Affinity Resins for the Isolation of Immunoglobulins G Obtained Using Biocatalytic Technology
Abstract
1. Introduction
2. Results and Discussion
2.1. BsrtA Ligand Design
- The design utilizes the natural sequence of protein A (domain B), which has a high affinity for the Fc fragment of immunoglobulins;
- The leader sequence has been introduced to increase the efficiency of recombinant protein biosynthesis;
- The recombinant protein contains two B domains of protein A followed by each other in order to increase the dynamic capacity of the resin obtained on its basis;
- Six-point mutations of asparagine were performed in the sequence of domain B to increase the resistance of the recombinant protein in alkaline conditions during the resin’s cleaning-in-place process;
- The recognition peptide sequence LPETG was introduced before the polyhistidine tag (HisTag), which is necessary for biocatalytic conjugation of the ligand matrix;
- The HisTag was introduced for affinity purification of the protein during ligand production.
2.2. Preparation of Conjugates
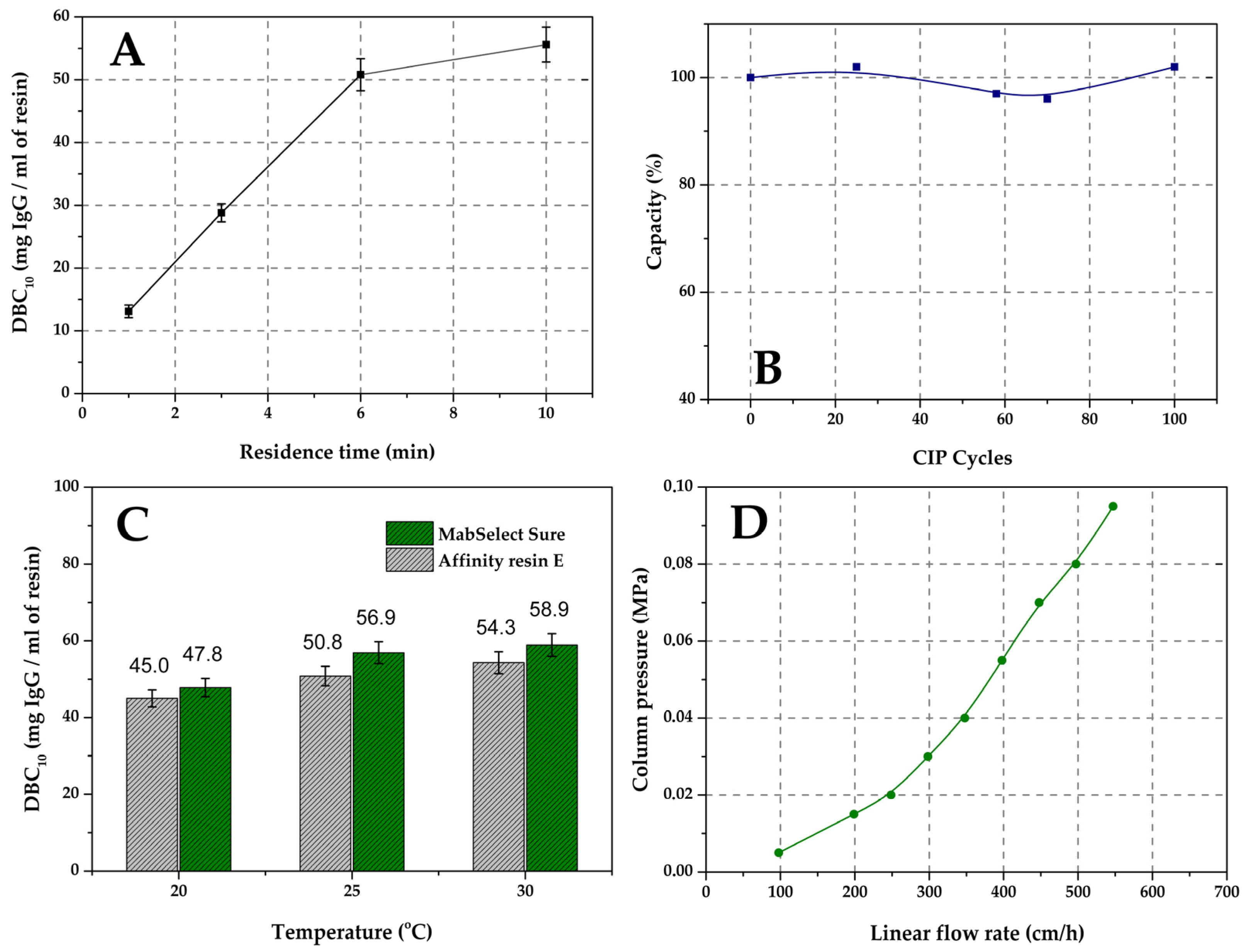
2.3. Characterization of Affinity Resin E
3. Materials and Methods
3.1. General
3.2. Chemistry
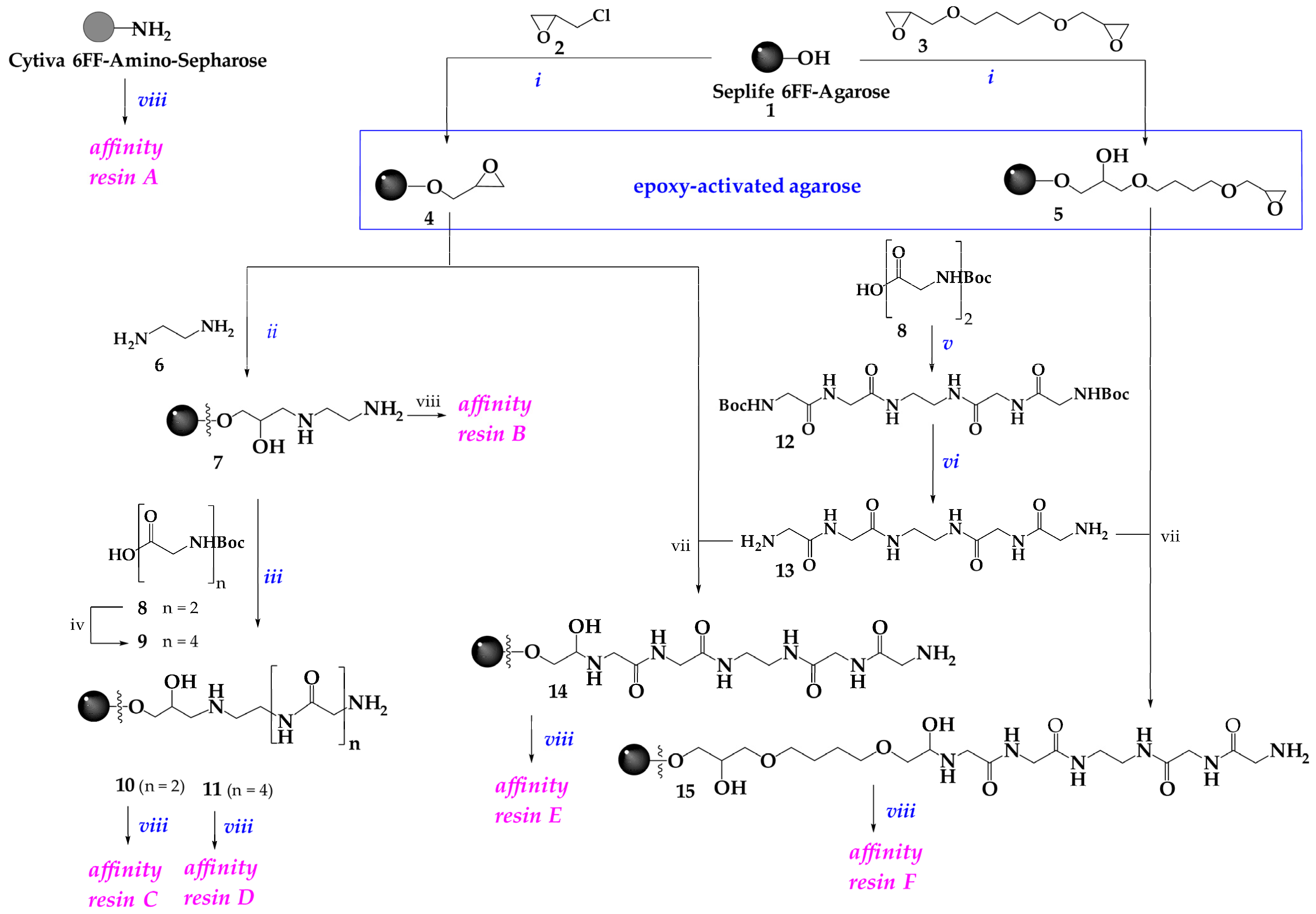
3.2.1. Epoxy-Activated Agarose Derivatives 4 and 5
3.2.2. Determination of the Number of Oxirane Groups Attached to the Matrix
3.2.3. Aminoethyl Agarose (7)
3.2.4. Boc-Gly-Gly-Gly-Gly-OH (9)
3.2.5. Boc-Gly-Gly-EDA-Gly-Gly-Boc (12)
3.2.6. Gly-Gly-EDA-Gly-Gly (13)
3.2.7. General Method for Preparation of polyGly-Seplife 6FF Agaroses 10 and 11
3.2.8. General Method for Preparation of Gly-Gly-EDA-Gly-Gly-Seplife 6FF-Agarose 14 and 15
3.2.9. Determination of the Number of Amino Groups on the Matrix
3.3. Biotechnology
3.3.1. Preparation of Recombinant Sortase A
3.3.2. Enzymatic Immobilization of the BsrtA Protein onto a Matrix Using Sortase A
3.4. Characterization of the Affinity Resins
3.4.1. Estimation of the Dynamic Binding Capacity of the Resins (DBC10) Using Model IgG Antibodies
3.4.2. Cleaning-in-Place Procedure (CIP)
3.4.3. Dependence of Pre-Column Pressure on the Flow Rate
