The Role of ctDNA in the Management of Non-Small-Cell Lung Cancer in the AI and NGS Era
Abstract
1. Introduction
2. Circulating Tumour DNA (ctDNA): Overview, Advantages, and Limitations
2.1. Introduction
2.2. Accuracy
2.3. Invasiveness
2.4. Tumour Heterogeneity
2.5. Cost
3. The Role of NGS in Molecular Analyses of ctDNA
3.1. Introduction
3.2. Clonal Haematopoiesis
3.3. NGS’s Reliability and Diagnostic Challenges
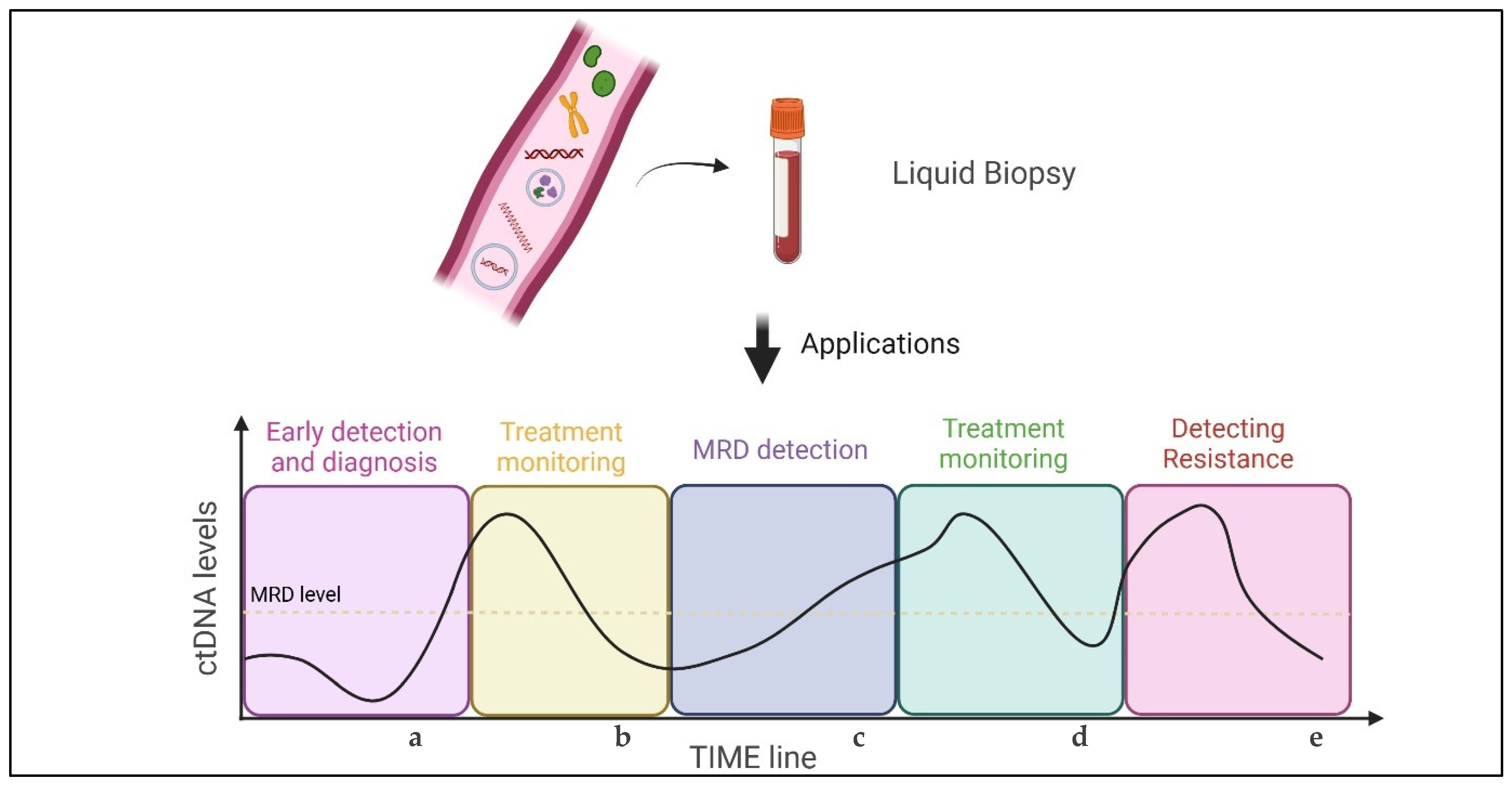
4. Prognostic and Predictive Value of ctDNA: A Brief Overview
4.1. Screening
4.2. Early-Stage Disease
4.3. Advanced NSCLC
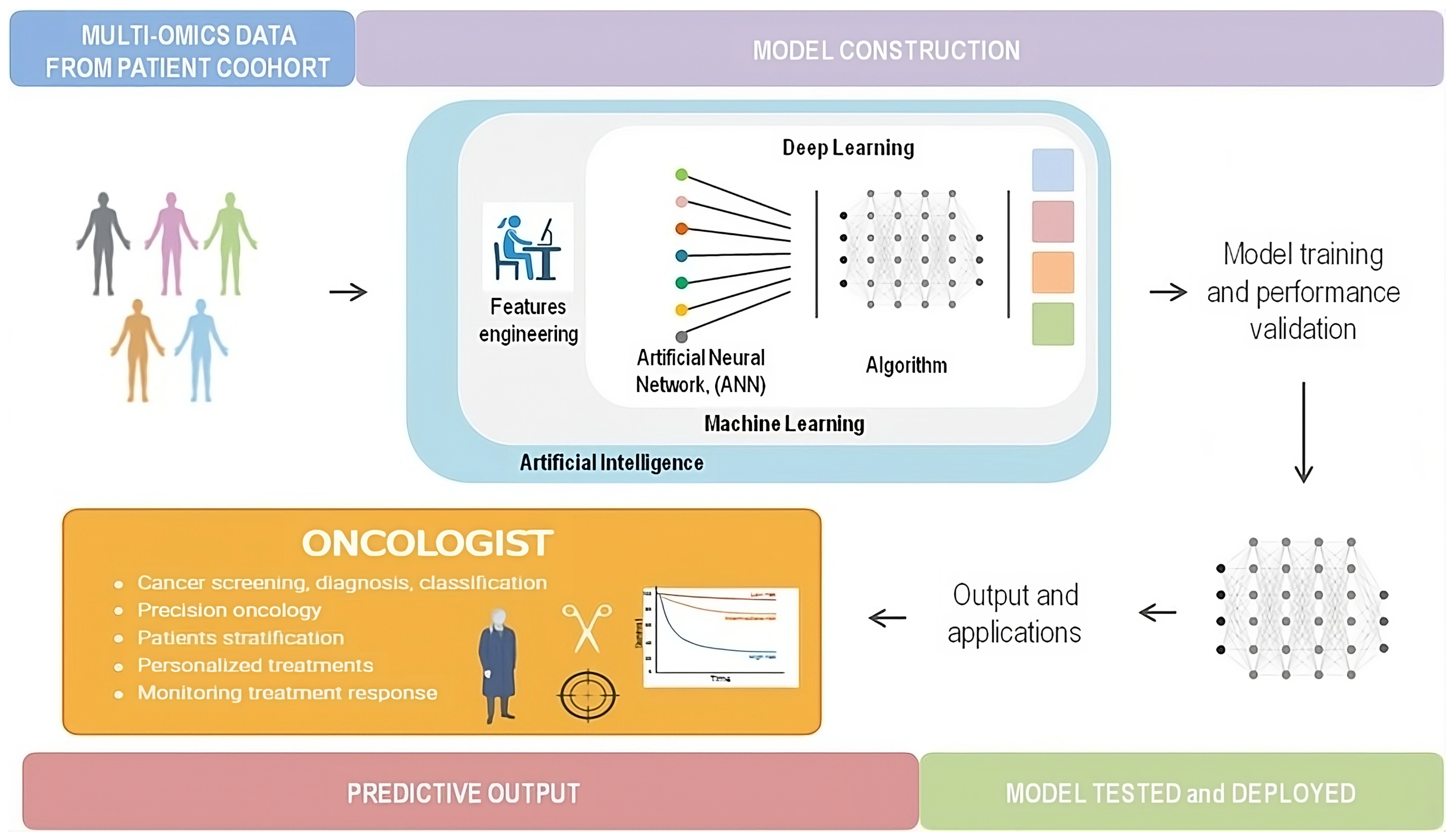
5. Liquid Biopsy and Artificial Intelligence
5.1. Definition
5.2. Integration of Artificial Intelligence into ctDNA Analysis: Advancing Precision Oncology
5.2.1. Screening and Risk Assessment
5.2.2. Early-Stage and MRD Monitoring
5.2.3. Advanced-Stage Treatment Monitoring
5.2.4. Radiomics
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hendriks, L.E.; Kerr, K.M.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Non-oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 358–376. [Google Scholar] [CrossRef] [PubMed]
- Herbreteau, G.; Vallée, A.; Charpentier, S.; Normanno, N.; Hofman, P.; Denis, M.G. Circulating free tumor DNA in non-small cell lung cancer (NSCLC): Clinical application and future perspectives. J. Thorac. Dis. 2019, 11, S113–S126. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Buono, M.; Pisapia, P.; Russo, G.; Tufano, R.; Pepe, F.; Rolfo, C.; Troncone, G. Circulating tumor DNA in cancer: Predictive molecular pathology meets mathematics. Crit. Rev. Oncol. Hematol. 2021, 163, 103394. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J. Circulating tumor DNA (ctDNA) as a biomarker for lung cancer: Early detection, monitoring and therapy prediction. Tumor Biol. 2024, 46, S283–S295. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Li, R.; Wen, H.; Zhou, Q.; Xu, C. Circulating tumor DNA (ctDNA)-based minimal residual disease in non-small cell lung cancer. Chin. Med. J. Pulm. Crit. Care Med. 2023, 1, 207–214. [Google Scholar] [CrossRef]
- Taus, Á.; Camacho, L.; Rocha, P.; Hardy-Werbin, M.; Pijuan, L.; Piquer, G.; López, E.; Dalmases, A.; Longarón, R.; Clavé, S.; et al. Dynamics of EGFR Mutation Load in Plasma for Prediction of Treatment Response and Disease Progression in Patients With EGFR-Mutant Lung Adenocarcinoma. Clin. Lung Cancer 2018, 19, 387–394.e2. [Google Scholar] [CrossRef]
- Goldberg, S.B.; Narayan, A.; Kole, A.J.; Decker, R.H.; Teysir, J.; Carriero, N.J.; Lee, A.; Nemati, R.; Nath, S.K.; Mane, S.M.; et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res. 2018, 24, 1872–1880. [Google Scholar] [CrossRef]
- Mok, T.; Wu, Y.-L.; Lee, J.S.; Yu, C.-J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef]
- Giroux Leprieur, E.; Herbretau, G.; Dumenil, C.; Julie, C.; Giraud, V.; Labrune, S.; Dumoulin, J.; Tisserand, J.; Emile, J.-F.; Blons, H.; et al. Circulating tumor DNA evaluated by Next-Generation Sequencing is predictive of tumor response and prolonged clinical benefit with nivolumab in advanced non-small cell lung cancer. OncoImmunology 2018, 7, e1424675. [Google Scholar] [CrossRef]
- Kato, K.; Uchida, J.; Kukita, Y.; Kumagai, T.; Nishino, K.; Inoue, T.; Kimura, M.; Imamura, F. Transient appearance of circulating tumor DNA associated with de novo treatment. Sci. Rep. 2016, 6, 38639. [Google Scholar] [CrossRef]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Li, R.-Y.; Liang, Z.-Y. Circulating tumor DNA in lung cancer: Real-time monitoring of disease evolution and treatment response. Chin. Med. J. 2020, 133, 2476–2485. [Google Scholar] [CrossRef] [PubMed]
- Lampignano, R.; Neumann, M.H.D.; Weber, S.; Kloten, V.; Herdean, A.; Voss, T.; Groelz, D.; Babayan, A.; Tibbesma, M.; Schlumpberger, M.; et al. Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Work Flows. Clin. Chem. 2020, 66, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Postel, M.; Roosen, A.; Laurent-Puig, P.; Taly, V.; Wang-Renault, S.-F. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: A cancer diagnostic perspective. Expert Rev. Mol. Diagn. 2018, 18, 7–17. [Google Scholar] [CrossRef]
