Thymic-Epithelial-Cell-Dependent Microenvironment Influences Proliferation and Apoptosis of Leukemic Cells
Abstract
1. Introduction
2. Results
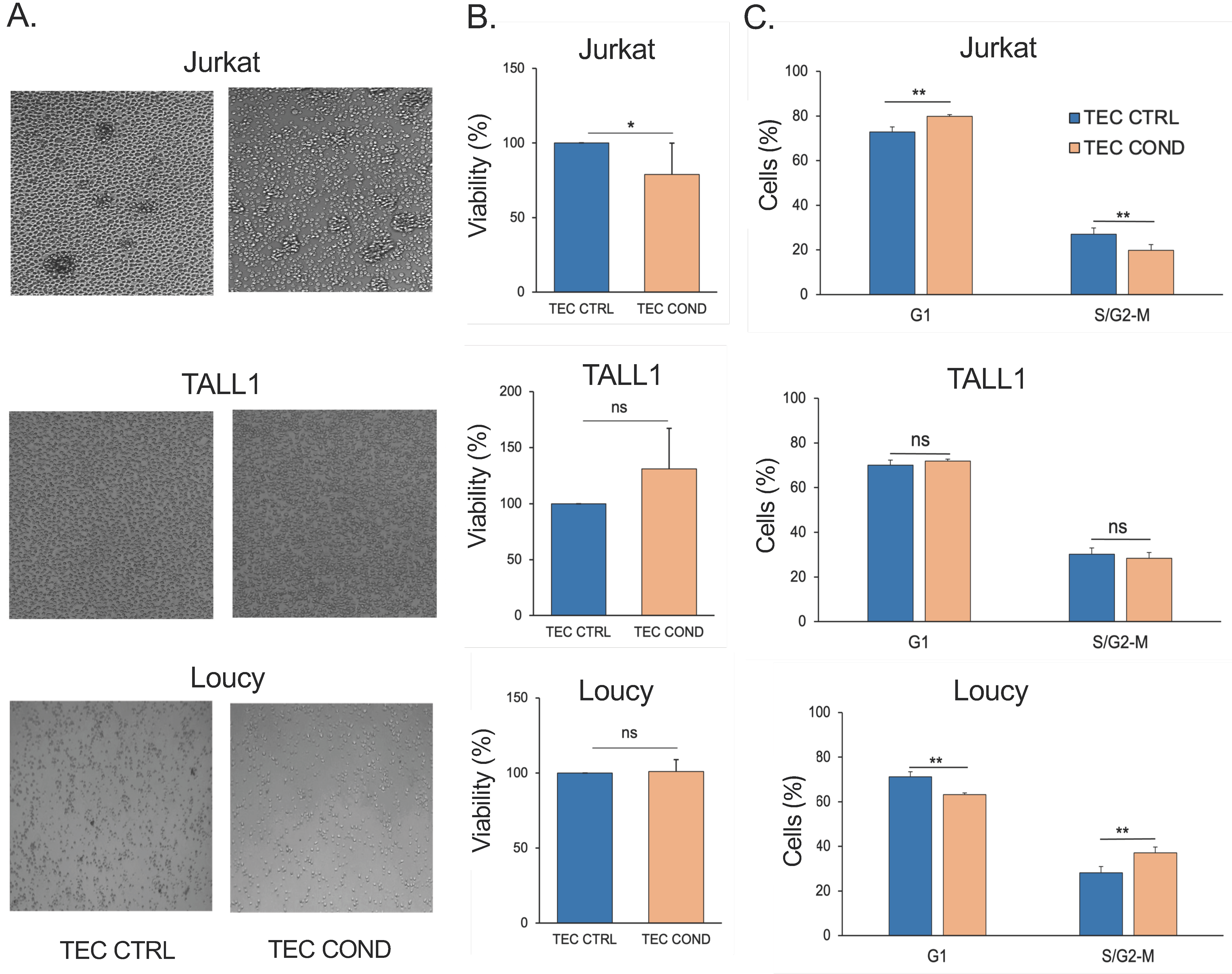
2.1. TEC-Conditioned Medium Provides a Microenvironment Influencing T-ALL Cell Viability
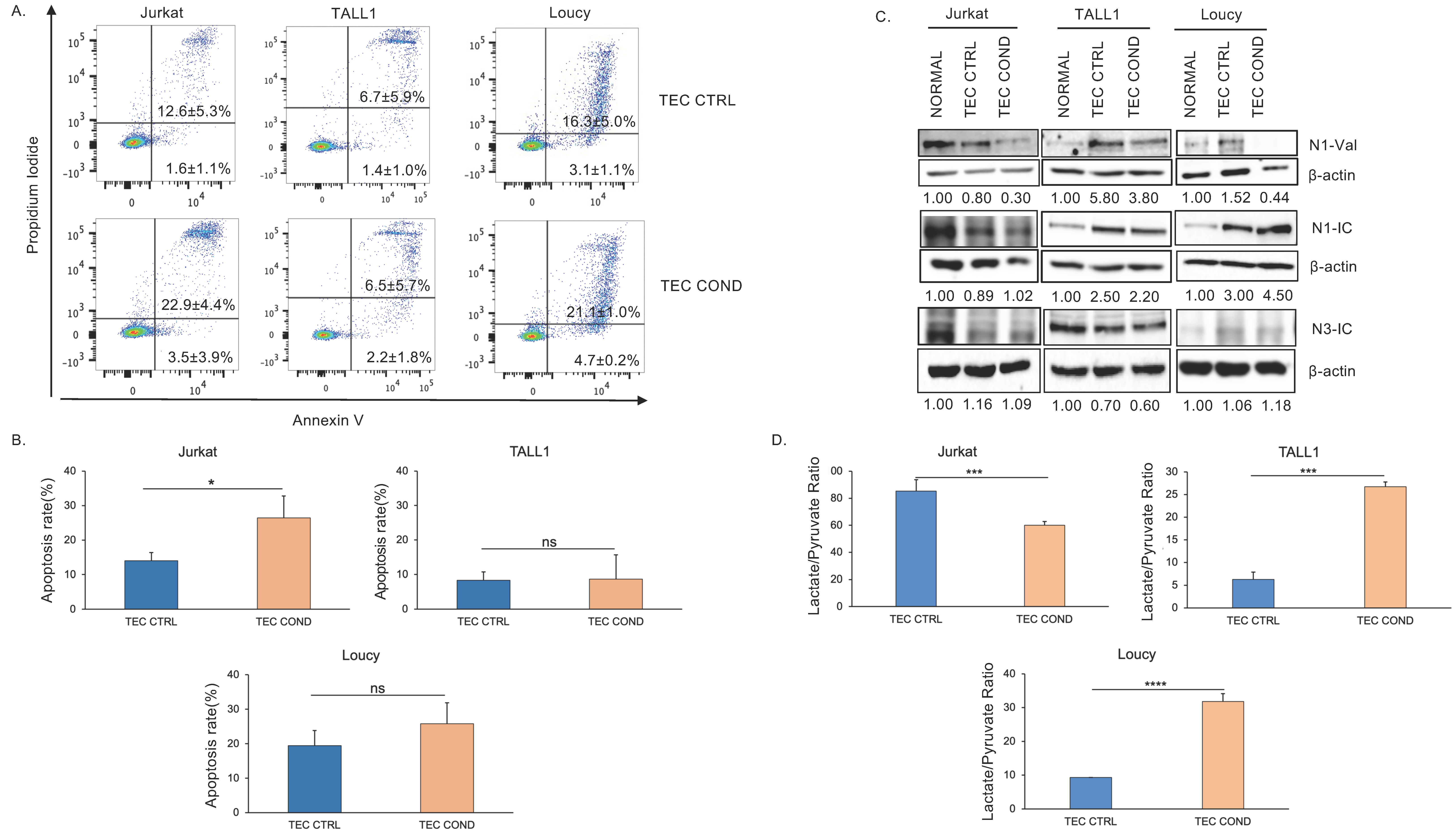
2.2. Jurkat, TALL1, and Loucy Cells Differ in Their Biological and Metabolic Profile when Cultured in a TEC-Conditioned Medium
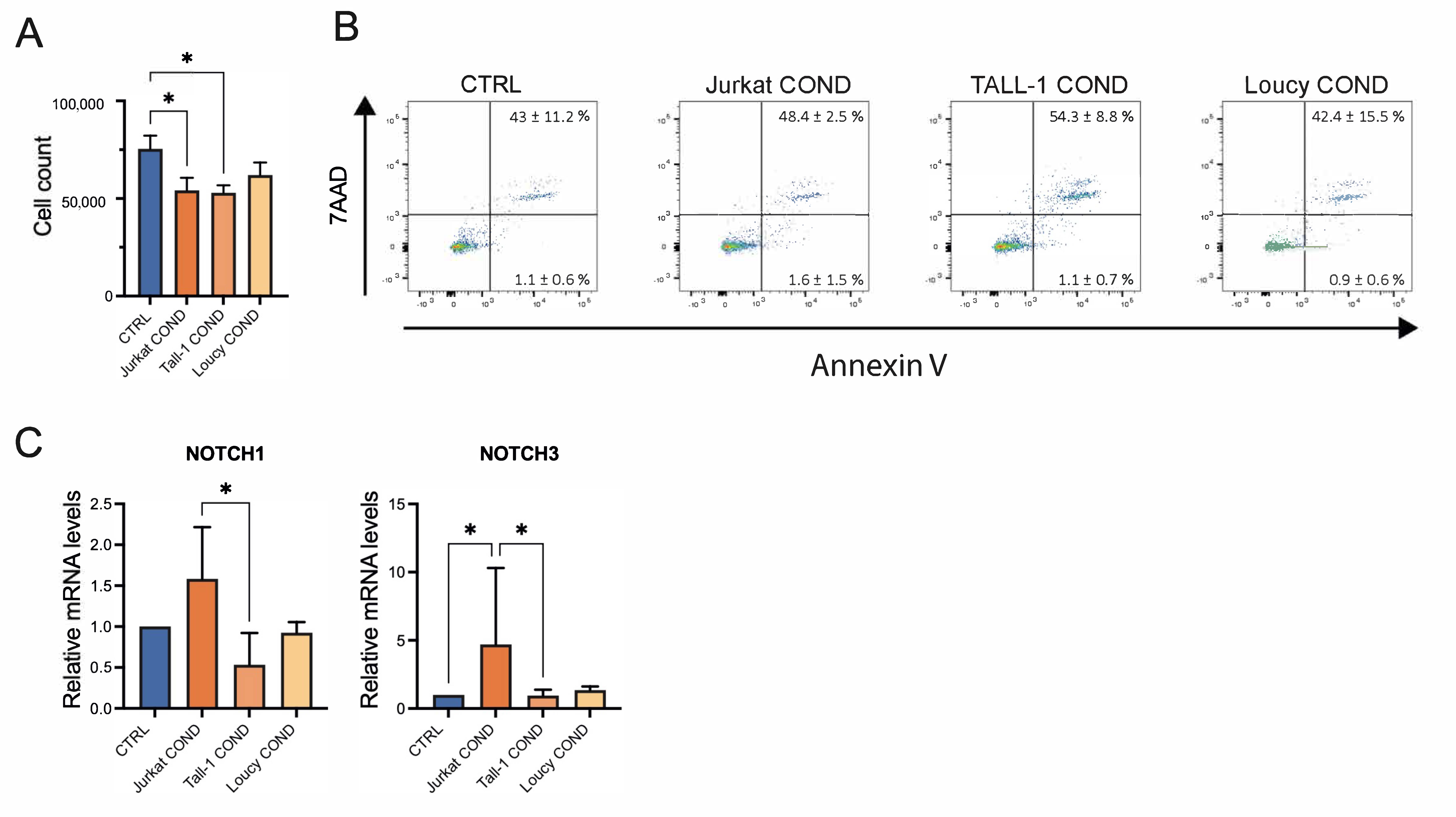
2.3. Leukemic Cells Conditioned Medium Regulates Survival and Gene Expression of TECs
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Generation of Human TEC Culture
4.2. Culture Conditions (Cell Viability and Morphology)
4.3. Western Blot Analysis
4.4. Flow Cytometry Analysis (Cell Cycle and Apoptosis)
4.5. Lactate and Pyruvate Assay
4.6. Gene Expression Analysis
4.7. Statistical Analysis
4.8. Patient Samples
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiaretti, S.; Li, X.; Gentleman, R.; Vitale, A.; Vignetti, M.; Mandelli, F.; Ritz, J.; Foa, R. Gene expression profile of adult T-cell acute lymphocytic leukemia identifies distinct subsets of patients with different response to therapy and survival. Blood 2004, 103, 2771–2778. [Google Scholar] [CrossRef]
- Shiraz, P.; Jehangir, W.; Agrawal, V. T-Cell Acute Lymphoblastic Leukemia-Current Concepts in Molecular Biology and Management. Biomedicines 2021, 9, 1621. [Google Scholar] [CrossRef]
- Fattizzo, B.; Rosa, J.; Giannotta, J.A.; Baldini, L.; Fracchiolla, N.S. The Physiopathology of T-Cell Acute Lymphoblastic Leukemia: Focus on Molecular Aspects. Front. Oncol. 2020, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Tsaouli, G.; Barbarulo, A.; Vacca, A.; Screpanti, I.; Felli, M.P. Molecular Mechanisms of Notch Signaling in Lymphoid Cell Lineages Development: NF-kappaB and Beyond. Adv. Exp. Med. Biol. 2020, 1227, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Grazioli, P.; Felli, M.P.; Screpanti, I.; Campese, A.F. The mazy case of Notch and immunoregulatory cells. J. Leukoc. Biol. 2017, 102, 361–368. [Google Scholar] [CrossRef]