3.4.4. Temperature Dependence of Dynamic Capacity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, O.; Qadan, M.; Ierapetritou, M. Economic Analysis of Batch and Continuous Biopharmaceutical Antibody Production: A Review. J. Pharm. Innov. 2020, 15, 182–200. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, H.; Crescioli, S.; Chenoweth, A.; Visweswaraiah, J.; Reichert, J.M. Antibodies to watch in 2023. MAbs 2023, 15, 2153410. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, T.; Miyahara, M.; Yamamoto, K. Monoclonal antibody purification using activated carbon as a replacement for Protein A affinity chromatography. J. Chromatogr. B 2018, 1102–1103, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Amritkar, V.; Adat, S.; Tejwani, V.; Rathore, A.; Bhambure, R. Engineering Staphylococcal Protein A for high-throughput affinity purification of monoclonal antibodies. Biotechnol. Adv. 2020, 44, 107632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Duan, Y.; Zeng, X. Improved Performance of Recombinant Protein A Immobilized on Agarose Beads by Site-Specific Conjugation. ACS Omega 2017, 2, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Clow, F.; Fraser, J.D.; Proft, T. Immobilization of proteins to biacore sensor chips using Staphylococcus aureus sortase A. Biotechnol. Lett. 2008, 30, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Pishesha, N.; Ingram, J.R.; Ploegh, H.L. Sortase A: A Model for Transpeptidation and Its Biological Applications. Annu. Rev. Cell Dev. Biol. 2018, 34, 163–188. [Google Scholar] [CrossRef]
- Jacobitz, A.W.; Kattke, M.D.; Wereszczynski, J.; Clubb, R.T. Sortase Transpeptidases: Structural Biology and Catalytic Mechanism. Adv. Protein Chem. Struct. Biol. 2017, 109, 223–264. [Google Scholar] [CrossRef]
- Chan, L.; Cross, H.F.; She, J.K.; Cavalli, G.; Martins, H.F.; Neylon, C. Covalent attachment of proteins to solid supports and surfaces via Sortase-mediated ligation. PLoS ONE 2007, 2, e1164. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Subramanian, S.; Boder, E.T. Sortase A as a novel molecular “stapler” for sequence-specific protein conjugation. Bioconjug. Chem. 2007, 18, 469–476. [Google Scholar] [CrossRef]
- Kuropka, B.; Royla, N.; Freund, C.; Krause, E. Sortase A mediated site-specific immobilization for identification of protein interactions in affinity purification-mass spectrometry experiments. Proteomics 2015, 15, 1230–1234. [Google Scholar] [CrossRef]
