Genome-Wide Identification and Analysis of BrTCP Transcription Factor Family Genes Involved in Cold Stress Tolerance in Winter Rapeseed (Brassica rapa L.)
Abstract
1. Introduction
2. Results
2.1. Identification and Protein Traits of TCPs Genes
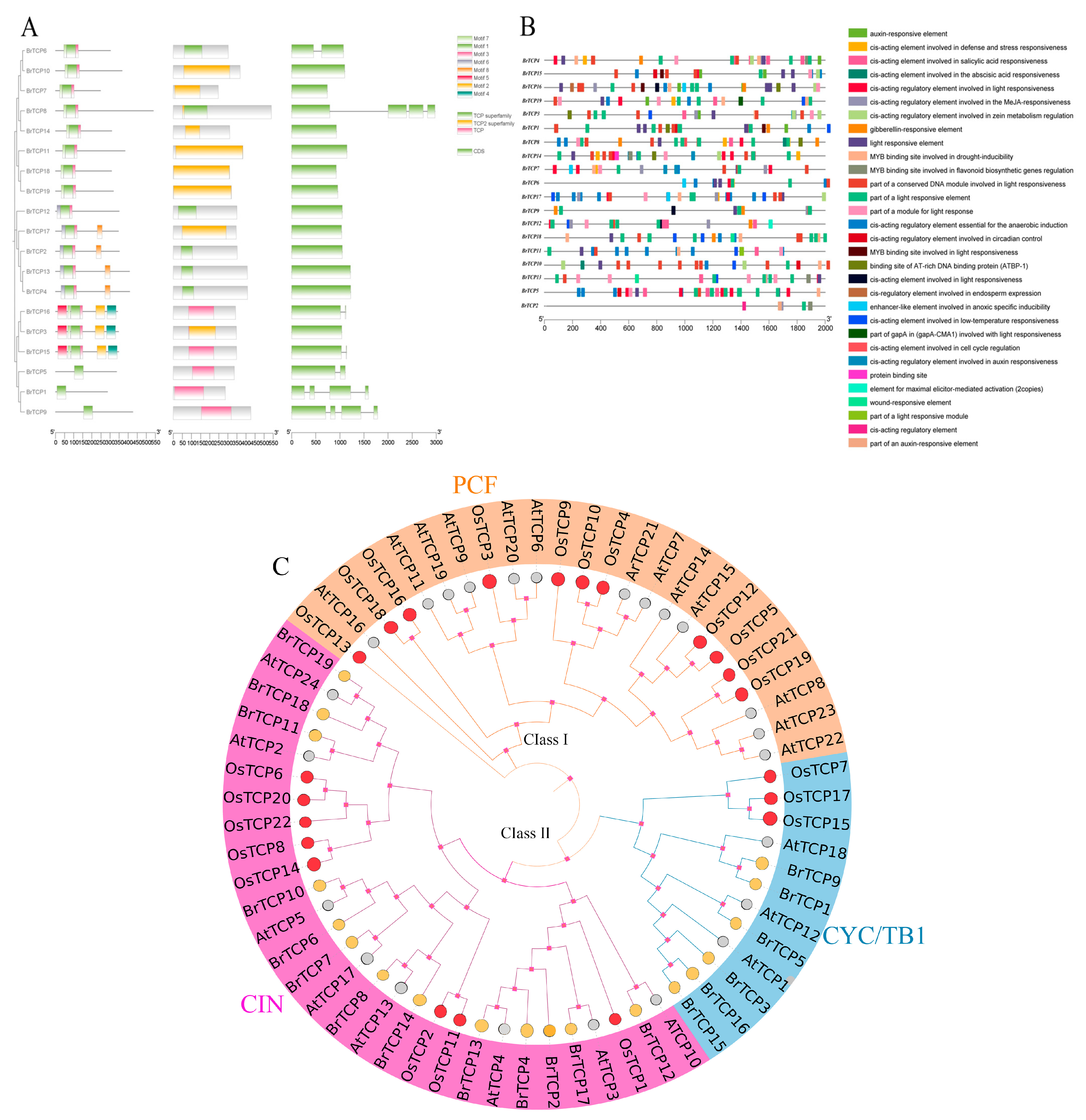
2.2. Gene Structure Analysis of TCPs Family in B. rapa
2.3. Analysis of Cis-Acting Elements of TCPs Gene in B. rapa
2.4. Phylogenetic Analysis of the Protein Sequence
2.5. Analysis of Collinearity and Localization of TCPs Genes in B. rapa
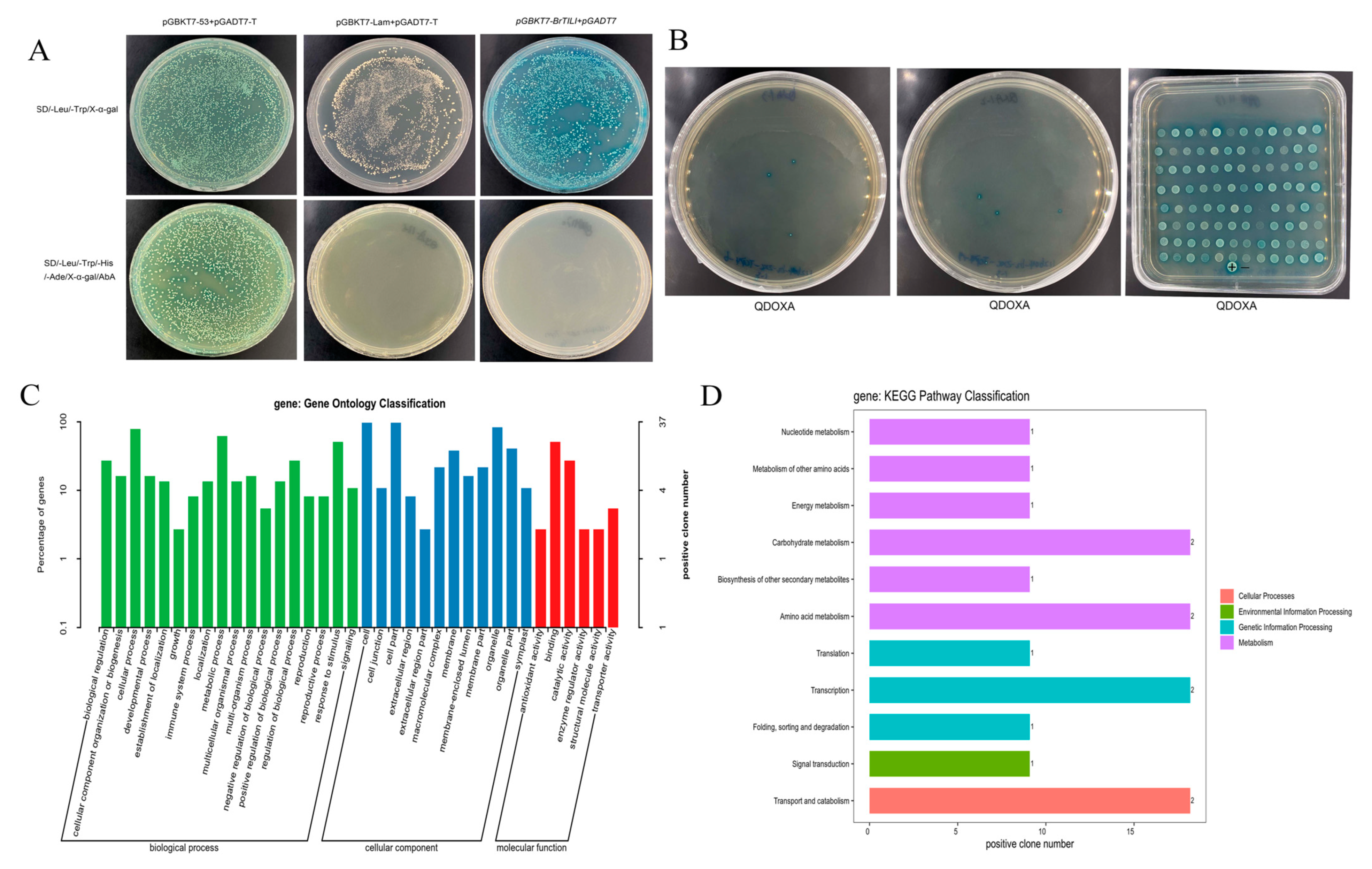
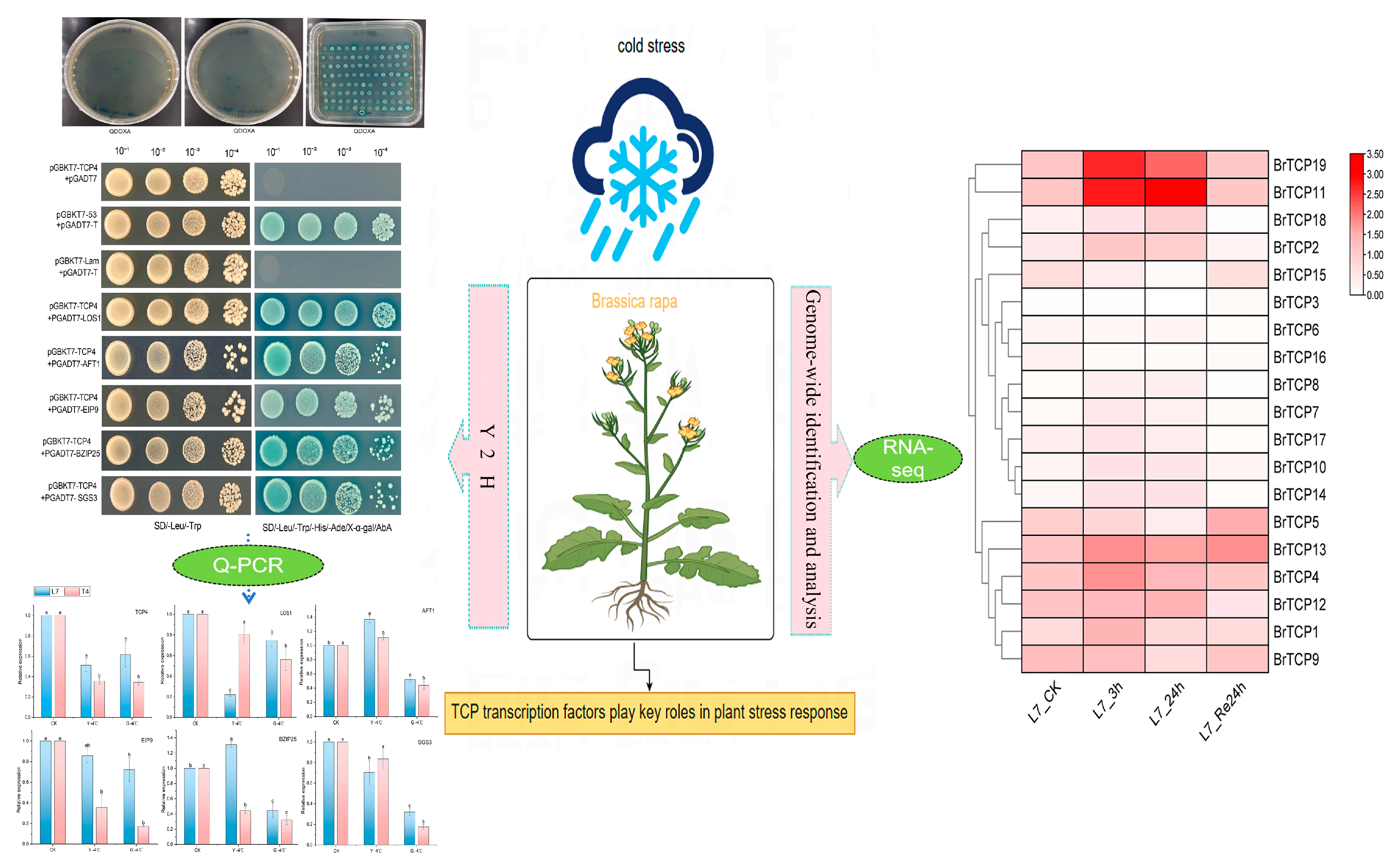
2.6. Validation of Self-Activation of BrTCP4 and Screening of Interacting Proteins
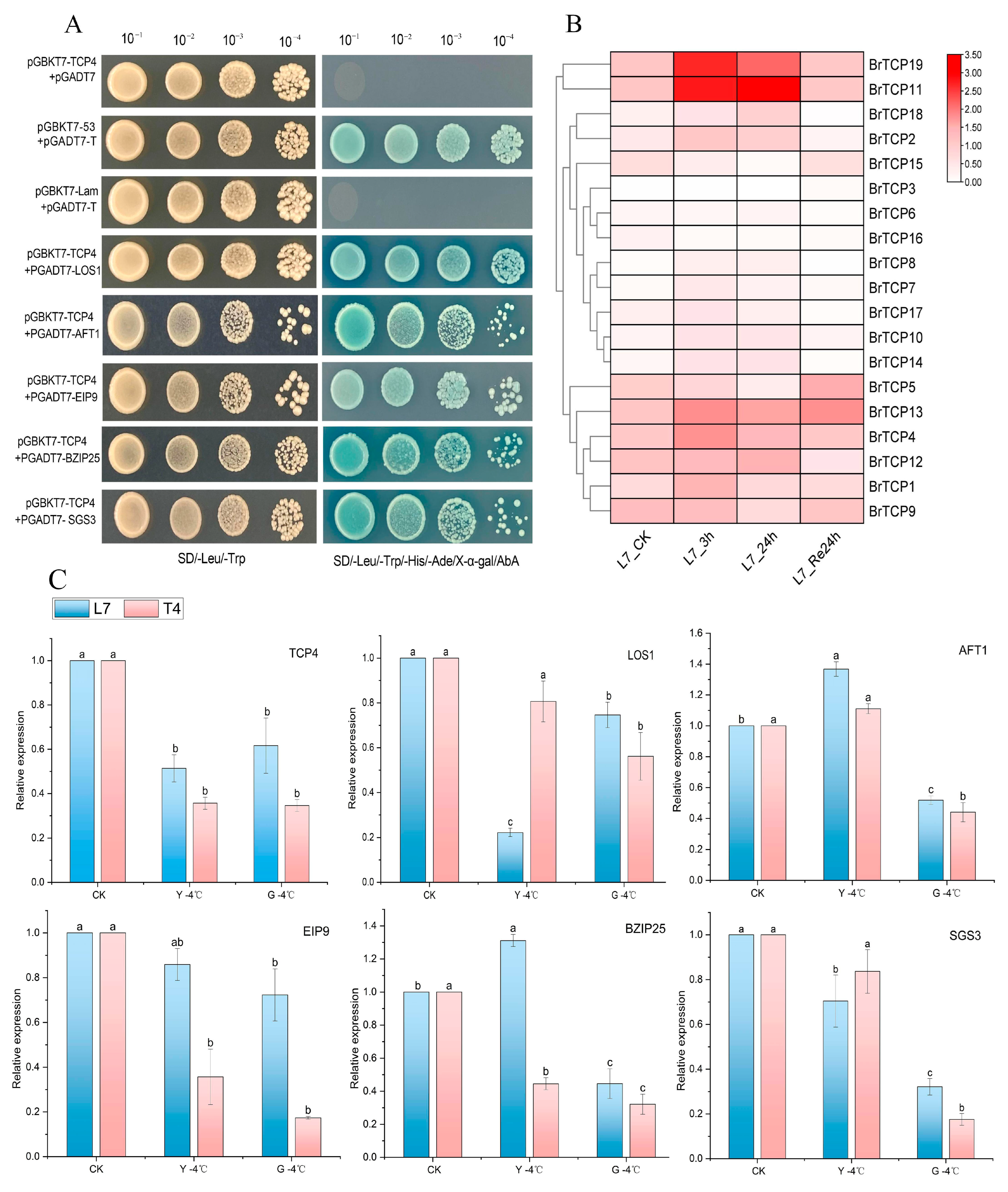
2.7. Y2H One-to-One Verification of Protein Interactions
2.8. Tissue Expression Analysis of BrTCP4 Interacting Proteins at Low Temperatures
2.9. Expression Analysis of the TCPs Gene Family Under Low-Temperature Stress
3. Discussion
4. Materials and Methods
4.1. Materials and Treatments
4.2. Genome-Wide Identification and Physicochemical Properties of TCP Genes
4.3. Analysis of TCPs Gene Family Structure, Motif, Domain, and Cis Elements
4.4. Phylogenetic Analysis of the TCP Proteins
4.5. Collinearity Analysis and Mapping of TCPs Genes on Chromosomes
4.6. Self-Activation and One-to-One Verification of BrTCP4
4.7. RNA Extraction and Real-Time Fluorescence Quantitative PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Qi, W.; Bai, J.; Li, H.; Fang, Y.; Xu, J.; Xu, Y.; Zeng, X.; Pu, Y.; Wang, W.; et al. Genome-Wide Identification and Analysis of the Ascorbate Peroxidase (APX) Gene Family of Winter Rapeseed (Brassica rapa L.) under Abiotic Stress. Front. Genet. 2021, 12, 753624. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Zhang, R.; Sun, W.; Zhang, J.; Wang, H. Study on climatic suitability for winter rapeseed planting in arid and cold regions in North China. Sci. Agric. Sin. 2014, 47, 2541–2551. [Google Scholar] [CrossRef]