- Binder, C.; Oberndorfer, F.; Müllauer, L. Core needle biopsy for screening detected lung cancer—Does it capture all in light of tumor heterogeneity?—A narrative review. Shanghai Chest 2021, 5, 40. [Google Scholar] [CrossRef]
- Andolfi, M.; Potenza, R.; Capozzi, R.; Liparulo, V.; Puma, F.; Yasufuku, K. The role of bronchoscopy in the diagnosis of early lung cancer: A review. J. Thorac. Dis. 2016, 8, 3329–3337. [Google Scholar] [CrossRef]
- Fernandes, M.G.O.; Cruz-Martins, N.; Machado, J.C.; Costa, J.L.; Hespanhol, V. The value of cell-free circulating tumour DNA profiling in advanced non-small cell lung cancer (NSCLC) management. Cancer Cell Int. 2021, 21, 675. [Google Scholar] [CrossRef]
- Qian, X.; Liu, J.; Sun, Y.; Wang, M.; Lei, H.; Luo, G.; Liu, X.; Xiong, C.; Liu, D.; Liu, J.; et al. Circulating cell-free DNA has a high degree of specificity to detect exon 19 deletions and the single-point substitution mutation L858R in non-small cell lung cancer. Oncotarget 2016, 7, 29154–29165. [Google Scholar] [CrossRef]
- Luo, J.; Shen, L.; Zheng, D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: A systematic review and meta-analysis. Sci. Rep. 2014, 4, 6269. [Google Scholar] [CrossRef]
- Qiu, M.; Wang, J.; Xu, Y.; Ding, X.; Li, M.; Jiang, F.; Xu, L.; Yin, R. Circulating Tumor DNA Is Effective for the Detection of EGFR Mutation in Non–Small Cell Lung Cancer: A Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2015, 24, 206–212. [Google Scholar] [CrossRef]
- Mao, C.; Yuan, J.-Q.; Yang, Z.-Y.; Fu, X.-H.; Wu, X.-Y.; Tang, J.-L. Blood as a Substitute for Tumor Tissue in Detecting EGFR Mutations for Guiding EGFR TKIs Treatment of Nonsmall Cell Lung Cancer: A Systematic Review and Meta-Analysis. Medicine 2015, 94, e775. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Huang, R.; Cao, Y. Detection of epidermal growth factor receptor mutations in peripheral blood circulating tumor DNA in patients with advanced non-small cell lung cancer: A PRISMA-compliant meta-analysis and systematic review. Medicine 2020, 99, e21965. [Google Scholar] [CrossRef] [PubMed]
- Passiglia, F.; Rizzo, S.; Di Maio, M.; Galvano, A.; Badalamenti, G.; Listì, A.; Gulotta, L.; Castiglia, M.; Fulfaro, F.; Bazan, V.; et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: A systematic review and meta-analysis. Sci. Rep. 2018, 8, 13379. [Google Scholar] [CrossRef]
- Franzi, S.; Seresini, G.; Borella, P.; Raviele, P.R.; Bonitta, G.; Croci, G.A.; Bareggi, C.; Tosi, D.; Nosotti, M.; Tabano, S. Liquid biopsy in non-small cell lung cancer: A meta-analysis of state-of-the-art and future perspectives. Front. Genet. 2023, 14, 1254839. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, X.; Wang, F.; Zhang, M.; Sun, B.; Yin, W.; Deng, S.; Wan, Y.; Lu, W. The Diagnostic Accuracy of Liquid Biopsy in EGFR-Mutated NSCLC: A Systematic Review and Meta-Analysis of 40 Studies. SLAS Technol. 2021, 26, 42–54. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; McCormack, R.; Webster, A.; Milenkova, T. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: A phase-IV, open-label, single-arm study. Br. J. Cancer 2014, 110, 55–62. [Google Scholar] [CrossRef]
- Reck, M.; Hagiwara, K.; Han, B.; Tjulandin, S.; Grohé, C.; Yokoi, T.; Morabito, A.; Novello, S.; Arriola, E.; Molinier, O.; et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J. Thorac. Oncol. 2016, 11, 1682–1689. [Google Scholar] [CrossRef]
- Papadimitrakopoulou, V.A.; Han, J.; Ahn, M.; Ramalingam, S.S.; Delmonte, A.; Hsia, T.; Laskin, J.; Kim, S.; He, Y.; Tsai, C.; et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non–small cell lung cancer. Cancer 2020, 126, 373–380. [Google Scholar] [CrossRef]
- Dal Maso, A.; Del Bianco, P.; Cortiula, F.; Nardo, G.; Zulato, E.; Bonanno, L.; Follador, A.; De Maglio, G.; Pasello, G.; Indraccolo, S. EGFR T790M testing through repeated liquid biopsy over time: A real-world multicentric retrospective experience. J. Thorac. Dis. 2022, 14, 3364–3375. [Google Scholar] [CrossRef]
