Visualized Nucleic Acid Hybridization Lateral Flow Strip Integrating with Microneedle for the Point-of-Care Authentication of Ophiocordyceps sinensis
Abstract
1. Introduction
2. Results
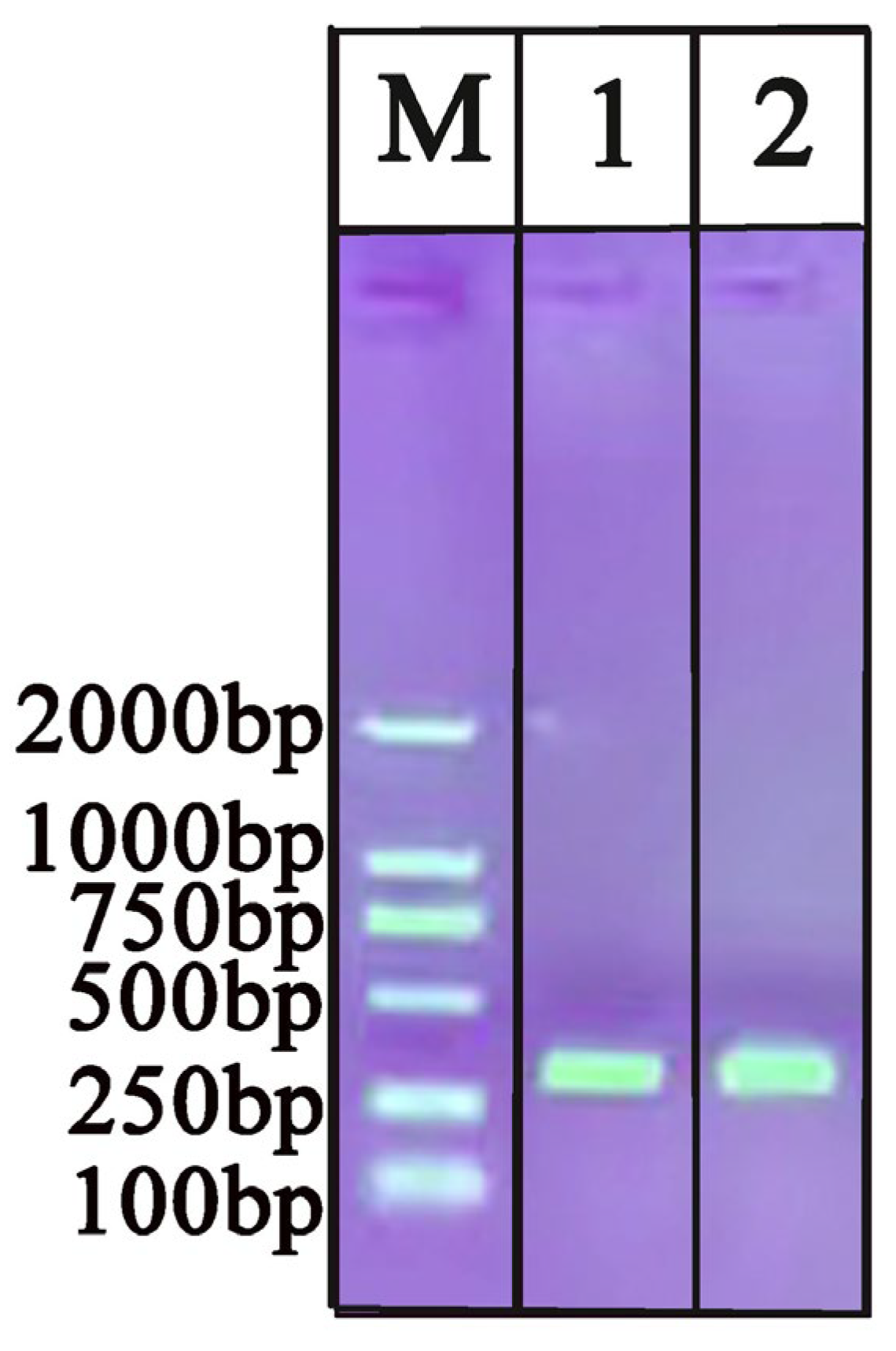
2.1. Evaluation of Extracted DNA by Commercial Kit and Tungsten