- Campese, A.F.; Bellavia, D.; Gulino, A.; Screpanti, I. Notch signalling at the crossroads of T cell development and leukemogenesis. Semin. Cell Dev. Biol. 2003, 14, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Zhdanovskaya, N.; Firrincieli, M.; Lazzari, S.; Pace, E.; Scribani Rossi, P.; Felli, M.P.; Talora, C.; Screpanti, I.; Palermo, R. Targeting Notch to Maximize Chemotherapeutic Benefits: Rationale, Advanced Strategies, and Future Perspectives. Cancers 2021, 13, 5106. [Google Scholar] [CrossRef]
- Bernasconi-Elias, P.; Hu, T.; Jenkins, D.; Firestone, B.; Gans, S.; Kurth, E.; Capodieci, P.; Deplazes-Lauber, J.; Petropoulos, K.; Thiel, P.; et al. Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies. Oncogene 2016, 35, 6077–6086. [Google Scholar] [CrossRef]
- Scupoli, M.T.; Donadelli, M.; Cioffi, F.; Rossi, M.; Perbellini, O.; Malpeli, G.; Corbioli, S.; Vinante, F.; Krampera, M.; Palmieri, M.; et al. Bone marrow stromal cells and the upregulation of interleukin-8 production in human T-cell acute lymphoblastic leukemia through the CXCL12/CXCR4 axis and the NF-kappaB and JNK/AP-1 pathways. Haematologica 2008, 93, 524–532. [Google Scholar] [CrossRef]
- Vadillo, E.; Dorantes-Acosta, E.; Pelayo, R.; Schnoor, M. T cell acute lymphoblastic leukemia (T-ALL): New insights into the cellular origins and infiltration mechanisms common and unique among hematologic malignancies. Blood Rev. 2018, 32, 36–51. [Google Scholar] [CrossRef]
- Matthijssens, F.; Sharma, N.D.; Nysus, M.; Nickl, C.K.; Kang, H.; Perez, D.R.; Lintermans, B.; Van Loocke, W.; Roels, J.; Peirs, S.; et al. RUNX2 regulates leukemic cell metabolism and chemotaxis in high-risk T cell acute lymphoblastic leukemia. J. Clin. Investig. 2021, 131, e141566. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, H.; Guo, W.; Zhou, X.; Shu, Y.; Liu, H.; Yang, L.; Tang, S.; Su, H.; Liu, Z.; et al. MiR-652-5p elevated glycolysis level by targeting TIGAR in T-cell acute lymphoblastic leukemia. Cell Death Dis. 2022, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Passaro, D.; Irigoyen, M.; Catherinet, C.; Gachet, S.; Da Costa De Jesus, C.; Lasgi, C.; Tran