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR -Mutated Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenegre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M.; et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Cole, R.; McWalter, G.; Walker, J.; Dearden, S.; Webster, A.; Milenkova, T.; et al. Gefitinib Treatment in EGFR Mutated Caucasian NSCLC: Circulating-Free Tumor DNA as a Surrogate for Determination of EGFR Status. J. Thorac. Oncol. 2014, 9, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.-H.; Ahn, M.-J.; Kim, D.-W.; Ramalingam, S.S.; Sequist, L.V.; Su, W.-C.; Kim, S.-W.; Kim, J.-H.; Planchard, D.; Felip, E.; et al. Osimertinib in Pretreated T790M-Positive Advanced Non–Small-Cell Lung Cancer: AURA Study Phase II Extension Component. J. Clin. Oncol. 2017, 35, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; Caramella, C.; Jovelet, C.; Lacroix, L.; Lawson, A.; Smalley, S.; Howarth, K.; Gale, D.; Green, E.; Plagnol, V.; et al. Osimertinib benefit inEGFR-mutant NSCLC patients withT790M-mutation detected by circulating tumour DNA. Ann. Oncol. 2017, 28, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Zaman, F.Y.; Subramaniam, A.; Afroz, A.; Samoon, Z.; Gough, D.; Arulananda, S.; Alamgeer, M. Circulating Tumour DNA (ctDNA) as a Predictor of Clinical Outcome in Non-Small Cell Lung Cancer Undergoing Targeted Therapies: A Systematic Review and Meta-Analysis. Cancers 2023, 15, 2425. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Tiseo, M.; Vivancos, A.; Kapp, J.; Serrano, M.J.; Tiemann, M. Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective. J. Mol. Pathol. 2021, 2, 255–273. [Google Scholar] [CrossRef]
- Uozu, S.; Imaizumi, K.; Yamaguchi, T.; Goto, Y.; Kawada, K.; Minezawa, T.; Okamura, T.; Akao, K.; Hayashi, M.; Isogai, S.; et al. Feasibility of tissue re-biopsy in non-small cell lung cancers resistant to previous epidermal growth factor receptor tyrosine kinase inhibitor therapies. BMC Pulm. Med. 2017, 17, 175. [Google Scholar] [CrossRef]
- Chabon, J.J.; Simmons, A.D.; Lovejoy, A.F.; Esfahani, M.S.; Newman, A.M.; Haringsma, H.J.; Kurtz, D.M.; Stehr, H.; Scherer, F.; Karlovich, C.A.; et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat. Commun. 2016, 7, 11815. [Google Scholar] [CrossRef]
- de Bruin, E.C.; McGranahan, N.; Mitter, R.; Salm, M.; Wedge, D.C.; Yates, L.; Jamal-Hanjani, M.; Shafi, S.; Murugaesu, N.; Rowan, A.J.; et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014, 346, 251–256. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Z.; Zhao, S.; Ye, D.; Cai, X.; Cheng, B.; Li, C.; Xiong, S.; Li, J.; Liang, H.; et al. Presence of allele frequency heterogeneity defined by ctDNA profiling predicts unfavorable overall survival of NSCLC. Transl. Lung Cancer Res. 2019, 8, 1045–1050. [Google Scholar] [CrossRef]
- Ezeife, D.A.; Spackman, E.; Juergens, R.A.; Laskin, J.J.; Agulnik, J.S.; Hao, D.; Laurie, S.A.; Law, J.H.; Le, L.W.; Kiedrowski, L.A.; et al. The economic value of liquid biopsy for genomic profiling in advanced non-small cell lung cancer. Ther. Adv. Med. Oncol. 2022, 14, 17588359221112696. [Google Scholar] [CrossRef] [PubMed]
- Marino, P.; Touzani, R.; Perrier, L.; Rouleau, E.; Kossi, D.S.; Zhaomin, Z.; Charrier, N.; Goardon, N.; Preudhomme, C.; Durand-Zaleski, I.; et al. Cost of cancer diagnosis using next-generation sequencing targeted gene panels in routine practice: A nationwide French study. Eur. J. Hum. Genet. 2018, 26, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.H.; Barnes, T.A.; Laskin, J.; Cheema, P.; Liu, G.; Iqbal, M.; Rothenstein, J.; Burkes, R.; Tsao, M.-S.; Leighl, N.B. The Perceived Value of Liquid Biopsy: Results From a Canadian Validation Study of Circulating Tumor DNA T790M Testing—Patient’s Willingness-to-Pay: A Brief Report. JTO Clin. Res. Rep. 2024, 5, 100615. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Laskin, J.; Laurie, S.; Agulnik, J.; Juergens, R.; Ezeife, D.; Law, J.; Le, L.; Kiedrowski, L.; Melosky, B.; et al. P89.03 Demonstrating VALUE of Liquid Biopsy for Lung Cancer in a Public Healthcare System. J. Thorac. Oncol. 2021, 16, S689. [Google Scholar] [CrossRef]