- Miagkikh, I.V.; Stepanenko, V.N.; Tereshin, M.N.; Melikhova, T.D.; Eletskaya, B.Z. Recombinant Analogue of b-Domain of the Bdpa-1 Protein a, Recombinant Plasmid for the Expression of the Bdpa-1 Protein, Escherichia coli Producer Strain Producing the Bdpa-1 Protein, Method for Producing the Bdpa-1 Protein, and Method for Creating an Affinity Sorbent Containing Bdpa-1 or a Fragment Thereof as a Ligand. Patent RU 2789032 C1, 27 January 2023. [Google Scholar]
- Hirakawa, H.; Ishikawa, S.; Nagamune, T. Design of Ca2+-independent Staphylococcus aureus sortase A mutants. Biotechnol. Bioeng. 2012, 109, 2955–2961. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Ishikawa, S.; Nagamune, T. Ca2+-independent sortase-A exhibits high selective protein ligation activity in the cytoplasm of Escherichia coli. Biotechnol. J. 2015, 10, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Krepper, W.; Satzer, P.; Beyer, B.M.; Jungbauer, A. Temperature dependence of antibody adsorption in protein A affinity chromatography. J. Chromatogr. A 2018, 1551, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhu, B.; Chen, T.; Zhang, X.; Dong, H. Recombinant protein A immobilized on cross-linked cellulose microspheres for immunoglobulin G adsorption from human plasma. BioResources 2016, 11, 4885–4898. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Zhang, X.; Han, N.; Wu, Y.; Wei, D. Oriented covalent immobilization of recombinant protein A on the glutaraldehyde activated agarose support. Int. J. Biol. Macromol. 2018, 120 Pt A, 100–108. [Google Scholar] [CrossRef]
- Matsumoto, I.; Mizuno, Y.; Seno, N. Activation of Sepharose with epichlorohydrin and subsequent immobilization of ligand for affinity adsorbent. J. Biochem. 1979, 85, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Uy, R.; Wold, F. 1,4-Butanediol diglycidyl ether coupling of carbohydrates to Sepharose: Affinity adsorbents for lectins and glycosidases. Anal. Biochem. 1977, 81, 98–107. [Google Scholar] [CrossRef]
- Yang, J.; Sun, L.; Guo, R.; Yang, H.; Feng, X.; Zhang, X. A Facile Route for Oriented Covalent Immobilization of Recombinant Protein A on Epoxy Agarose Gels: In Situ Generation of Heterofunctional Amino-Epoxy Supports. ChemistrySelect 2018, 3, 10320–10324. [Google Scholar] [CrossRef]