Quang, C.; Ghysdael, J. CXCR4 Is Required for Leukemia-Initiating Cell Activity in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2015, 27, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Ferrandino, F.; Bernardini, G.; Tsaouli, G.; Grazioli, P.; Campese, A.F.; Noce, C.; Ciuffetta, A.; Vacca, A.; Besharat, Z.M.; Bellavia, D.; et al. Intrathymic Notch3 and CXCR4 combinatorial interplay facilitates T-cell leukemia propagation. Oncogene 2018, 37, 6285–6298. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Anderson, G. Thymic Epithelial Cells. Annu. Rev. Immunol. 2017, 35, 85–118. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Leon, M.J.; Mosquera, M.; Cela, C.; Alcain, J.; Zuklys, S.; Hollander, G.; Toribio, M.L. Abrogation of Notch Signaling in Embryonic TECs Impacts Postnatal mTEC Homeostasis and Thymic Involution. Front. Immunol. 2022, 13, 867302. [Google Scholar] [CrossRef]
- Toribio, M.L.; Gonzalez-Garcia, S. Notch Partners in the Long Journey of T-ALL Pathogenesis. Int. J. Mol. Sci. 2023, 24, 1383. [Google Scholar] [CrossRef]
- Dagklis, A.; Demeyer, S.; De Bie, J.; Radaelli, E.; Pauwels, D.; Degryse, S.; Gielen, O.; Vicente, C.; Vandepoel, R.; Geerdens, E.; et al. Hedgehog pathway activation in T-cell acute lymphoblastic leukemia predicts response to SMO and GLI1 inhibitors. Blood 2016, 128, 2642–2654. [Google Scholar] [CrossRef]
- Tosello, V.; Bongiovanni, D.; Liu, J.; Pan, Q.; Yan, K.K.; Saccomani, V.; Van Trimpont, M.; Pizzi, M.; Mazzoni, M.; Dei Tos, A.P.; et al. Cross-talk between GLI transcription factors and FOXC1 promotes T-cell acute lymphoblastic leukemia dissemination. Leukemia 2021, 35, 984–1000. [Google Scholar] [CrossRef]
- Klein, L.; Kyewski, B.; Allen, P.M.; Hogquist, K.A. Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 2014, 14, 377–391. [Google Scholar] [CrossRef]
- Anderson, G.; Jenkinson, E.J. Lymphostromal interactions in thymic development and function. Nat. Rev. Immunol. 2001, 1, 31–40. [Google Scholar] [CrossRef]