- Foundation Medicine. FDA Approves Foundation Medicine’s FoundationOne® Liquid CDx, a Comprehensive Pan-Tumor Liquid Biopsy Test with Multiple Companion Diagnostic Indications for Patients with Advanced Cancer. 2020. Available online: https://www.foundationmedicine.com/press-releases/fda-approves-foundation-medicine%27s-foundationone%C2%AEliquid-cdx%2C-a-comprehensive-pan-tumor-liquid-biopsy-test-with-multiple-companion-diagnostic-indications-for-patients-with-advanced-cancer (accessed on 27 August 2020).
- FDA Approves First Liquid Biopsy Next-Generation Sequencing Companion Diagnostic Test. Published online 8 November 2020. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test (accessed on 7 August 2020).
- Malapelle, U.; Pisapia, P.; Pepe, F.; Russo, G.; Buono, M.; Russo, A.; Gomez, J.; Khorshid, O.; Mack, P.C.; Rolfo, C.; et al. The evolving role of liquid biopsy in lung cancer. Lung Cancer 2022, 172, 53–64. [Google Scholar] [CrossRef]
- Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.-M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life 2021, 11, 890. [Google Scholar] [CrossRef]
- What Are UMIs and Why Are They Used in High-Throughput Sequencing? Available online: https://dnatech.genomecenter.ucdavis.edu/faqs/what-are-umis-and-why-are-they-used-in-high-throughput-sequencing/ (accessed on 5 November 2024).
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Chan, H.T.; Chin, Y.M.; Nakamura, Y.; Low, S.-K. Clonal Hematopoiesis in Liquid Biopsy: From Biological Noise to Valuable Clinical Implications. Cancers 2020, 12, 2277. [Google Scholar] [CrossRef]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef]
- Li, X.-M.; Li, W.-F.; Lin, J.-T.; Yan, H.-H.; Tu, H.-Y.; Chen, H.-J.; Wang, B.-C.; Wang, Z.; Zhou, Q.; Zhang, X.-C.; et al. Predictive and Prognostic Potential of TP53 in Patients With Advanced Non–Small-Cell Lung Cancer Treated With EGFR-TKI: Analysis of a Phase III Randomized Clinical Trial (CTONG 0901). Clin. Lung Cancer 2021, 22, 100–109.e3. [Google Scholar] [CrossRef] [PubMed]
- Deveson, I.W.; Gong, B.; Lai, K.; LoCoco, J.S.; Richmond, T.A.; Schageman, J.; Zhang, Z.; Novoradovskaya, N.; Willey, J.C.; Jones, W.; et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat. Biotechnol. 2021, 39, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.J.; Stone, E.; Baldwin, D.R.; Vliegenthart, R.; Lee, P.; Fintelmann, F.J. Lung cancer screening. The Lancet 2023, 401, 390–408. [Google Scholar] [CrossRef] [PubMed]
- Marinello, A.; Tagliamento, M.; Pagliaro, A.; Conci, N.; Cella, E.; Vasseur, D.; Remon, J.; Levy, A.; Dall’Olio, F.G.; Besse, B. Circulating tumor DNA to guide diagnosis and treatment of localized and locally advanced non-small cell lung cancer. Cancer Treat. Rev. 2024, 129, 102791. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Vázquez, T.A.; Arce, K.M.; Corassa, M.; Mack, P.C.; Gray, J.E.; Pellini, B. ctDNA for the Evaluation and Management of EGFR-Mutant Non-Small Cell Lung Cancer. Cancers 2024, 16, 940. [Google Scholar] [CrossRef]
- Bayle, A.; Bonastre, J.; Chaltiel, D.; Latino, N.; Rouleau, E.; Peters, S.; Galotti, M.; Bricalli, G.; Besse, B.; Giuliani, R. ESMO study on the availability and accessibility of biomolecular technologies in oncology in Europe. Ann. Oncol. 2023, 34, 934–945. [Google Scholar] [CrossRef]
- Abbosh, C.; Hodgson, D.; Doherty, G.J.; Gale, D.; Black, J.R.M.; Horn, L.; Reis-Filho, J.S.; Swanton, C. Implementing circulating tumor DNA as a prognostic biomarker in resectable non-small cell lung cancer. Trends Cancer 2024, 10, 643–654. [Google Scholar] [CrossRef]
- García-Pardo, M.; Makarem, M.; Li, J.J.N.; Kelly, D.; Leighl, N.B. Integrating circulating-free DNA (cfDNA) analysis into clinical practice: Opportunities and challenges. Br. J. Cancer 2022, 127, 592–602. [Google Scholar] [CrossRef]
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; Esfahani, M.S.; Moding, E.J.; Stehr, H.; Schroers-Martin, J.; Nabet, B.Y.; Chen, B.; Chaudhuri, A.A.; et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020, 580, 245–251. [Google Scholar] [CrossRef]
- Xia, L.; Mei, J.; Kang, R.; Deng, S.; Chen, Y.; Yang, Y.; Feng, G.; Deng, Y.; Gan, F.; Lin, Y.; et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non–Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1). Clin. Cancer Res. 2022, 28, 3308–3317. [Google Scholar] [CrossRef]