2.2. Characterization of AuNPs and Verification the Formation of AuNPs-SH Probes
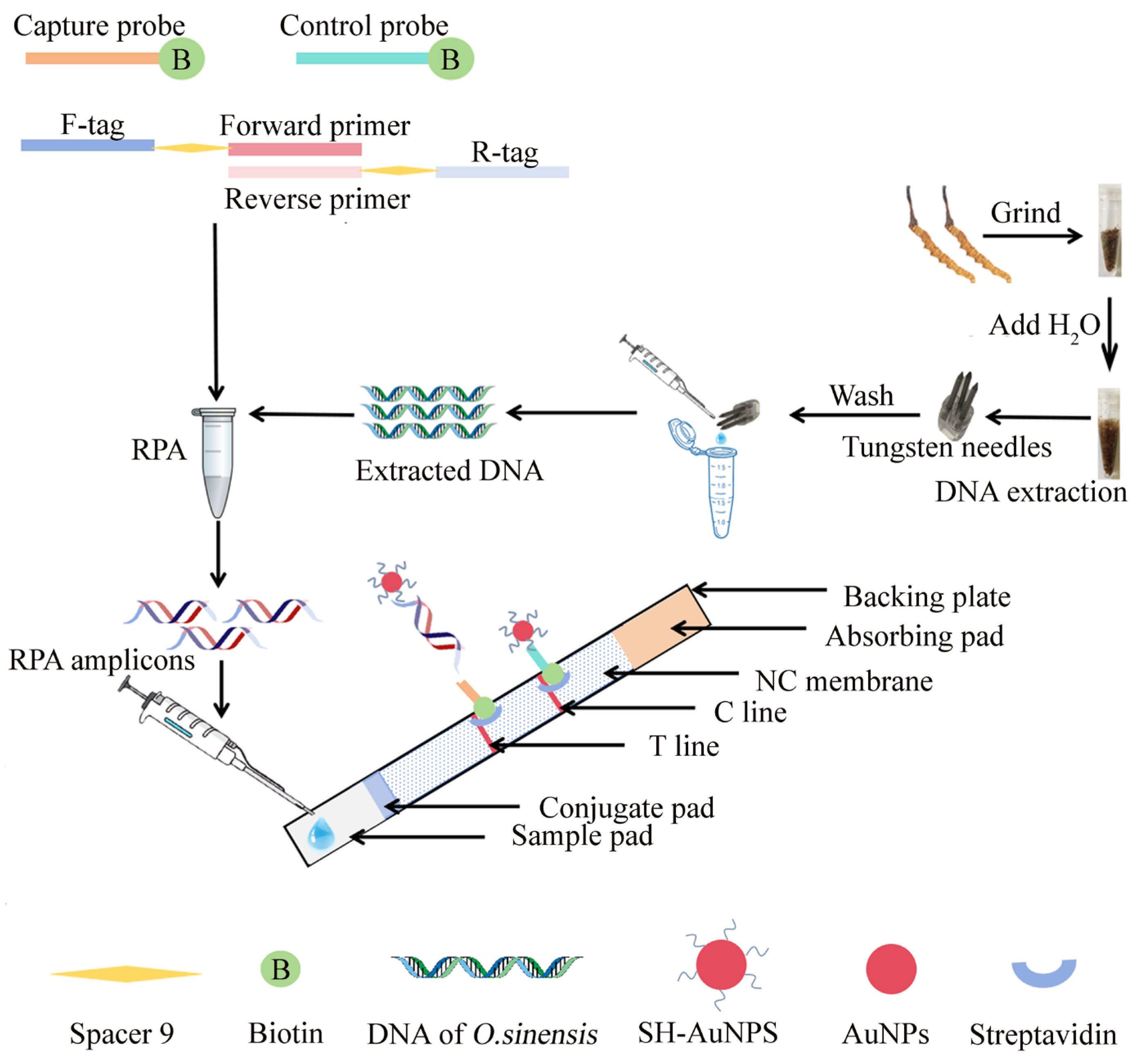
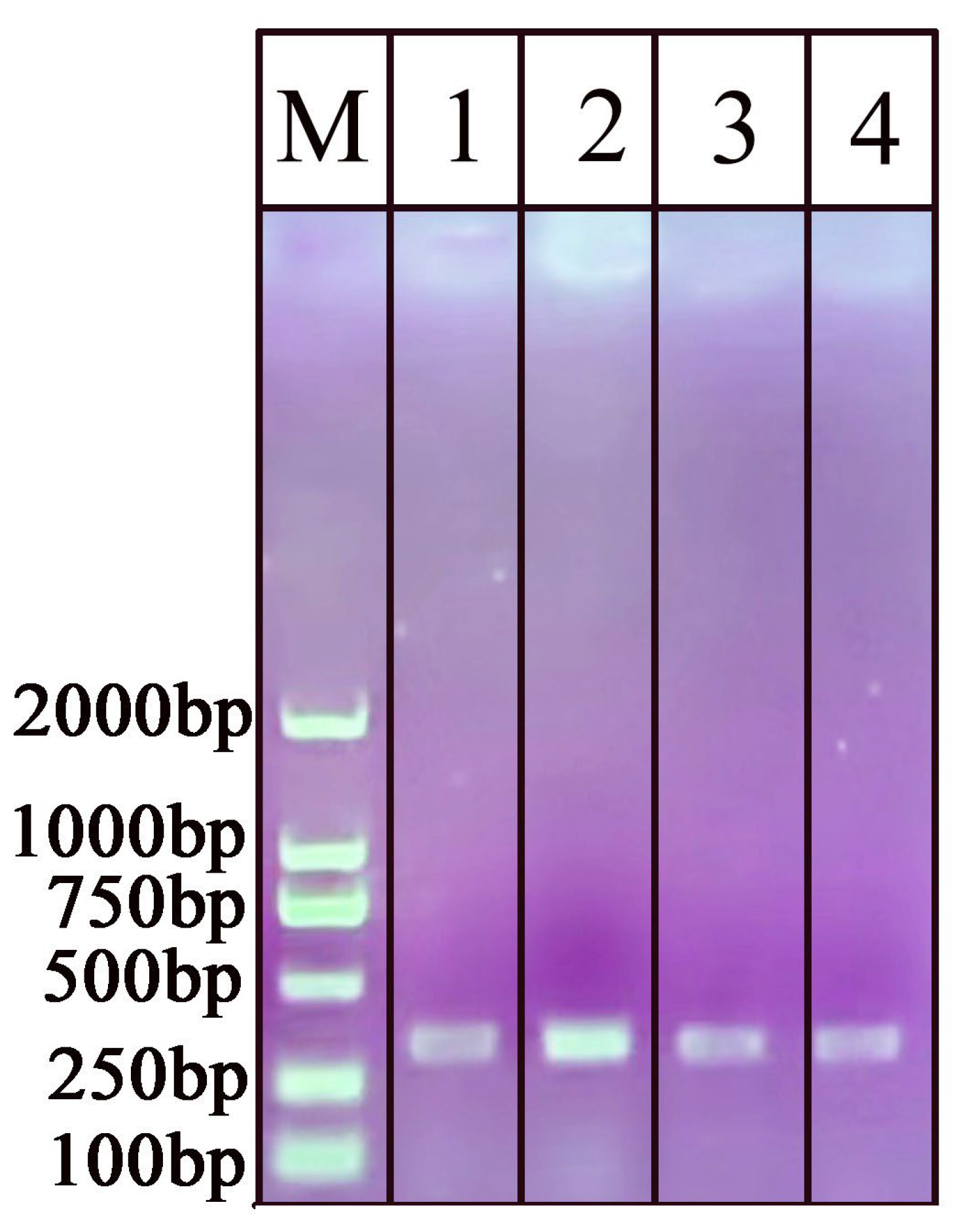
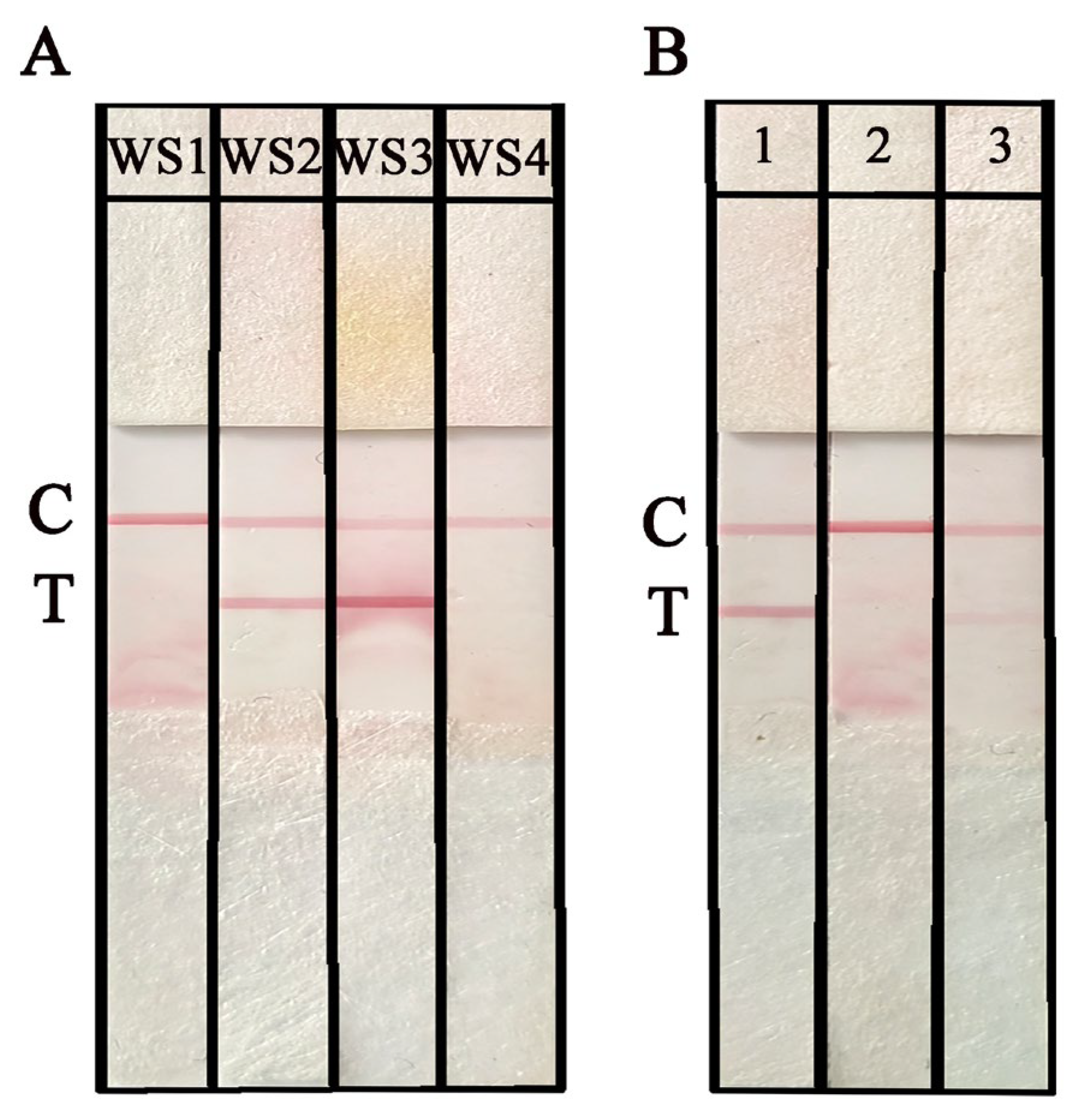
2.3. Primer Design and NAH-LFS Validation