- Pitt, L.A.; Tikhonova, A.N.; Hu, H.; Trimarchi, T.; King, B.; Gong, Y.; Sanchez-Martin, M.; Tsirigos, A.; Littman, D.R.; Ferrando, A.A.; et al. CXCL12-Producing Vascular Endothelial Niches Control Acute T Cell Leukemia Maintenance. Cancer Cell 2015, 27, 755–768. [Google Scholar] [CrossRef]
- Tsaouli, G.; Ferretti, E.; Bellavia, D.; Vacca, A.; Felli, M.P. Notch/CXCR4 Partnership in Acute Lymphoblastic Leukemia Progression. J. Immunol. Res. 2019, 2019, 5601396. [Google Scholar] [CrossRef] [PubMed]
- Lucas, B.; White, A.J.; Parnell, S.M.; Henley, P.M.; Jenkinson, W.E.; Anderson, G. Progressive Changes in CXCR4 Expression That Define Thymocyte Positive Selection Are Dispensable for Both Innate and Conventional alphabetaT-cell Development. Sci. Rep. 2017, 7, 5068. [Google Scholar] [CrossRef]
- Hernandez-Lopez, C.; Varas, A.; Sacedon, R.; Jimenez, E.; Munoz, J.J.; Zapata, A.G.; Vicente, A. Stromal cell-derived factor 1/CXCR4 signaling is critical for early human T-cell development. Blood 2002, 99, 546–554. [Google Scholar] [CrossRef]
- Koch, U.; Fiorini, E.; Benedito, R.; Besseyrias, V.; Schuster-Gossler, K.; Pierres, M.; Manley, N.R.; Duarte, A.; Macdonald, H.R.; Radtke, F. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J. Exp. Med. 2008, 205, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; Campese, A.F.; Vacca, A.; Gulino, A.; Screpanti, I. Notch3, another Notch in T cell development. Semin. Immunol. 2003, 15, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Velardi, E.; Tsai, J.J.; Holland, A.M.; Wertheimer, T.; Yu, V.W.; Zakrzewski, J.L.; Tuckett, A.Z.; Singer, N.V.; West, M.L.; Smith, O.M.; et al. Sex steroid blockade enhances thymopoiesis by modulating Notch signaling. J. Exp. Med. 2014, 211, 2341–2349. [Google Scholar] [CrossRef]
- Rosichini, M.; Catanoso, M.; Screpanti, I.; Felli, M.P.; Locatelli, F.; Velardi, E. Signaling Crosstalks Drive Generation and Regeneration of the Thymus. Front. Immunol. 2022, 13, 920306. [Google Scholar] [CrossRef]
- Hozumi, K.; Mailhos, C.; Negishi, N.; Hirano, K.; Yahata, T.; Ando, K.; Zuklys, S.; Hollander, G.A.; Shima, D.T.; Habu, S. Delta-like 4 is indispensable in thymic environment specific for T cell development. J. Exp. Med. 2008, 205, 2507–2513. [Google Scholar] [CrossRef]