- Gale, D.; Heider, K.; Ruiz-Valdepenas, A.; Hackinger, S.; Perry, M.; Marsico, G.; Rundell, V.; Wulff, J.; Sharma, G.; Knock, H.; et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 2022, 33, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Huang, Q.; Yin, W.; Tan, S.; Chen, C.; Liu, W.; Tang, J.; Wang, X.; Zhang, B.; Zou, M.; et al. Circulating Tumor DNA as a Prognostic Biomarker in Localized Non-small Cell Lung Cancer. Front. Oncol. 2020, 10, 561598. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Cascone, T.; Awad, M.M.; Spicer, J.D.; He, J.; Lu, S.; Sepesi, B.; Tanaka, F.; Taube, J.M.; Cornelissen, R.; Havel, L.; et al. Perioperative Nivolumab in Resectable Lung Cancer. N. Engl. J. Med. 2024, 390, 1756–1769. [Google Scholar] [CrossRef] [PubMed]
- Provencio Pulla, M.; Awad, M.; Cascone, T.; Spicer, J.D.; He, J.; Lu, S.; Alexandru, A.; Watanabe, Y.; Cornelissen, R.; Koch, L.D.O.; et al. LBA50 Perioperative nivolumab (NIVO) v placebo (PBO) in patients (pts) with resectable NSCLC: Clinical update from the phase III CheckMate 77T study. Ann. Oncol. 2024, 35, S1239–S1240. [Google Scholar] [CrossRef]
- Heymach, J.V.; Harpole, D.; Mitsudomi, T.; Taube, J.M.; Galffy, G.; Hochmair, M.; Winder, T.; Zukov, R.; Garbaos, G.; Gao, S.; et al. Perioperative Durvalumab for Resectable Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2023, 389, 1672–1684. [Google Scholar] [CrossRef]
- Reck, M.; Gale, D.; Harpole, D.; Taube, J.M.; Mitsudomi, T.; Hochmair, M.J.; Winder, T.; Zhu, Z.; Lai, Z.; Stewart, R.; et al. LBA59 Associations of ctDNA clearance and pathological response with neoadjuvant treatment in patients with resectable NSCLC from the phase III AEGEAN trial. Ann. Oncol. 2023, 34, S1300. [Google Scholar] [CrossRef]
- Waldeck, S.; Mitschke, J.; Wiesemann, S.; Rassner, M.; Andrieux, G.; Deuter, M.; Mutter, J.; Lüchtenborg, A.; Kottmann, D.; Titze, L.; et al. Early assessment of circulating tumor DNA after curative-intent resection predicts tumor recurrence in early-stage and locally advanced non-small-cell lung cancer. Mol. Oncol. 2022, 16, 527–537. [Google Scholar] [CrossRef]
- Zografos, E.; Dimitrakopoulos, F.-I.; Koutras, A. Prognostic Value of Circulating Tumor DNA (ctDNA) in Oncogene-Driven NSCLC: Current Knowledge and Future Perspectives. Cancers 2022, 14, 4954. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Liu, S.-Y.; Gao, W.; Liu, S.-Y.M.; Yan, H.-H.; Ji, L.; Chen, Y.; Gong, Y.; Lu, H.-L.; Lin, J.-T.; et al. Longitudinal Undetectable Molecular Residual Disease Defines Potentially Cured Population in Localized Non–Small Cell Lung Cancer. Cancer Discov. 2022, 12, 1690–1701. [Google Scholar] [CrossRef]
- Hendriks, L.E.; Kerr, K.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Dziadziuszko, R.; Peters, S.; Mok, T.; Camidge, D.R.; Gadgeel, S.M.; Ou, S.-H.I.; Konopa, K.; Noé, J.; Nowicka, M.; Bordogna, W.; et al. Circulating Cell-free DNA as a Prognostic Biomarker in Patients with Advanced ALK + Non–small Cell Lung Cancer in the Global Phase III ALEX Trial. Clin. Cancer Res. 2022, 28, 1800–1808. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; Lacroix, L.; Jovelet, C.; Caramella, C.; Howarth, K.; Plagnol, V.; Rosenfeld, N.; Morris, C.; Mezquita, L.; Pannet, C.; et al. Real-World Utility of an Amplicon-Based Next-Generation Sequencing Liquid Biopsy for Broad Molecular Profiling in Patients With Advanced Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2019, 3, 1–14. [Google Scholar] [CrossRef]
- Page, R.D.; Drusbosky, L.M.; Dada, H.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; et al. Clinical Outcomes for Plasma-Based Comprehensive Genomic Profiling Versus Standard-of-Care Tissue Testing in Advanced Non–Small Cell Lung Cancer. Clin. Lung Cancer 2022, 23, 72–81. [Google Scholar] [CrossRef]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non–small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Thompson, J.C.; Aggarwal, C.; Wong, J.; Nimgaonkar, V.; Hwang, W.-T.; Andronov, M.; Dibardino, D.M.; Hutchinson, C.T.; Ma, K.C.; Lanfranco, A.; et al. Plasma Genotyping at the Time of Diagnostic Tissue Biopsy Decreases Time-to-Treatment in Patients With Advanced NSCLC—Results From a Prospective Pilot Study. JTO Clin. Res. Rep. 2022, 3, 100301. [Google Scholar] [CrossRef]