- Ma, L.; Coulter, J.A.; Liu, L.; Zhao, Y.; Chang, Y.; Pu, Y.; Zeng, X.; Xu, Y.; Wu, J.; Fang, Y.; et al. Transcriptome Analysis Reveals Key Cold-Stress-Responsive Genes in Winter Rapeseed (Brassica rapa L.). Int. J. Mol. Sci. 2019, 20, 1071. [Google Scholar] [CrossRef]
- Fang, Y.; Coulter, J.A.; Wu, J.; Liu, L.; Li, X.; Dong, Y.; Ma, L.; Pu, Y.; Sun, B.; Niu, Z.; et al. Identification of differentially expressed genes involved in amino acid and lipid accumulation of winter turnip rape (Brassica rapa L.) in response to cold stress. PLoS ONE 2021, 16, e0245494. [Google Scholar] [CrossRef]
- Hou, L.; Wu, Q.; Zhu, X.; Li, X.; Fan, X.; Hui, M.; Ye, Q.; Liu, G.; Liu, X. Transcription Factor VvDREB2A from Vitis vinifera Improves Cold Tolerance. Int. J. Mol. Sci. 2023, 24, 9381. [Google Scholar] [CrossRef]
- Carlo, D.; Daniele, S. TRANSPARENT: A Python tool for designing transcription factor regulatory networks. Soft Comput. 2023, 27, 6261–6266. [Google Scholar]
- Wang, Q.; Xu, G.; Zhao, X.; Zhang, Z.; Wang, X.; Liu, X.; Xiao, W.; Fu, X.; Chen, X.; Gao, D.; et al. Transcription factor TCP20 regulates peach bud endodormancy by inhibiting DAM5/DAM6 and interacting with ABF2. J. Exp. Bot. 2020, 71, 1585–1597. [Google Scholar] [CrossRef]
- Kieffer, M.; Master, V.; Waites, R.; Davies, B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011, 68, 147–158. [Google Scholar] [CrossRef]
- Hur, Y.-S.; Kim, J.; Kim, S.; Son, O.; Kim, W.-Y.; Kim, G.-T.; Ohme-Takagi, M.; Cheon, C.-I. Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development. Genes 2019, 10, 644. [Google Scholar] [CrossRef]
- Xie, Y.G.; Ma, Y.Y.; Bi, P.P.; Wei, W.; Liu, J.; Hu, Y.; Gou, Y.J.; Zhu, D.; Wen, Y.Q.; Feng, J.Y. Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol. Biochem. 2020, 146, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Hu, Y.; Cui, M.Y.; Han, Y.T.; Gao, K.; Feng, J.Y. Identification and Transcript Analysis of the TCP Transcription Factors in the Diploid Woodland Strawberry Fragaria vesca. Front. Plant Sci. 2016, 7, 1937. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Wei, H.; Xu, X.; Ghuge, S.A.; Jia, D.; Liu, G.; Wang, Y.; Yuan, Y. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genom. 2019, 20, 786. [Google Scholar] [CrossRef] [PubMed]