- Sison, E.A.; Magoon, D.; Li, L.; Annesley, C.E.; Romagnoli, B.; Douglas, G.J.; Tuffin, G.; Zimmermann, J.; Brown, P. POL5551, a novel and potent CXCR4 antagonist, enhances sensitivity to chemotherapy in pediatric ALL. Oncotarget 2015, 6, 30902–30918. [Google Scholar] [CrossRef] [PubMed]
- Scupoli, M.T.; Vinante, F.; Krampera, M.; Vincenzi, C.; Nadali, G.; Zampieri, F.; Ritter, M.A.; Eren, E.; Santini, F.; Pizzolo, G. Thymic epithelial cells promote survival of human T-cell acute lymphoblastic leukemia blasts: The role of interleukin-7. Haematologica 2003, 88, 1229–1237. [Google Scholar] [PubMed]
- Ghezzo, M.N.; Fernandes, M.T.; Pacheco-Leyva, I.; Rodrigues, P.M.; Machado, R.S.; Araujo, M.A.S.; Kalathur, R.K.; Futschik, M.E.; Alves, N.L.; Dos Santos, N.R. FoxN1-dependent thymic epithelial cells promote T-cell leukemia development. Carcinogenesis 2018, 39, 1463–1476. [Google Scholar] [CrossRef] [PubMed]
- Wu, I.; Shin, S.C.; Cao, Y.; Bender, I.K.; Jafari, N.; Feng, G.; Lin, S.; Cidlowski, J.A.; Schleimer, R.P.; Lu, N.Z. Selective glucocorticoid receptor translational isoforms reveal glucocorticoid-induced apoptotic transcriptomes. Cell Death Dis. 2013, 4, e453. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Severson, E.; Pear, W.S.; Liu, X.S.; Aster, J.C.; Blacklow, S.C. The common oncogenomic program of NOTCH1 and NOTCH3 signaling in T-cell acute lymphoblastic leukemia. PLoS ONE 2017, 12, e0185762. [Google Scholar] [CrossRef] [PubMed]
- Go, S.; Kramer, T.T.; Verhoeven, A.J.; Oude Elferink, R.P.J.; Chang, J.C. The extracellular lactate-to-pyruvate ratio modulates the sensitivity to oxidative stress-induced apoptosis via the cytosolic NADH/NAD(+) redox state. Apoptosis 2021, 26, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Chonghaile, T.N.; Roderick, J.E.; Glenfield, C.; Ryan, J.; Sallan, S.E.; Silverman, L.B.; Loh, M.L.; Hunger, S.P.; Wood, B.; DeAngelo, D.J.; et al. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer Discov. 2014, 4, 1074–1087. [Google Scholar] [CrossRef]
- Zhang, M.; Lin, X.; Yang, Z.; Li, X.; Zhou, Z.; Love, P.E.; Huang, J.; Zhao, B. Metabolic regulation of T cell development. Front. Immunol. 2022, 13, 946119. [Google Scholar] [CrossRef]