- Swalduz, A.; Curcio, H.; Ambasager, B.; Le Moel, G.; Debieuvre, D.; Dot, J.M.; Duruisseaux, M.; Fournel, P.; Odier, L.; Demolombe, S.; et al. LIBELULE: A randomized phase III study to evaluate the clinical relevance of early liquid biopsy (LB) in patients with suspicious metastatic lung cancer. J. Clin. Oncol. 2023, 41, 9019. [Google Scholar] [CrossRef]
- García-Pardo, M.; Czarnecka-Kujawa, K.; Law, J.H.; Salvarrey, A.M.; Fernandes, R.; Fan, Z.J.; Waddell, T.K.; Yasufuku, K.; Liu, G.; Donahoe, L.L.; et al. Association of Circulating Tumor DNA Testing Before Tissue Diagnosis With Time to Treatment Among Patients With Suspected Advanced Lung Cancer: The ACCELERATE Nonrandomized Clinical Trial. JAMA Netw. Open 2023, 6, e2325332. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Lee, J.S.; Thongprasert, S.; Yu, C.-J.; Zhang, L.; Ladrera, G.; Srimuninnimit, V.; Sriuranpong, V.; Sandoval-Tan, J.; Zhu, Y.; et al. Intercalated combination of chemotherapy and erlotinib for patients with advanced stage non-small-cell lung cancer (FASTACT-2): A randomised, double-blind trial. Lancet Oncol. 2013, 14, 777–786. [Google Scholar] [CrossRef]
- Mok, T.; Camidge, D.R.; Gadgeel, S.M.; Rosell, R.; Dziadziuszko, R.; Kim, D.-W.; Pérol, M.; Ou, S.-H.I.; Ahn, J.S.; Shaw, A.T.; et al. Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Ann. Oncol. 2020, 31, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.; Wu, Y.-L.; Ahn, M.-J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.M.E.; et al. Osimertinib or Platinum–Pemetrexed in EGFR T790M–Positive Lung Cancer. N. Engl. J. Med. 2017, 376, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.E.; Ahn, M.-J.; Oxnard, G.R.; Shepherd, F.A.; Imamura, F.; Cheng, Y.; Okamoto, I.; Cho, B.C.; Lin, M.-C.; Wu, Y.-L.; et al. Early Clearance of Plasma Epidermal Growth Factor Receptor Mutations as a Predictor of Outcome on Osimertinib in Advanced Non–Small Cell Lung Cancer; Exploratory Analysis from AURA3 and FLAURA. Clin. Cancer Res. 2023, 29, 3340–3351. [Google Scholar] [CrossRef] [PubMed]
- Anagnostou, V.; Ho, C.; Nicholas, G.; Juergens, R.A.; Sacher, A.; Fung, A.S.; Wheatley-Price, P.; Laurie, S.A.; Levy, B.; Brahmer, J.R.; et al. ctDNA response after pembrolizumab in non-small cell lung cancer: Phase 2 adaptive trial results. Nat. Med. 2023, 29, 2559–2569. [Google Scholar] [CrossRef]
- Pellini, B.; Madison, R.W.; Childress, M.A.; Miller, S.T.; Gjoerup, O.; Cheng, J.; Huang, R.S.P.; Krainock, M.; Gupta, P.; Zou, W.; et al. Circulating Tumor DNA Monitoring on Chemo-immunotherapy for Risk Stratification in Advanced Non–Small Cell Lung Cancer. Clin. Cancer Res. 2023, 29, 4596–4605. [Google Scholar] [CrossRef]
- Piotrowska, Z.; Ahn, M.-J.; Pang, Y.-K.; How, S.H.; Sang-We, K.; Voon, P.J.; Cortinovis, D.L.; De Castro Carpeno, J.; Tiseo, M.; Rodriguez Abreu, D.; et al. 360P ELIOS: A multicentre, molecular profiling study of patients (pts) with epidermal growth factor receptor-mutated (EGFRm) advanced NSCLC treated with first-line (1L) osimertinib. Ann. Oncol. 2022, 33, S1581–S1582. [Google Scholar] [CrossRef]
- Berko, E.R.; Witek, G.M.; Matkar, S.; Petrova, Z.O.; Wu, M.A.; Smith, C.M.; Daniels, A.; Kalna, J.; Kennedy, A.; Gostuski, I.; et al. Circulating tumor DNA reveals mechanisms of lorlatinib resistance in patients with relapsed/refractory ALK-driven neuroblastoma. Nat. Commun. 2023, 14, 2601. [Google Scholar] [CrossRef]
- Hua, G.; Zhang, X.; Zhang, M.; Wang, Q.; Chen, X.; Yu, R.; Bao, H.; Liu, J.; Wu, X.; Shao, Y.; et al. Real-world circulating tumor DNA analysis depicts resistance mechanism and clonal evolution in ALK inhibitor-treated lung adenocarcinoma patients. ESMO Open 2022, 7, 100337. [Google Scholar] [CrossRef]
- Shimizu, H.; Nakayama, K.I. Artificial intelligence in oncology. Cancer Sci. 2020, 111, 1452–1460. [Google Scholar] [CrossRef]
- Fiste, O.; Gkiozos, I.; Charpidou, A.; Syrigos, N.K. Artificial Intelligence-Based Treatment Decisions: A New Era for NSCLC. Cancers 2024, 16, 831. [Google Scholar] [CrossRef]