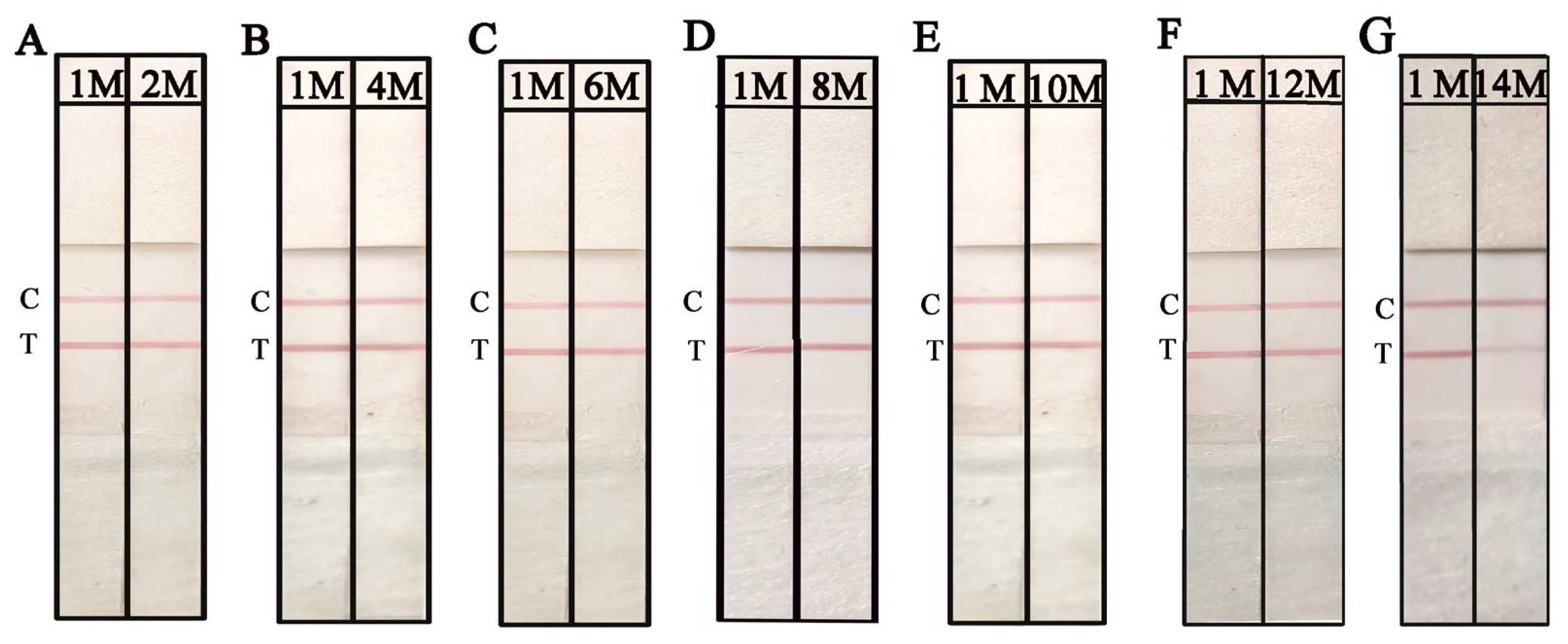
2.4. Optimization of RPA and the NAH-LFS Stability Under Room Temperature
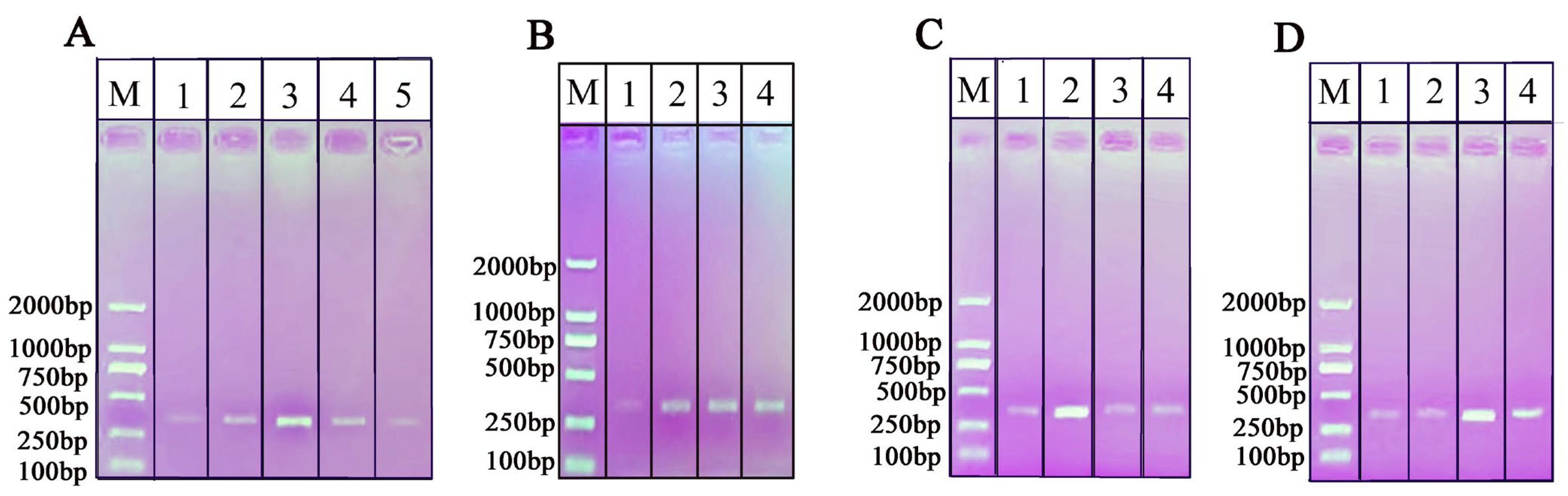
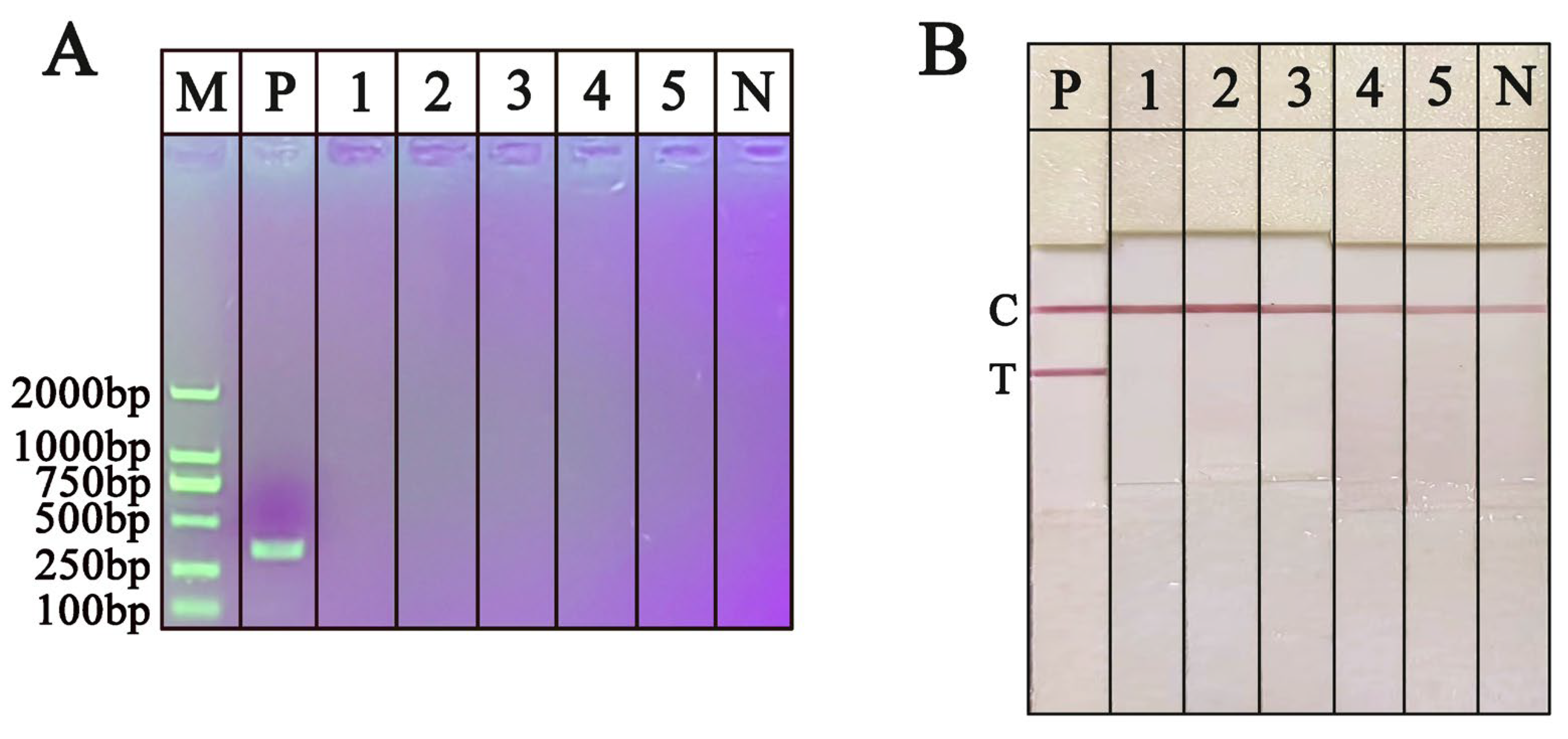
2.5. Specificity of the RPA-NAH-LFS Assay