- Jiu, S.; Xu, Y.; Wang, J.; Wang, L.; Wang, S.; Ma, C.; Guan, L.; Abdullah, M.; Zhao, M.; Xu, W.; et al. Genome-Wide Identification, Characterization, and Transcript Analysis of the TCP Transcription Factors in Vitis vinifera. Front. Genet. 2019, 10, 1276. [Google Scholar] [CrossRef]
- Wang, S.T.; Sun, X.L.; Hoshino, Y.; Yu, Y.; Jia, B.; Sun, Z.W.; Sun, M.Z.; Duan, X.B.; Zhu, Y.M. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 2014, 9, e91357. [Google Scholar] [CrossRef]
- Yin, Z.; Li, Y.; Zhu, W.; Fu, X.; Han, X.; Wang, J.; Lin, H.; Ye, W. Identification, Characterization, and Expression Patterns of TCP Genes and microRNA319 in Cotton. Int. J. Mol. Sci. 2018, 19, 3655. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Tyagi, A.K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015, 5, 9998. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- Lei, N.; Yu, X.; Li, S.; Zeng, C.; Zou, L.; Liao, W.; Peng, M. Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress. Sci. Rep. 2017, 7, 10016. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Howarth, D.G.; Donoghue, M.J. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9101–9106. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Y.; Han, Y.; Yu, S.; Wang, S.; Liu, Y.; Lin, X. Identification and expression analysis of TCP family members in tobacco (Nicotiana tabacum L.). Chin. J. Biotechnol. 2024, 40, 226–238. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Y.Q.; Ding, A.M.; Chen, H.; Xia, F.; Wang, W.F.; Sun, Y.H. Genome-wide analysis of TCP family in tobacco. Genet. Mol. Res. 2016, 15, 10-4238. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Z.; Jia, C.; Miao, H.; Zhang, J.; Liu, J.; Xu, B.; Jin, Z. Genome-Wide Identification and Transcript Analysis of TCP Gene Family in Banana (Musa acuminata L.). Biochem. Genet. 2022, 60, 204–222. [Google Scholar] [CrossRef]