- Luchini, C.; Pea, A.; Scarpa, A. Artificial intelligence in oncology: Current applications and future perspectives. Br. J. Cancer 2022, 126, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Bahado-Singh, R.; Vlachos, K.T.; Aydas, B.; Gordevicius, J.; Radhakrishna, U.; Vishweswaraiah, S. Precision Oncology: Artificial Intelligence and DNA Methylation Analysis of Circulating Cell-Free DNA for Lung Cancer Detection. Front. Oncol. 2022, 12, 790645. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Park, J.; Oh, S.; Jeong, B.-H.; Byun, Y.; Shin, S.H.; Im, Y.; Cho, J.H.; Cho, E.-H. Deep learning model integrating cfDNA methylation and fragment size profiles for lung cancer diagnosis. Sci. Rep. 2024, 14, 14797. [Google Scholar] [CrossRef]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 5060. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.H.; Cha, S.; Lee, H.Y.; Shin, S.-H.; Kim, Y.J.; Park, D.; Han, K.Y.; Oh, Y.J.; Park, W.-Y.; Ahn, M.-J.; et al. Machine learning model for circulating tumor DNA detection in chronic obstructive pulmonary disease patients with lung cancer. Transl. Lung Cancer Res. 2024, 13, 112–125. [Google Scholar] [CrossRef]
- Widman, A.J.; Shah, M.; Frydendahl, A.; Halmos, D.; Khamnei, C.C.; Øgaard, N.; Rajagopalan, S.; Arora, A.; Deshpande, A.; Hooper, W.F.; et al. Ultrasensitive plasma-based monitoring of tumor burden using machine-learning-guided signal enrichment. Nat. Med. 2024, 30, 1655–1666. [Google Scholar] [CrossRef]
- Altorki, N.K.; McGraw, T.E.; Borczuk, A.C.; Saxena, A.; Port, J.L.; Stiles, B.M.; Lee, B.E.; Sanfilippo, N.J.; Scheff, R.J.; Pua, B.B.; et al. Neoadjuvant durvalumab with or without stereotactic body radiotherapy in patients with early-stage non-small-cell lung cancer: A single-centre, randomised phase 2 trial. Lancet Oncol. 2021, 22, 824–835. [Google Scholar] [CrossRef]
- Socinski, M.A.; Jotte, R.M.; Cappuzzo, F.; Orlandi, F.; Stroyakovskiy, D.; Nogami, N.; Rodríguez-Abreu, D.; Moro-Sibilot, D.; Thomas, C.A.; Barlesi, F.; et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N. Engl. J. Med. 2018, 378, 2288–2301. [Google Scholar] [CrossRef]
- Assaf, Z.J.F.; Zou, W.; Fine, A.D.; Socinski, M.A.; Young, A.; Lipson, D.; Freidin, J.F.; Kennedy, M.; Polisecki, E.; Nishio, M.; et al. A longitudinal circulating tumor DNA-based model associated with survival in metastatic non-small-cell lung cancer. Nat. Med. 2023, 29, 859–868. [Google Scholar] [CrossRef]
- Ding, H.; Xu, X.S.; Yang, Y.; Yuan, M. Improving Prediction of Survival and Progression in Metastatic Non–Small Cell Lung Cancer After Immunotherapy Through Machine Learning of Circulating Tumor DNA. JCO Precis. Oncol. 2024, 8, e2300718. [Google Scholar] [CrossRef]
- Cucchiara, F.; Del Re, M.; Valleggi, S.; Romei, C.; Petrini, I.; Lucchesi, M.; Crucitta, S.; Rofi, E.; De Liperi, A.; Chella, A.; et al. Integrating Liquid Biopsy and Radiomics to Monitor Clonal Heterogeneity of EGFR-Positive Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 593831. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Dong, D.; She, Y.; Zhou, C.; Fang, M.; Zhu, Y.; Zhang, H.; Huang, Z.; Jiang, T.; Tian, J.; et al. Predicting response to immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden radiomic biomarker. J. Immunother. Cancer 2020, 8, e000550. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, B.; LaRiviere, M.J.; Cohen, E.A.; Buckingham, T.H.; Yee, S.S.; Black, T.A.; Chien, A.L.; Noël, P.; Hwang, W.-T.; Katz, S.I.; et al. Combining radiomic phenotypes of non-small cell lung cancer with liquid biopsy data may improve prediction of response to EGFR inhibitors. Sci. Rep. 2021, 11, 9984. [Google Scholar] [CrossRef]
- Soo, R.A.; De Marinis, F.; Han, J.-Y.; Ho, J.C.-M.; Martin, E.; Servidio, L.; Sandelin, M.; Popat, S. TARGET: A Phase II, Open-Label, Single-Arm Study of 5-Year Adjuvant Osimertinib in Completely Resected EGFR-Mutated Stage II to IIIB NSCLC Post Complete Surgical Resection. Clin. Lung Cancer 2024, 25, 80–84. [Google Scholar] [CrossRef]
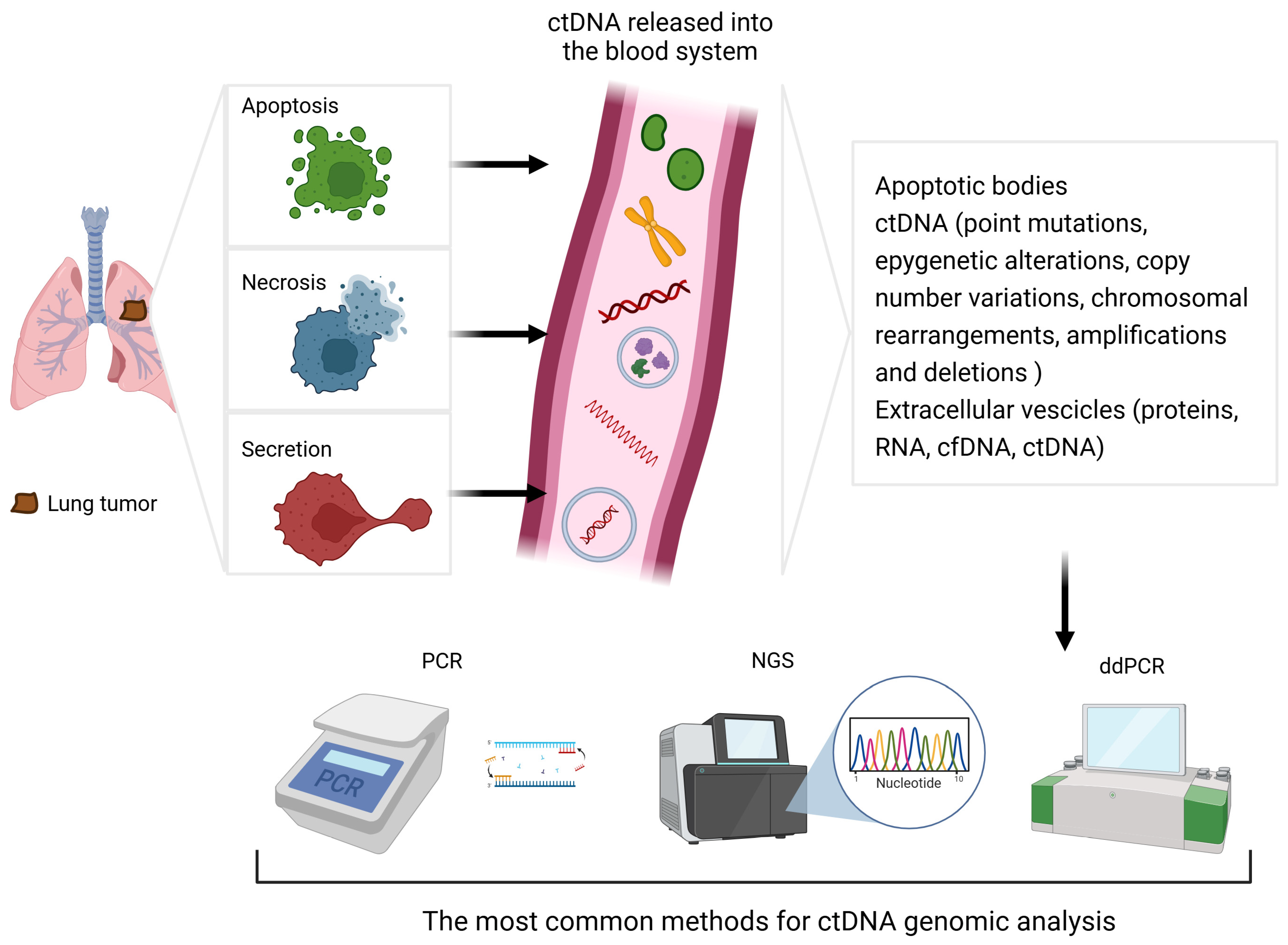

| Method | Mechanism | Advantages | Drawbacks | Detection Limit |