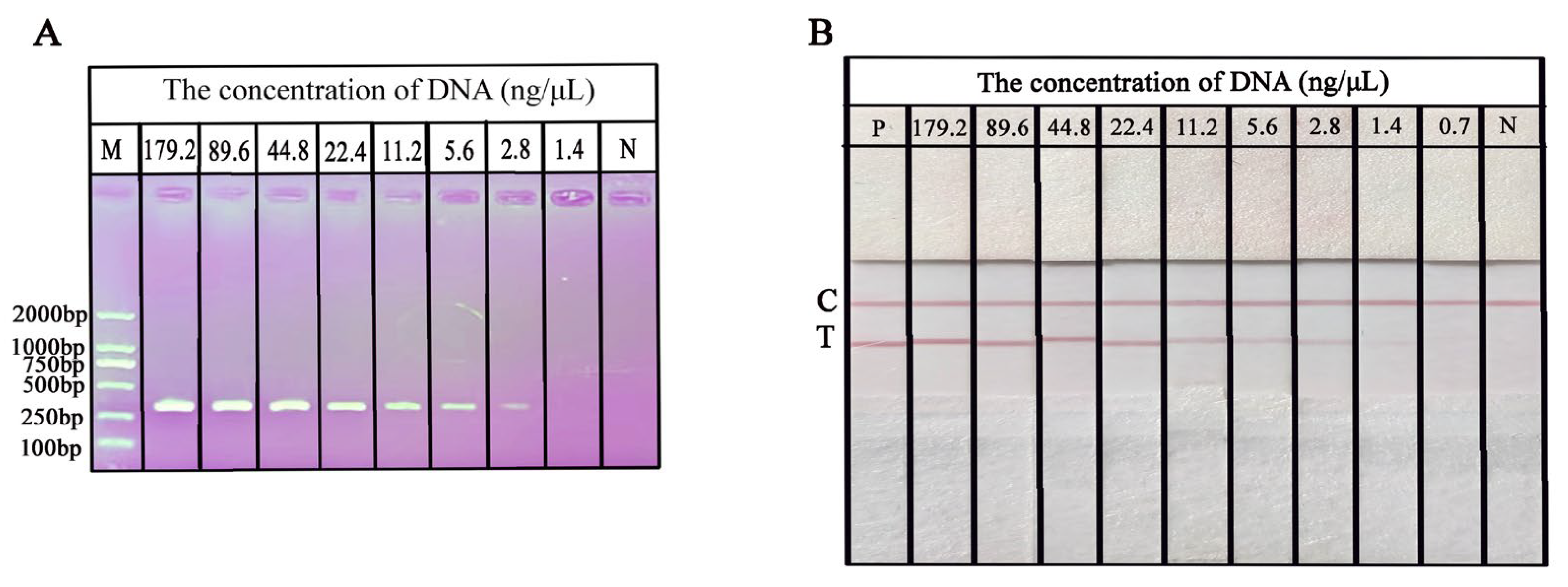
2.6. Sensitivity of the RPA-NAH-LFS Assay
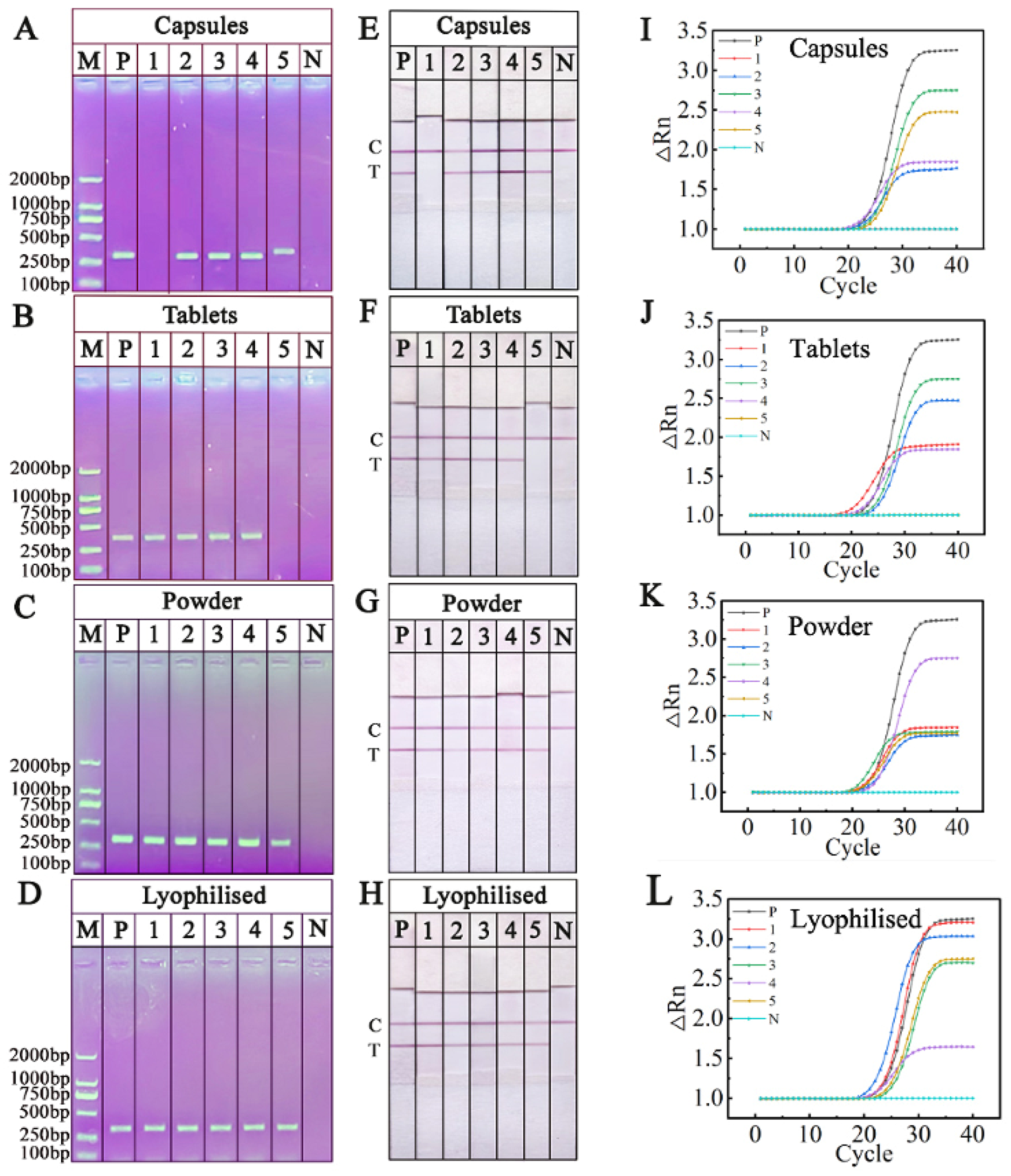
2.7. Detection Commercial O. sinensis Samples by RPA-NAH-LFS
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Sample Preparation and DNA Extraction
4.3. Synthesis of AuNPs and Conjunction with SH-Probes
- SH probes (10 μL 100 μM) were first reduced by adding them into TCEP (10 μL, 1 mM) and adjusted the solution pH to 3.0 with citrate–HCl buffer. The mixture was then incubated at room temperature for 1 h.