- Sun, X.; Wang, C.; Xiang, N.; Li, X.; Yang, S.; Du, J.; Yang, Y.; Yang, Y. Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 2017, 15, 1284–1294. [Google Scholar] [CrossRef]
- Chong, X.; Liu, Y.; Li, P.; Wang, Y.; Zhou, T.; Chen, H.; Wang, H. Heterologous Expression of Chrysanthemum TCP Transcription Factor CmTCP13 Enhances Salinity Tolerance in Arabidopsis. Plants 2024, 13, 2118. [Google Scholar] [CrossRef]
- Ding, S.; Cai, Z.; Du, H.; Wang, H. Genome-Wide Analysis of TCP Family Genes in Zea mays L. Identified a Role for ZmTCP42 in Drought Tolerance. Int. J. Mol. Sci. 2019, 20, 2762. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Dong, Z.; Hao, Y.; Zhao, Y.; Tang, W.; Wang, X.; Li, J.; Wang, L.; Hu, Y.; Guan, X.; Gu, F.; et al. Genome-Wide Analysis of the TCP Transcription Factor Gene Family in Pepper (Capsicum annuum L.). Plants 2024, 13, 641. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Wang, H.W.; Cao, Y.N.; Kan, S.L.; Liu, Y.Y. Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 2022, 13, 994567. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Li, C.; Shen, J.; Zhuang, B.; Wen, Y. Genome-wide identification and analysis of the TCP gene family in eggplant (Solanum melongena L.). Chin. J. Biotechnol. 2022, 38, 2974–2988. [Google Scholar] [CrossRef]
- Calkhoven, C.F.; Ab, G. Multiple steps in the regulation of transcription-factor level and activity. Biochem. J. 1996, 317 Pt 2, 329–342. [Google Scholar] [CrossRef]
- Rueda-Romero, P.; Barrero-Sicilia, C.; Gómez-Cadenas, A.; Carbonero, P.; Oñate-Sánchez, L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 2012, 63, 1937–1949. [Google Scholar] [CrossRef]
- Viola, I.L.; Uberti Manassero, N.G.; Ripoll, R.; Gonzalez, D.H. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 2011, 435, 143–155. [Google Scholar] [CrossRef]
