Biochip-Based Identification of Mycobacterial Species in Russia
Abstract
1. Introduction
2. Results
2.1. The Development of Biochip Test for Species Identification
2.2. The Spectra of Mycobacterium Species Identified in Clinical Isolates
2.3. Regional Variation of Mycobacterial Species
2.4. Whole-Genome Sequencing of Novel Mycobacterial Species
3. Discussion
4. Materials and Methods
4.1. Biochip Design and Fabrication
4.2. Processing of Clinical Isolates
4.3. Phylogenetic Analysis and Target Sequencing
4.4. Whole-Genome Sequencing and Bioinformatic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dahl, V.N.; Mølhave, M.; Fløe, A.; van Ingen, J.; Schön, T.; Lillebaek, T.; Andersen, A.B.; Wejse, C. Global trends of pulmonary infections with nontuberculous mycobacteria: A systematic review. Int. J. Infect. Dis. 2022, 125, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Falkinham, J.O. Ecology of Nontuberculous Mycobacteria. Microorganisms 2021, 9, 2262. [Google Scholar] [CrossRef] [PubMed]
- Loebinger, M.R.; Quint, J.K.; van der Laan, R.; Obradovic, M.; Chawla, R.; Kishore, A.; van Ingen, J. Risk Factors for Nontuberculous Mycobacterial Pulmonary Disease. Chest 2023, 164, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.A.; Pomerantz, M.; Bishop, A.; Weyant, M.J.; Mitchell, J.D. Lady Windermere revisited: Treatment with thoracoscopic lobectomy/segmentectomy for right middle lobe and lingular bronchiectasis associated with non-tuberculous mycobacterial disease☆. Eur. J. Cardio-Thorac. Surg. 2011, 40, 671–675. [Google Scholar] [CrossRef]
- Chan, E.D.; Iseman, M.D. Slender, Older Women Appear to Be More Susceptible to Nontuberculous Mycobacterial Lung Disease. Gend. Med. 2010, 7, 5–18. [Google Scholar] [CrossRef]
- Brode, S.K.; Jamieson, F.B.; Ng, R.; Campitelli, M.A.; Kwong, J.C.; Paterson, J.M.; Li, P.; Marchand-Austin, A.; Bombardier, C.; Marras, T.K. Increased risk of mycobacterial infections associated with anti-rheumatic medications. Thorax 2015, 70, 677–682. [Google Scholar] [CrossRef]
- Yoon, E.C.; Lee, H.; Yoon, H.-Y. Inhaled Corticosteroids and the Risk of Nontuberculous Mycobacterial Infection in Chronic Airway Disease: A Nationwide Population-Based Study. Tuberc. Respir. Dis. 2024, 87, 473–482. [Google Scholar] [CrossRef]
- Wu, U.-I.; Holland, S.M. Host susceptibility to non-tuberculous mycobacterial infections. Lancet Infect. Dis. 2015, 15, 968–980. [Google Scholar] [CrossRef]
- Ma, X.; Li, X.Y.; Liu, J.-W. Demographic and Clinical Features of Nontuberculous Mycobacteria Infection Resulting from Cosmetic Procedures: A Systematic Review. Int. J. Infect. Dis. 2024, 149, 107259. [Google Scholar] [CrossRef]
- Nie, C.; Zhang, J.; Ji, H.; Li, X.; Luo, J.; Fu, A.; Ge, Y.; Liu, T.; Chen, T. Multiple Postoperative Lung Infections after Thymoma Surgery Diagnosed as Nontuberculous Mycobacterial Infection. Clin. Lab. 2024, 70, 1579–1582. [Google Scholar] [CrossRef]
- Lake, M.A.; Ambrose, L.R.; Lipman, M.C.I.; Lowe, D.M. “Why me, why now?” Using clinical immunology and epidemiology to explain who gets nontuberculous mycobacterial infection. BMC Med. 2016, 14, 54. [Google Scholar] [CrossRef]