- Kaiser, E.; Colescott, R.L.; Bossinger, C.D.; Cook, P.I. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 1970, 34, 595–598. [Google Scholar] [CrossRef] [PubMed]
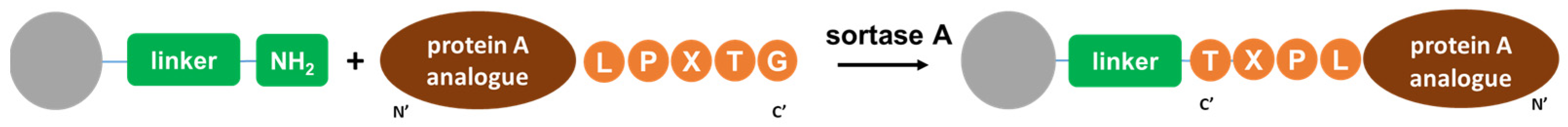

| Affinity Resin | Matrix | Epoxy Activation | Linker | Number of Amino Groups, µmol/mL of Matrix with Linker | DBC10 * with a Residence Time of 6 min, mg IgG/mL of Resin |
|---|---|---|---|---|---|
| A | GE Amino- Sepharose 6F | – | – | 17–22 | 22.7 |
| B | Seplife 6FF | ECH | -EDA | 18.3 | 40.6 |
| C | Seplife 6FF | ECH | -EDA-Gly-Gly | 16.1 | 41.4 |
| D | Seplife 6FF | ECH | -EDA-Gly-Gly Gly-Gly | 18.0 | 44.3 |
| E | Seplife 6FF | ECH | -Gly-Gly-EDA-Gly-Gly | 17.5 | 50.8 |
| F | Seplife 6FF | BDDE | -Gly-Gly-EDA-Gly-Gly | 15.0 | 36.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tereshin, M.N.; Melikhova, T.D.; Eletskaya, B.Z.; Ksenofontova, O.B.; Pantyushenko, P.V.; Berzina, M.Y.; Ivanov, I.; Myagkikh, I.V.; Stepanenko, V.N. Affinity Resins for the Isolation of Immunoglobulins G Obtained Using Biocatalytic Technology. Int. J. Mol. Sci. 2024, 25, 1367. https://doi.org/10.3390/ijms25031367
Tereshin MN, Melikhova TD, Eletskaya BZ, Ksenofontova OB, Pantyushenko PV, Berzina MY, Ivanov I, Myagkikh IV, Stepanenko VN. Affinity Resins for the Isolation of Immunoglobulins G Obtained Using Biocatalytic Technology. International Journal of Molecular Sciences. 2024; 25(3):1367. https://doi.org/10.3390/ijms25031367
Chicago/Turabian StyleTereshin, Mikhail N., Tatiana D. Melikhova, Barbara Z. Eletskaya, Olga B. Ksenofontova, Pavel V. Pantyushenko, Maria Ya. Berzina, Igor Ivanov, Igor V. Myagkikh, and Vasiliy N. Stepanenko. 2024. "Affinity Resins for the Isolation of Immunoglobulins G Obtained Using Biocatalytic Technology" International Journal of Molecular Sciences 25, no. 3: 1367. https://doi.org/10.3390/ijms25031367
APA StyleTereshin, M. N., Melikhova, T. D., Eletskaya, B. Z., Ksenofontova, O. B., Pantyushenko, P. V., Berzina, M. Y., Ivanov, I., Myagkikh, I. V., & Stepanenko, V. N. (2024). Affinity Resins for the Isolation of Immunoglobulins G Obtained Using Biocatalytic Technology. International Journal of Molecular Sciences, 25(3), 1367. https://doi.org/10.3390/ijms25031367