- Davière, J.M.; Wild, M.; Regnault, T.; Baumberger, N.; Eisler, H.; Genschik, P.; Achard, P. Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 2014, 24, 1923–1928. [Google Scholar] [CrossRef]
- Liu, D.K.; Zhang, C.; Zhao, X.; Ke, S.; Li, Y.; Zhang, D.; Zheng, Q.; Li, M.H.; Lan, S.; Liu, Z.J. Genome-wide analysis of the TCP gene family and their expression pattern in Cymbidium goeringii. Front. Plant Sci. 2022, 13, 1068969. [Google Scholar] [CrossRef]
- Wen, H.; Chen, Y.; Du, H.; Zhang, L.; Zhang, K.; He, H.; Pan, J.; Cai, R.; Wang, G. Genome-Wide Identification and Characterization of the TCP Gene Family in Cucumber (Cucumis sativus L.) and Their Transcriptional Responses to Different Treatments. Genes 2020, 11, 1379. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, T.; Tian, C.; Sun, S.; Li, J.; Chen, M. Transcription factors in rice: A genome-wide comparative analysis between monocots and eudicots. Plant Mol. Biol. 2005, 59, 191–203. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, J.; Li, C.; Deng, X.; Wang, T.; Dong, L. Genome-wide analysis of TCP transcription factor family in sunflower and identification of HaTCP1 involved in the regulation of shoot branching. BMC Plant Biol. 2023, 23, 222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, C.; Yang, D.; Wang, Y.; Yang, Y.; Sun, X. Genome-Wide Analysis of the TCP Transcription Factor Genes in Dendrobium catenatum Lindl. Int. J. Mol. Sci. 2021, 22, 10269. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.; Guerringue, Y.; Ranty, B.; Pouzet, C.; Jauneau, A.; Robe, E.; Mazars, C.; Galaud, J.P.; Aldon, D. Specific TCP transcription factors interact with and stabilize PRR2 within different nuclear sub-domains. Plant Sci. 2019, 287, 110197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, S.Q.; Li, J.Y.; Hu, X.B.; Gu, Y.; Wang, J.H. Genome-wide identification and expression analysis of TCP gene family of Andrographis paniculata under abiotic stress. China J. Chin. Mater. Medica 2024, 49, 379–388. [Google Scholar] [CrossRef]