- Peirs, S.; Matthijssens, F.; Goossens, S.; Van de Walle, I.; Ruggero, K.; de Bock, C.E.; Degryse, S.; Cante-Barrett, K.; Briot, D.; Clappier, E.; et al. ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia. Blood 2014, 124, 3738–3747. [Google Scholar] [CrossRef]
- Zhang, C.; Quinones, A.; Le, A. Metabolic reservoir cycles in cancer. Semin. Cancer Biol. 2022, 86, 180–188. [Google Scholar] [CrossRef]
- Issa, N.; Bjeije, H.; Wilson, E.R.; Krishnan, A.; Dunuwille, W.M.B.; Parsons, T.M.; Zhang, C.R.; Han, W.; Young, A.L.; Ren, Z.; et al. KDM6B protects T-ALL cells from NOTCH1-induced oncogenic stress. Leukemia 2023, 37, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Herranz, D.; Ambesi-Impiombato, A.; Sudderth, J.; Sanchez-Martin, M.; Belver, L.; Tosello, V.; Xu, L.; Wendorff, A.A.; Castillo, M.; Haydu, J.E.; et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat. Med. 2015, 21, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gordon, J.; Chen, E.L.Y.; Xiao, S.; Wu, L.; Zuniga-Pflucker, J.C.; Manley, N.R. NOTCH1 signaling establishes the medullary thymic epithelial cell progenitor pool during mouse fetal development. Development 2020, 147, dev178988. [Google Scholar] [CrossRef]
- Tasca, A.; Helmstadter, M.; Brislinger, M.M.; Haas, M.; Mitchell, B.; Walentek, P. Notch signaling induces either apoptosis or cell fate change in multiciliated cells during mucociliary tissue remodeling. Dev. Cell 2021, 56, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Green, H.; Kehinde, O.; Thomas, J. Growth of cultured human epidermal cells into multiple epithelia suitable for grafting. Proc. Natl. Acad. Sci. USA 1979, 76, 5665–5668. [Google Scholar] [CrossRef]
- Rosichini, M.; Bordoni, V.; Silvestris, D.A.; Mariotti, D.; Matusali, G.; Cardinale, A.; Zambruno, G.; Condorelli, A.G.; Flamini, S.; Genah, S.; et al. SARS-CoV-2 infection of thymus induces loss of function that correlates with disease severity. J. Allergy Clin. Immunol. 2023, 151, 911–921. [Google Scholar] [CrossRef]
