey, S.; Ng, C.K.; Gygi, S.P.; Shimazaki, K.; Assmann, S.M. Transcriptional co-activator PGC-1a drives the formation of slow-twitch muscle fibres. Nature 2002, 418, 793–797. [Google Scholar] [CrossRef]
- Andres, A.M.; Tucker, K.C.; Thomas, A.; Taylor, D.J.; Sengstock, D.; Jahania, S.M.; Dabir, R.; Pourpirali, S.; Brown, J.A.; Westbrook, D.G.; et al. Mitophagy and mitochondrial biogenesis in atrial tissue of patients undergoing heart surgery with cardiopulmonary bypass. JCI Insight 2017, 2, e89303. [Google Scholar] [CrossRef] [PubMed]
- Popov, L. Mitochondrial biogenesis: An update. J. Cell. Mol. Med. 2020, 24, 4892–4899. [Google Scholar] [CrossRef]
- Picca, A.; Lezza, A.M. Regulation of mitochondrial biogenesis through TFAM-mitochondrial DNA interactions: Useful insights from aging and calorie restriction studies. Mitochondrion 2015, 25, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Woldt, E.; Sebti, Y.; Solt, L.A.; Duhem, C.; Lancel, S.; Eeckhoute, J.; Hesselink, M.K.C.; Paquet, C.; Delhaye, S.; Shin, Y.; et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 2013, 19, 1039–1046. [Google Scholar] [CrossRef]
- Twig, G.; Hyde, B.; Shirihai, O.S. Mitochondrial fusion, fission and autophagy as a quality control axis: The bioenergetic view. Biochim. Biophys. Acta (BBA) Bioenerg. 2008, 1777, 1092–1097. [Google Scholar] [CrossRef]
- Twig, G.; Elorza, A.; Molina, A.J.A.; Mohamed, H.; Wikstrom, J.D.; Walzer, G.; Stiles, L.; Haigh, S.E.; Katz, S.; Las, G.; et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008, 27, 433–446. [Google Scholar] [CrossRef]
- Chen, H.; Vermulst, M.; Wang, Y.E.; Chomyn, A.; Prolla, T.A.; McCaffery, J.M.; Chan, D.C. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 2010, 141, 280–289. [Google Scholar] [CrossRef]
- Favaro, G.; Romanello, V.; Varanita, T.; Desbats, M.A.; Morbidoni, V.; Tezze, C.; Albiero, M.; Canato, M.; Gherardi, G.; De Stefani, D.; et al. DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat. Commun. 2019, 10, 2576. [Google Scholar] [CrossRef] [PubMed]
- Romanello, V.; Guadagnin, E.; Gomes, L.; Roder, I.; Sandri, C.; Petersen, Y.; Milan, G.; Masiero, E.; Del Piccolo, P.; Foretz, M.; et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010, 29, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- Touvier, T.; De Palma, C.; Rigamonti, E.; Scagliola, A.; Incerti, E.; Mazelin, L.; Thomas, J.-L.; D’Antonio, M.; Politi, L.; Schaeffer, L.; et al. Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. 2015, 6, e1663. [Google Scholar] [CrossRef]
- Gomes, L.C.; Di Benedetto, G.; Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011, 13, 589–598. [Google Scholar] [CrossRef]
- Mitra, K.; Wunder, C.; Roysam, B.; Lin, G.; Lippincott-Schwartz, J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. USA 2009, 106, 11960–11965. [Google Scholar] [CrossRef]
- Tondera, D.; Grandemange, S.; Jourdain, A.; Karbowski, M.; Mattenberger, Y.; Herzig, S.; Da Cruz, S.; Clerc, P.; Raschke, I.; Merkwirth, C.; et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009, 28, 1589–1600. [Google Scholar] [CrossRef]