- Liu, H.; Lyu, H.M.; Zhu, K.; Van de Peer, Y.; Max Cheng, Z.M. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef]
- Yan, H.; Jiang, C.; Li, X.; Sheng, L.; Dong, Q.; Peng, X.; Li, Q.; Zhao, Y.; Jiang, H.; Cheng, B. PIGD: A database for intronless genes in the Poaceae. BMC Genom. 2014, 15, 832. [Google Scholar] [CrossRef]
- Pan, J.; Ju, Z.; Ma, X.; Duan, L.; Jia, Z. Genome-wide characterization of TCP family and their potential roles in abiotic stress resistance of oat (Avena sativa L.). Front. Plant Sci. 2024, 15, 1382790. [Google Scholar] [CrossRef]
- Sang, Y.; Liu, Q.; Lee, J.; Ma, W.; McVey, D.S.; Blecha, F. Expansion of amphibian intronless interferons revises the paradigm for interferon evolution and functional diversity. Sci. Rep. 2016, 6, 29072. [Google Scholar] [CrossRef]
- Bentolila, S.; Chateigner-Boutin, A.L.; Hanson, M.R. Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Physiol. 2005, 139, 2006–2016. [Google Scholar] [CrossRef][Green Version]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Chae, K.; Isaacs, C.G.; Reeves, P.H.; Maloney, G.S.; Muday, G.K.; Nagpal, P.; Reed, J.W. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012, 71, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Fahim, A.M.; Wu, J.; Liu, L.; Pu, Y.; Ma, L.; Fang, Y.; Zhang, D.; Yang, G.; Wang, W.; et al. Systematic Analysis of the BrHAT Gene Family and Physiological Characteristics of Brassica rapa L. Treated with Histone Acetylase and Deacetylase Inhibitors under Low Temperature. Int. J. Mol. Sci. 2024, 25, 9200. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.J.; Choi, Y.D. Methyl jasmonate as a vital substance in plants. Trends Genet. 2003, 19, 409–413. [Google Scholar] [CrossRef]
- Mundy, J.; Yamaguchi-Shinozaki, K.; Chua, N.H. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. USA 1990, 87, 1406–1410. [Google Scholar] [CrossRef]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef]