|---|---|---|---|---|
| PCR | Non-selective DNA amplification using the polymerase I enzyme | Low cost Ready-to-use kits | Detection of known and/or expected mutations only | 0.1–1% [2] |
| ddPCR | Amplification of DNA segments of interest within thousands of nanolitre-sized droplets | Low cost High sensitivity Quantitative analysis (copies/mL) | Detection of known and/or expected mutations only | 0.01–0.1%, up to 0.001% [14] |
| NGS | Analysis of the DNA sequence by identifying each individual nucleotide in rapid succession | Systematic sequencing Detection of any DNA alteration | High cost Longer time to yield results Variable sensitivity | 0.01–2%, based on computational power [2] |
| Metanalysis | Year | Technique | Pooled Sensitivity | Pooled Specificity | %NGS | NGS Sensitivity | NGS Specificity |
|---|---|---|---|---|---|---|---|
| Luo et al. [19] | 2014 | PCR/HRM | 67% (52–80%) | 93,5% (89–96%) | |||
| Mao et al. [21] | 2015 | PCR/NGS | 61% (50–71%) | 90% (85–94%) | 15% (222/14840) | 70% (46–87%) | 90% (49–99%) |
| Qiu et al. [20] | 2015 | PCR/HRM | 62% (51–72%) | 96% (93–98%) | |||
| Qian et al. [18] | 2016 | PCR/HRM | 60% (57–62%) | 94% (93–95%) | |||
| Passiglia et al. [23] | 2018 | PCR/ddPCR/NGS | 67% (64–70%) | 80% (77–83%) | 10% (169/1639) | 87% (76–95%) | 89% (82–94%) |
| Zhou et al. [22] | 2020 | PCR/ddPCR/NGS | 70% (63–75%) | 98% (96–99%) | NE | 80% (64–96%) | 98% (96–100%) |
| Wang et al. [25] | 2021 | PCR/HRM7NGS | 68% (60–75%) | 98% (95–99%) | NE | 79% (NE) | 98% (NE) |
| Franzi et al. [24] | 2023 | PCR/ddPCR/NGS | 59% (41–75%) | 96% (92–97%) | 39% (668/1711) | 62% (46–76%) | 95% (89–98%) |
| Method | Mechanism | Accuracy |
|---|---|---|
| UMIs | Selective analysis of DNA fragments bound to predefined nucleotide sequences | Detection limit: 0.1~0.5% [49] |
| TAM-SEQ | Selective amplification of DNA fragments using predefined primers | Detection limit: 0.25~2% Sensitivity: 94–97% [50] |
| CAPP-SEQ | Amplification of selected DNA fragments based on databases and bioinformatics algorithms | Detection limit: down to 0.02% Sensitivity: up to 100% [51] |
| Reference | Stage | No. Patients | Pretreatment Positive ctDNA | Post-Treatment Positive ctDNA |
|---|---|---|---|---|
| Chabon et al. [62] | I–III | 85 | RFS, HR = 3.4 (p = 0.026) FFM, HR = 6.0 (p < 0.001) | RFS, HR = 4.5 (p < 0.001) |
| Xia et al. [63] | I–III | 427 | RFS, HR = 4.2 (p < 0.001) | RFS, HR = 11.1 (p < 0.001) |
| Gale et al. [64] | I–III | 88 | RFS, HR = 3.1 (p = 0.003) OS, HR = 3.0 (p = 0.01) | RFS, HR = 14.8 (p < 0.001) OS, HR = 5.5 (p < 0.001) |
| Peng et al. [65] | I–IVr | 77 | RFS, HR = 3.6 (p < 0.001) OS, HR = 4.8 (p = 0.0013) | RFS, HR = 2.9 (p = 0.0035) OS, HR = 3.0 (p = 0.0086) |
| Chaudhuri et al. [66] | I–III | 40 | NE | RFS HR = 16.3 (p = 0.001) OS HR = 10.9 (p < 0.001) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, J.; Membrino, A.; Zanchetta, C.; Rizzato, S.; Cortiula, F.; Rossetto, C.; Pelizzari, G.; Aprile, G.; Macerelli, M. The Role of ctDNA in the Management of Non-Small-Cell Lung Cancer in the AI and NGS Era. Int. J. Mol. Sci. 2024, 25, 13669. https://doi.org/10.3390/ijms252413669
Costa J, Membrino A, Zanchetta C, Rizzato S, Cortiula F, Rossetto C, Pelizzari G, Aprile G, Macerelli M. The Role of ctDNA in the Management of Non-Small-Cell Lung Cancer in the AI and NGS Era. International Journal of Molecular Sciences. 2024; 25(24):13669. https://doi.org/10.3390/ijms252413669
Chicago/Turabian StyleCosta, Jacopo, Alexandro Membrino, Carol Zanchetta, Simona Rizzato, Francesco Cortiula, Ciro Rossetto, Giacomo Pelizzari, Giuseppe Aprile, and Marianna Macerelli. 2024. "The Role of ctDNA in the Management of Non-Small-Cell Lung Cancer in the AI and NGS Era" International Journal of Molecular Sciences 25, no. 24: 13669. https://doi.org/10.3390/ijms252413669
APA StyleCosta, J., Membrino, A., Zanchetta, C., Rizzato, S., Cortiula, F., Rossetto, C., Pelizzari, G., Aprile, G., & Macerelli, M. (2024). The Role of ctDNA in the Management of Non-Small-Cell Lung Cancer in the AI and NGS Era. International Journal of Molecular Sciences, 25(24), 13669. https://doi.org/10.3390/ijms252413669