- Drazen, J.C.; Dugan, B.; Friedman, J.R. Red muscle proportions and enzyme activities in deep-sea demersal fishes. J. Fish Biol. 2013, 83, 1592–1612. [Google Scholar] [CrossRef] [PubMed]
- Teulier, L.; Thoral, E.; Queiros, Q.; McKenzie, D.J.; Roussel, D.; Dutto, G.; Gasset, E.; Bourjea, J.; Saraux, C. Muscle bioenergetics of two emblematic Mediterranean fish species: Sardina pilchardus and Sparus aurata. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2019, 235, 174–179. [Google Scholar] [CrossRef] [PubMed]



| Protein Name | Cat. Num | Company |
|---|---|---|
| Hsp60 | GB11243 | Servicebio, Wuhan, China |
| Tfam | 22586-1-AP | Proteinintech, Tokyo, Japan |
| GAPDH | GB11243 | Servicebio, Wuhan, China |
| OPA1 | 612606 | BD, Franklin Lakes, NJ, USA |
| MFN2 | ab56889 | Abcam, Cambridge, UK |
| Gene | Sequences (5′-3′) | Application |
|---|---|---|
| nd1 | TACTAGCCGTAGCTTTCTTA | mtDNA copy number analysis |
| GGTCGGACTGGTTCTTT | ||
| β-actin | CCAGAAAGACAGCTACGTTGG | |
| GCAACTCTCAGCTCGTTGTAG | ||
| coi | TGATTGGAGGCTTTGGGA | mitochondrial-encoded genes |
| CAGAAGGAGGAAGGATGG | ||
| cytb | ACGATTCTTTGCCTTCCA | |
| AATAGGGCGAGTGTTGC | ||
| fis1 | GCAAATACACGGAGGACAT | mitochondrial fission |
| CTTCAGGGCTTTCTCGTAG | ||
| drp1 | CATTTCAAACCCAAACTCCA | |
| CGCATCTGCCACCAAC | ||
| mfn2 | GCCACTCAAGCATTTCGTC | mitochondrial fusion |
| CTCGTCATTCTTGTGAGTATCT | ||
| opa1 | CCGCACACAGTTAAAATATCAG | |
| TGGTCCTGGGTGTTGTAG | ||
| atg5 | CGAGCCTTACTATCTGTTGC | mitochondrial autophagy |
| GTCCCTTCATACTCAAACCA | ||
| pink1 | CTGGTGGTCAGTGACTTTGGC | |
| GCGTCCGCTTTGCTGTAG | ||
| pgc-1α | GGCGACATTGCACCGAGTT | mitochondrial biogenesis |
| GCCCTTGGCGTCGTTATTT | ||
| tfam | AAGCGTCCTCGCTCAC | |
| TTAGGTCTTCCCGTCCA | ||
| rpl19 | GTCTCATCATCCGCAAACC | reference gene |
| TCTCAGGCATACGAGCATT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Wang, H.; Zeng, X.; Liu, S.; Zhuang, Z. Mitochondrial Homeostasis Regulating Mitochondrial Number and Morphology Is a Distinguishing Feature of Skeletal Muscle Fiber Types in Marine Teleosts. Int. J. Mol. Sci. 2024, 25, 1512. https://doi.org/10.3390/ijms25031512
Li B, Wang H, Zeng X, Liu S, Zhuang Z. Mitochondrial Homeostasis Regulating Mitochondrial Number and Morphology Is a Distinguishing Feature of Skeletal Muscle Fiber Types in Marine Teleosts. International Journal of Molecular Sciences. 2024; 25(3):1512. https://doi.org/10.3390/ijms25031512
Chicago/Turabian StyleLi, Busu, Huan Wang, Xianghui Zeng, Shufang Liu, and Zhimeng Zhuang. 2024. "Mitochondrial Homeostasis Regulating Mitochondrial Number and Morphology Is a Distinguishing Feature of Skeletal Muscle Fiber Types in Marine Teleosts" International Journal of Molecular Sciences 25, no. 3: 1512. https://doi.org/10.3390/ijms25031512
APA StyleLi, B., Wang, H., Zeng, X., Liu, S., & Zhuang, Z. (2024). Mitochondrial Homeostasis Regulating Mitochondrial Number and Morphology Is a Distinguishing Feature of Skeletal Muscle Fiber Types in Marine Teleosts. International Journal of Molecular Sciences, 25(3), 1512. https://doi.org/10.3390/ijms25031512