- Sun, W.C.; Liu, H.-Q.; Liu, Z.G.; Wu, J.-Y.; Li, X.C.; Fang, Y.; Zeng, X.C.; Xu, Y.Z.; Zhang, Y.-H.; Dong, Y. Critical Index Analysis of Safe Over-wintering Rate of Winter Rapeseed (Brassica rapa) in Cold and Arid Region in North China. Crop J. 2016, 42, 609–618. [Google Scholar]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomics Data. Methods Mol. Biol. 2016, 1374, 115–140. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zhao, Y.; Ma, L.; Wu, J.; Zeng, R.; Jiao, J.; Li, R.; Ma, W.; Lian, Y.; Wang, W.; et al. Cloning and functional analysis of the BrCUC2 gene in Brassica rapa L. Front. Plant Sci. 2023, 14, 1274567. [Google Scholar] [CrossRef]

| Gene Name | Sequence ID | Chromosomes | Number of Amino Acids | Molecular Weight/kDa | PI | Instability Index | Aliphatic Index | Hydrophilia | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| BrTCP15 | Bra004097.1 | A07 | 347 | 39,379.98 | 5.83 | 49.42 | 64.32 | −0.788 | Nucleus |
| BrTCP18 | Bra010789.1 | A08 | 308 | 34,410.52 | 6.61 | 64.29 | 62.08 | −0.921 | Nucleus |
| BrTCP13 | Bra027284.1 | A05 | 406 | 44,035.42 | 6.99 | 64.44 | 54.16 | −0.791 | Nucleus |
| BrTCP19 | Bra032365.1 | A09 | 318 | 35,327.53 | 7.8 | 51.7 | 61.7 | −0.929 | Nucleus |
| BrTCP6 | Bra029344.1 | A02 | 301 | 33,796.16 | 6.5 | 47.54 | 64.12 | −0.791 | Nucleus |
| BrTCP8 | Bra001032.1 | A03 | 538 | 59,433.64 | 8.61 | 47.33 | 72.27 | −0.354 | Nucleus |
| BrTCP10 | Bra012990.1 | A03 | 366 | 40,698.22 | 6.18 | 56 | 59.15 | −0.904 | Nucleus |
| BrTCP4 | Bra001579.1 | A03 | 407 | 44,472.92 | 7.49 | 67.78 | 54.96 | −0.817 | Nucleus |
| BrTCP11 | Bra012600.1 | A03 | 382 | 42,262.93 | 7.12 | 45.39 | 52.15 | −0.979 | Nucleus |
| BrTCP14 | Bra039158.1 | A05 | 309 | 34,451.32 | 7.21 | 56.7 | 64.3 | −0.706 | Nucleus |
| BrTCP2 | Bra021586.1 | A01 | 350 | 38,203.22 | 6.81 | 75.02 | 56.91 | −0.719 | Nucleus |
| BrTCP1 | Bra037579.1 | A01 | 285 | 32,660.73 | 9.48 | 43.63 | 56.11 | −0.974 | Nucleus |
| BrTCP7 | Bra005967.1 | A03 | 246 | 27,367.45 | 6.49 | 51.36 | 76.54 | −0.518 | Nucleus |
| BrTCP16 | Bra004212.1 | A07 | 341 | 38,786.27 | 6.14 | 57.44 | 64.05 | −0.874 | Nucleus |