- A total of 1330 μL AuNPs was injected into the above SH probe solution. After 10 min of reaction, citrate-HCl (30 μL, 10 mM) was added to AuNPs-SH probe solution to adjust the pH to 3.0. Then, NaCl (10 μL, 2.0 M) was added sequentially and allowed to react for additional 10 min.
- The pH of the AuNPs-SH probe solution was adjusted to 7.5 by HEPES buffer (100 μL, 500 mM).
- The AuNPs-SH probes were incubated for 1 h before being centrifuged at 2000× g for 10 min. The supernatant was removed, and the pellet was resuspended in 90 μL buffer (1 mM Tris-HCl, 10% sucrose, 0.5% Tween-20, and 1% BSA, pH of 7.6). The AuNPs-SH probe were sprayed onto the conjugate pad at 20 μL/cm.
4.4. Design of PCR/RPA Primers and NAH-LFS Probes
- SH probes on AuNPs, which are reverse complementary to the tag sequence of forward primers;
- biotin-labeled capture probe, which are reverse complementary to tag sequence of reverse primers; and
- biotin-labeled control probe, which complementarily hybridize with SH probes.
4.5. PCR and AGE Testing
4.6. Assembly and Application of the NAH-LFS
4.7. Optimizing RPA Conditions and NAH-LFS Working Conditions
4.7.1. Optimizing RPA Conditions
- Reaction temperature and incubation time: RPA amplification reactions were conducted at temperatures ranging from 37 to 41 °C and varying incubation times between 5 and 20 min. These parameters were systematically varied to determine the optimal conditions for efficient amplification.
- Primer concentration: The concentration of the primers in the RPA reaction mixture was optimized within the range of 1–1.6 μL (10 μM) to determine the ideal amount that promotes robust amplification.
- Magnesium Acetate (MgAcO) concentration: The MgAcO concentration in the reaction mixture was varied from 2 μL (280 mM) to 3.5 μL (280 mM) to identify the optimal concentration that facilitates efficient amplification.
4.7.2. Optimizing NAH-LFS Working Conditions
4.8. Specificity and Sensitivity of the RPA-NAH-LFS Assay
4.9. Detection of Commercial O. sinensis Products
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Subedi, A.; Hamal, S.; Joshi, A. A Review on Ophiocordyceps sinensis. Science 2023, 7, 44–48. [Google Scholar] [CrossRef]
- Jin, G.S.; Wang, X.L.; Li, Y.; Wang, W.J.; Yang, R.H.; Ren, S.Y.; Yao, Y.J. Development of conventional and nested PCR assays for the detection of Ophiocordyceps sinensis. J. Basic. Microbiol. 2013, 53, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, H.H.; Kim, J.; Jung, J.; Moon, A.; Jeong, C.S.; Kang, H.; Cho, H. Anti-tumor effect of Cordyceps militaris in HCV-infected human hepatocarcinoma 7.5 cells. J. Microbiol. 2015, 53, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, Z.; Li, X. Moderate warming will expand the suitable habitat of Ophiocordyceps sinensis and expand the area of O. sinensis with high adenosine content. Sci. Total Environ. 2021, 787, 147605. [Google Scholar] [CrossRef]
- Xu, J.; Huang, Y.; Chen, X.X.; Zheng, S.C.; Chen, P.; Mo, M.H. The mechanisms of pharmacological activities of Ophiocordyceps sinensis fungi. Phytother. Res. 2016, 30, 1572–1583. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhou, L.W.; Yang, Z.L.; Bau, T.; Li, T.H.; Dai, Y.C. Resource diversity of Chinese macrofungi: Edible, medicinal and poisonous species. Fungal Divers. 2019, 98, 1–76. [Google Scholar] [CrossRef]
- Guo, M.; Jiang, W.; Yu, J.; Pang, X. Investigating the authenticity of Ophiopogonis Radix and its Chinese patent medicines by using a nucleotide signature. J. Ethnopharmacol. 2020, 261, 113134. [Google Scholar] [CrossRef]
- Au, D.; Wang, L.; Yang, D.; Mok, D.K.; Chan, A.S.; Xu, H. Application of microscopy in authentication of valuable Chinese medicine I—Cordyceps sinensis, its counterfeits, and related products. Microsc. Res. Tech. 2012, 75, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Wang, Y.; Xue, Z.; Chen, L.; Qiu, Y.; Cao, J.; Peng, C.; Guo, J. Proteomic identification of marker proteins and its application to authenticate Ophiocordyceps sinensis. 3 Biotech 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, S.; Li, J.; Wei, F.; Cheng, X.; Zhang, G.; Ma, S.; Liu, B.J.M. Identification of Ophiocordyceps sinensis and its artificially cultured Ophiocordyceps mycelia by ultra-performance liquid chromatography/Orbitrap fusion mass spectrometry and chemometrics. Int. J. Agric. Biol. Eng. 2018, 23, 1013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, B.; Men, X.H.; Xu, Z.W.; Wu, H.; Qin, X.T.; Xu, F.; Teng, Y.; Yuan, S.J.; Jin, L.Q.; et al. Proteome sequencing and analysis of Ophiocordyceps sinensis at different culture periods. BMC Genom. 2020, 21, 886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.L.; Yang, X.F.; Wang, D.; Lei, S.R.; Guo, L.A.; Liu, W.J.; Song, J. A simple and effective method to discern the true commercial Chinese cordyceps from counterfeits. Sci. Rep. 2020, 10, 2974. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.W.; Huang, L.F.; Hu, W.; He, Y.B.; Wong, K.P. Analysis of the Main Nucleosides in Cordyceps sinensis by LC/ESI-MS. Molecules 2010, 15, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.T.; Shaw, P.C.J.F.C. DNA-based techniques for authentication of processed food and food supplements. Food Chem. 2018, 240, 767–774. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, Q.; Cheng, P.; Zhu, M.; Zhang, H.; Liu, J.; Zuo, M.; Huang, C.; Wu, C.; Sun, Z. Whole-genome sequencing and analysis of the Chinese herbal plant Gelsemium elegans. Acta Pharm. Sin. B 2020, 10, 374–382. [Google Scholar] [CrossRef]