- Giuli, M.V.; Diluvio, G.; Giuliani, E.; Franciosa, G.; Di Magno, L.; Pignataro, M.G.; Tottone, L.; Nicoletti, C.; Besharat, Z.M.; Peruzzi, G.; et al. Notch3 contributes to T-cell leukemia growth via regulation of the unfolded protein response. Oncogenesis 2020, 9, 93. [Google Scholar] [CrossRef]
- Paik, M.J.; Cho, E.Y.; Kim, H.; Kim, K.R.; Choi, S.; Ahn, Y.H.; Lee, G. Simultaneous clinical monitoring of lactic acid, pyruvic acid and ketone bodies in plasma as methoxime/tert-butyldimethylsilyl derivatives by gas chromatography-mass spectrometry in selected ion monitoring mode. Biomed. Chromatogr. 2008, 22, 450–453. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, S.K.; Zhdanovskaya, N.; Sergio, I.; Cardinale, A.; Rosichini, M.; Varricchio, C.; Pace, E.; Capalbo, C.; Locatelli, F.; Macone, A.; et al. Thymic-Epithelial-Cell-Dependent Microenvironment Influences Proliferation and Apoptosis of Leukemic Cells. Int. J. Mol. Sci. 2024, 25, 1412. https://doi.org/10.3390/ijms25031412
Patel SK, Zhdanovskaya N, Sergio I, Cardinale A, Rosichini M, Varricchio C, Pace E, Capalbo C, Locatelli F, Macone A, et al. Thymic-Epithelial-Cell-Dependent Microenvironment Influences Proliferation and Apoptosis of Leukemic Cells. International Journal of Molecular Sciences. 2024; 25(3):1412. https://doi.org/10.3390/ijms25031412
Chicago/Turabian StylePatel, Sandesh Kumar, Nadezda Zhdanovskaya, Ilaria Sergio, Antonella Cardinale, Marco Rosichini, Claudia Varricchio, Eleonora Pace, Carlo Capalbo, Franco Locatelli, Alberto Macone, and et al. 2024. "Thymic-Epithelial-Cell-Dependent Microenvironment Influences Proliferation and Apoptosis of Leukemic Cells" International Journal of Molecular Sciences 25, no. 3: 1412. https://doi.org/10.3390/ijms25031412
APA StylePatel, S. K., Zhdanovskaya, N., Sergio, I., Cardinale, A., Rosichini, M., Varricchio, C., Pace, E., Capalbo, C., Locatelli, F., Macone, A., Velardi, E., Palermo, R., & Felli, M. P. (2024). Thymic-Epithelial-Cell-Dependent Microenvironment Influences Proliferation and Apoptosis of Leukemic Cells. International Journal of Molecular Sciences, 25(3), 1412. https://doi.org/10.3390/ijms25031412