| BrTCP3 | Bra034010.1 | A02 | 346 | 39,308.67 | 5.5 | 48.92 | 65.03 | −0.823 | Nucleus |
| BrTCP9 | Bra001710.1 | A03 | 425 | 48,457.23 | 8.37 | 54.8 | 62.64 | −0.86 | Nucleus |
| BrTCP17 | Bra030952.1 | A08 | 345 | 37,595.34 | 6.74 | 66.6 | 54.12 | −0.786 | Nucleus |
| BrTCP5 | Bra038350.1 | A02 | 335 | 38,247.78 | 8.58 | 57.63 | 62.3 | −0.881 | Nucleus |
| BrTCP12 | Bra018280.1 | A05 | 349 | 38,727.41 | 6.34 | 50.44 | 51.15 | −0.87 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Ma, L.; Zeng, X.; Xu, Y.; Tao, X.; Fahim, A.M.; Liu, L.; Wu, J.; Yang, G.; Pu, Y.; et al. Genome-Wide Identification and Analysis of BrTCP Transcription Factor Family Genes Involved in Cold Stress Tolerance in Winter Rapeseed (Brassica rapa L.). Int. J. Mol. Sci. 2024, 25, 13592. https://doi.org/10.3390/ijms252413592
Xu Y, Ma L, Zeng X, Xu Y, Tao X, Fahim AM, Liu L, Wu J, Yang G, Pu Y, et al. Genome-Wide Identification and Analysis of BrTCP Transcription Factor Family Genes Involved in Cold Stress Tolerance in Winter Rapeseed (Brassica rapa L.). International Journal of Molecular Sciences. 2024; 25(24):13592. https://doi.org/10.3390/ijms252413592
Chicago/Turabian StyleXu, Yanxia, Li Ma, Xiucun Zeng, Yaozhao Xu, Xiaolei Tao, Abbas Muhammad Fahim, Lijun Liu, Junyan Wu, Gang Yang, Yuanyuan Pu, and et al. 2024. "Genome-Wide Identification and Analysis of BrTCP Transcription Factor Family Genes Involved in Cold Stress Tolerance in Winter Rapeseed (Brassica rapa L.)" International Journal of Molecular Sciences 25, no. 24: 13592. https://doi.org/10.3390/ijms252413592
APA StyleXu, Y., Ma, L., Zeng, X., Xu, Y., Tao, X., Fahim, A. M., Liu, L., Wu, J., Yang, G., Pu, Y., Fan, T., Wang, W., & Sun, W. (2024). Genome-Wide Identification and Analysis of BrTCP Transcription Factor Family Genes Involved in Cold Stress Tolerance in Winter Rapeseed (Brassica rapa L.). International Journal of Molecular Sciences, 25(24), 13592. https://doi.org/10.3390/ijms252413592