- Xu, R.; Wei, S.; Zhou, G.; Ren, J.; Liu, Z.; Tang, S.; Cheung, P.C.; Wu, X. Multiplex TaqMan locked nucleic acid real-time PCR for the differential identification of various meat and meat products. Meat Sci. 2018, 137, 41–46. [Google Scholar] [CrossRef]
- Liu, H.; Cao, T.; Chen, H.; Zhang, J.; Li, W.; Zhang, Y.; Liu, H.J. Two-color lateral flow nucleic acid assay combined with double-tailed recombinase polymerase amplification for simultaneous detection of chicken and duck adulteration in mutton. J. Food Compos. Anal. 2023, 118, 105209. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.J.B. Bioelectronics, Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef]
- Yusop, M.H.M.; Bakar, M. Review on halal forensic: A focus on DNA-based methods for pork authentication. Food Res. 2020, 4, 2347–2354. [Google Scholar] [CrossRef]
- Yeh, E.C.; Lee, L.P. One-step digital plasma separation for molecular diagnostics. In Proceedings of the 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences, MicroTAS 2013, Freiburg, Germany, 27–31 October 2013; pp. 1323–1325. [Google Scholar]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.; Stetten, F. Correction: Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2020, 145, 1950–1960. [Google Scholar] [CrossRef] [PubMed]
- Yusop, M.H.M.; Bakar, M.F.A.; Kamarudin, K.R.; Mokhtar, N.F.K.; Hossain, M.A.M.; Johan, M.R. Rapid Detection of Porcine DNA in Meatball Using Recombinase Polymerase Amplification Couple with Lateral Flow Immunoassay for Halal Authentication. Molecules 2022, 27, 8122. [Google Scholar] [CrossRef] [PubMed]
- Bougadi, E.T.; Kalogianni, D.P. Paper based DNA biosensor for food authenticity testing. Food Chem. 2020, 322, 126758. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Hu, X.; Chen, Y.; Jiang, W.; Liu, X.; Liu, J.; Zhu, L.; Zeng, H.; Liu, H. A dual-RPA based lateral flow strip for sensitive, on-site detection of CP4-EPSPS and Cry1Ab/Ac genes in genetically modified crops. Food Sci. Hum. Wellness 2024, 13, 183–190. [Google Scholar] [CrossRef]
- Xu, Y.; Wei, Y.; Cheng, N.; Huang, K.; Wang, W.; Zhang, L.; Xu, W.; Luo, Y. Nucleic acid biosensor synthesis of an all-in-one universal blocking linker recombinase polymerase amplification with a peptide nucleic acid-based lateral flow device for ultrasensitive detection of food pathogens. Anal. Chem. 2018, 90, 708–715. [Google Scholar] [CrossRef]
- Chatnarin, S.; Thirabunyanon, M. Potential bioactivities via anticancer, antioxidant, and immunomodulatory properties of cultured mycelial enriched β-D-glucan polysaccharides from a novel fungus Ophiocordyceps sinensis OS8. Front. Immunol. 2023, 14, 1150287. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Ye, Y.; Han, R. Fruiting body production of the medicinal Chinese caterpillar mushroom, Ophiocordyceps sinensis (Ascomycetes), in artificial medium. Int. J. Med. Mushrooms 2015, 17, 1107–1112. [Google Scholar] [CrossRef]
- Winkler, D. Caterpillar fungus (Ophiocordyceps sinensis) production and sustainability on the Tibetan Plateau and in the Himalayas. Asian Med. 2009, 5, 291–316. [Google Scholar] [CrossRef]
- Osathanunkul, M.; Osathanunkul, K.; Wongwanakul, S.; Osathanunkul, R.; Madesis, P. Multiuse of Bar-HRM for Ophiocordyceps sinensis identification and authentication. Sci. Rep. 2018, 8, 12770. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.Y.; Gao, Z.T.; Han, J.P.; Xiang, L. Detection of Ophiocordyceps sinensis and its common adulterates using species-specific primers. Front. Microbiol. 2017, 8, 1179. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Vu, M.T.; Nguyen, M.H.; Duong, H.A.; Mai, T.D.; Pham, H.V. A rapid and simple dual-channeled capillary electrophoresis with contactless conductivity detection method for the determination of adenosine, cordycepin, and inosine in Ophiocordyceps sinensis-based products. Food Anal. Methods 2021, 14, 1779–1787. [Google Scholar] [CrossRef]
- Duan, H.; Tong, X.; Cui, R.; Han, L.; Huang, G. On-site identification of Ophiocordyceps sinensis using multispectral imaging and chemometrics. Int. J. Agric. Biol. Eng. 2020, 13, 166–170. [Google Scholar] [CrossRef]
- Zhang, X.; Servos, M.R.; Liu, J. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J. Am. Chem. Soc. 2012, 134, 7266–7269. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.Y.; Wei, X.M.; Gao, Z.T.; Han, J.P. Rapid authentication of Ginkgo biloba herbal products using the recombinase polymerase amplification assay. Sci. Rep. 2018, 8, 8002. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, B.; Xiang, L.; Xiong, C.; Shi, Y.; Wu, L.; Meng, X.; Dong, G.; Xie, Y.; Sun, W. A novel onsite and visual molecular technique to authenticate saffron (Crocus sativus) and its adulterants based on recombinase polymerase amplification. Food Control 2019, 100, 117–121. [Google Scholar] [CrossRef]
- Liu, H.; Cao, T.; Wang, J.; Yuan, Y.; Li, H.; He, K.; Chen, H.; Wang, L. Analysis, Accurate and simultaneous detection of pork and horse meat adulteration by double tailed recombinase polymerase amplification integrated with SERS based two-color lateral flow nucleic acid hybridization strip. J. Food Compos. Anal. 2024, 134, 106562. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Multiplex assay of viruses integrating recombinase polymerase amplification, barcode—Anti-barcode pairs, blocking anti-primers, and lateral flow assay. Anal. Chem. 2021, 93, 13641–13650. [Google Scholar] [CrossRef]
- Batra, A.R.; Dike, C.C.; Mantri, N.; Ball, A.S. Recombinase polymerase amplification-lateral flow assay (RPA-LFA) as a rapid and sensitive test for Escherichia coli O157:H7 detection in food and beverage: A comparative study. Food Control 2024, 155, 110076. [Google Scholar] [CrossRef]
- Rodrigues, V.; Honrado, M.; Santos, J.; Pinto, M.A.; Amaral, J.S. Development of a loop-mediated isothermal amplification assay for the rapid detection of Styphnolobium japonicum (L.) Schott as an adulterant of Ginkgo biloba (L.). Phytomedicine 2024, 128, 155322. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Cao, R.; Xu, W.; Ma, Y.; Li, W.; Zhang, Y.; Liu, H. A cost-effective method for the rapid detection of chicken adulteration in meat using recombinase polymerase amplification combined with nucleic acid hybridization lateral flow strip. J. Food Compos. Anal. 2022, 111, 104602. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, J.; Na, W.; Sung, G.H.; Baek, S.K.; Kim, Y.K.; Kim, G.R.; Hu, H.J.; Park, J.H. Biotechnology, Development of a microneedle swab for acquisition of genomic DNA from buccal cells. Front. Bioeng. Biotech. 2022, 10, 829648. [Google Scholar] [CrossRef]
- Xiao, B.; Zhao, R.; Wang, N.; Zhang, J.; Sun, X.; Huang, F.; Chen, A. Integrating microneedle DNA extraction to hand-held microfluidic colorimetric LAMP chip system for meat adulteration detection. Food Chem. 2023, 411, 135508. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.B.; Zang, Y.X.; Du, X.J.; Li, P.; Wang, S. Development of an isothermal amplification-based assay for the rapid visual detection of Salmonella bacteria. J. Dairy Sci. 2017, 100, 7016–7025. [Google Scholar] [CrossRef]

| Method | A260/230 | A260/280 | A260 | DNA Concentration | Time | Installation |
|---|---|---|---|---|---|---|
| Tungsten | 0.518 ± 0.03 | 1.323 ± 0.13 | 1.004 | 50.2 ± 1.27 ng/µL | 1 min | / |
| Commercial kit | 0.683 ± 0.06 | 1.741 ± 0.16 | 1.430 | 71.5 ± 2.59 ng/µL | 50 min | Water bath, Centrifuge, Vortex oscillator |
| Name | Sequences (5′→3′) | Tag Sequence (5′→3′) | Function |
|---|---|---|---|
| Primer 1 | TTGTAGAAAACGGGGCAGGA | / | PCR |
| CCGCAGGGTCGAAAAATGAAG | / | ||
| Primer 2 | TAGAAAACGGGGCAGGAACA | ATTTTTCACTGGGTTTATAGT-spacer9 | PCR/RPA |
| TTGTAGAAAACGGGGCAGGA | TCGAGTGACAGCTAATGTGTGATT-spacer9 | ||
| Primer 3 | TTGGTGAACCAGCGGAGGGATCATT | / | PCR |
| GCTTGCTTCTTGACTGAGAGGTGCC | / | ||
| Primer 4 | ATTAAGTCGTGGAAATG | / | PCR |
| GATCAGGAATAGTGGGA | / | ||
| SH-probe | ACTATAAACCCAGTGAAAAAT | -SH | NAH-LFS |
| Control probe | ATTTTTCAGGGTTTTATAGT | Biotin | NAH-LFS |
| Capture probe | AATCACACATTAGCTGTCACTCGA | Biotin | NAH-LFS |
| Method | Time | Cost | ||
|---|---|---|---|---|
| Amplification | Detection | Equipment | Reagent | |
| PCR-AGE | 120 min | 45 min | Thermal cycling equipment | 1.00 $ |
| PCR-ICTS | 120 min | 5–15 min | Thermal cycling equipment | 1.50 $ |
| RPA-ICTS | 10 min | 5 min | Water bath | 6.92 $ |
| RPA-NAH-LFS | 10 min | 5 min | Water bath | 6.42 $ |
| No. | Sample | From | Test Results a |
|---|---|---|---|
| 1 | O. sinensis capsule 1 | China Beijing Tang | + |
| 2 | O. sinensis capsule 2 | Mannings | − |
| 3 | O. sinensis capsule 3 | Vitahealth | + |
| 4 | O. sinensis capsule 4 | Alibaba Health | + |
| 5 | O. sinensis capsule 5 | Wei shiya | + |
| 6 | O. sinensis tablet 1 | China Beijing Tang | + |
| 7 | O. sinensis tablet 2 | Mannings | + |
| 8 | O. sinensis tablet 3 | Vitahealth | + |
| 9 | O. sinensis tablet 4 | Alibaba Health | + |
| 10 | O. sinensis tablet 5 | Wei shiya | − |
| 11 | O. sinensis powder 1 | China Beijing Tang | + |
| 12 | O. sinensis powder 2 | Mannings | + |
| 13 | O. sinensis powder 3 | Vitahealth | + |
| 14 | O. sinensis powder 4 | Alibaba Health | + |
| 15 | O. sinensis powder 5 | Wei shiya | + |
| 16 | lyophilized O. sinensis 1 | China Beijing Tang | + |
| 17 | lyophilized O. sinensis 2 | Mannings | + |
| 18 | lyophilized O. sinensis 3 | Vitahealth | + |
| 19 | lyophilized O. sinensis 4 | Alibaba Health | + |
| 20 | lyophilized O. sinensis 5 | Wei shiya | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Wang, X.; Tian, H.; Yuan, Y.; Wang, J.; Cheng, Y.; Sun, L.; Chen, H.; Song, X. Visualized Nucleic Acid Hybridization Lateral Flow Strip Integrating with Microneedle for the Point-of-Care Authentication of Ophiocordyceps sinensis. Int. J. Mol. Sci. 2024, 25, 13599. https://doi.org/10.3390/ijms252413599
Liu H, Wang X, Tian H, Yuan Y, Wang J, Cheng Y, Sun L, Chen H, Song X. Visualized Nucleic Acid Hybridization Lateral Flow Strip Integrating with Microneedle for the Point-of-Care Authentication of Ophiocordyceps sinensis. International Journal of Molecular Sciences. 2024; 25(24):13599. https://doi.org/10.3390/ijms252413599
Chicago/Turabian StyleLiu, Haibin, Xinyue Wang, Hang Tian, Yi Yuan, Jing Wang, Yani Cheng, Linyao Sun, Hongshuo Chen, and Xiaoming Song. 2024. "Visualized Nucleic Acid Hybridization Lateral Flow Strip Integrating with Microneedle for the Point-of-Care Authentication of Ophiocordyceps sinensis" International Journal of Molecular Sciences 25, no. 24: 13599. https://doi.org/10.3390/ijms252413599
APA StyleLiu, H., Wang, X., Tian, H., Yuan, Y., Wang, J., Cheng, Y., Sun, L., Chen, H., & Song, X. (2024). Visualized Nucleic Acid Hybridization Lateral Flow Strip Integrating with Microneedle for the Point-of-Care Authentication of Ophiocordyceps sinensis. International Journal of Molecular Sciences, 25(24), 13599. https://doi.org/10.3390/ijms252413599