- Prieto, M.D.; Alam, M.E.; Franciosi, A.N.; Quon, B.S. Global burden of nontuberculous mycobacteria in the cystic fibrosis population: A systematic review and meta-analysis. ERJ Open Res. 2022, 9, 00336-2022. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Ponnuswamy, A.; Capstick, T.G.; Chen, C.; McCabe, D.; Hurst, R.; Morrison, L.; Moore, F.; Gallardo, M.; Keane, J.; et al. Non-tuberculous mycobacterial pulmonary disease (NTM-PD): Epidemiology, diagnosis and multidisciplinary management. Clin. Med. 2024, 24, 100017. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, K.; Singh, S. Non-Tuberculous Mycobacteria in TB-Endemic Countries: Are We Neglecting the Danger? PLoS Neglected Trop. Dis. 2010, 4, e615. [Google Scholar] [CrossRef] [PubMed]
- Maiga, M.; Siddiqui, S.; Diallo, S.; Diarra, B.; Traoré, B.; Shea, Y.R.; Zelazny, A.M.; Dembele, B.P.P.; Goita, D.; Kassambara, H.; et al. Failure to Recognize Nontuberculous Mycobacteria Leads to Misdiagnosis of Chronic Pulmonary Tuberculosis. PLoS ONE 2012, 7, e36902. [Google Scholar] [CrossRef] [PubMed]
- Behra, P.R.K.; Pettersson, B.M.F.; Ramesh, M.; Das, S.; Dasgupta, S.; Kirsebom, L.A. Comparative genome analysis of mycobacteria focusing on tRNA and non-coding RNA. BMC Genom. 2022, 23, 704. [Google Scholar] [CrossRef]
- Wengenack, N.L.; Brown-Elliott, B.A.; Parrish, N.M.; Salfinger, M.; Turenne, C.Y.; Wallace, R.J.; Zelazny, A.M. This is giving me a complex: A practical attempt to streamline nontuberculous mycobacteria nomenclature for clinical purposes. J. Clin. Microbiol. 2024, 62, e0153123. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kinjo, T.; Motooka, D.; Nabeya, D.; Jung, N.; Uechi, K.; Horii, T.; Iida, T.; Fujita, J.; Nakamura, S. Comprehensive subspecies identification of 175 nontuberculous mycobacteria species based on 7547 genomic profiles. Emerg. Microbes Infect. 2019, 8, 1043–1053. [Google Scholar] [CrossRef]
- Hoefsloot, W.; van Ingen, J.; Andrejak, C.; Ängeby, K.; Bauriaud, R.; Bemer, P.; Beylis, N.; Boeree, M.J.; Cacho, J.; Chihota, V.; et al. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: An NTM-NET collaborative study. Eur. Respir. J. 2013, 42, 1604–1613. [Google Scholar] [CrossRef]
- Yan, J.; Zheng, R.; Zhang, Z.; Shi, J.; Yan, T.; Liu, H.; Li, F. Swollen Necrotic Lymphadenitis Infected with Mycobacterium paracondontium in an AIDS Patient: A Case Report and Literature Review. Infect. Drug Resist. 2024, 17, 3475–3482. [Google Scholar] [CrossRef]
- Fukano, H.; Kazumi, Y.; Sakagami, N.; Fujiwara, N.; Ato, M.; Mitarai, S.; Hoshino, Y. Mycobacterium kiyosense sp. nov., a scotochromogenic slow-glowing species isolated from respiratory specimens. Int. J. Syst. Evol. Microbiol. 2023, 73, 005917. [Google Scholar] [CrossRef] [PubMed]
- Tortoli, E. Microbiological Features and Clinical Relevance of New Species of the Genus Mycobacterium. Clin. Microbiol. Rev. 2014, 27, 727–752. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, M.; Li, D.; Xu, M.; Ao, Y.; Lin, L. Applications and advances in molecular diagnostics: Revolutionizing non-tuberculous mycobacteria species and subspecies identification. Front. Public Health 2024, 12, 1410672. [Google Scholar] [CrossRef] [PubMed]
- Markanović, M.; Makek, M.J.; Glodić, G.; Kuliš, T.; Mareković, I. Evaluation and clinical impact of MALDI Biotyper Mycobacteria Library v6.0 for identification of nontuberculous mycobacteria by MALDI-TOF mass spectrometry. J. Mass Spectrom. 2023, 58, e4915. [Google Scholar] [CrossRef] [PubMed]
- Padilla, E.; González, V.; Manterola, J.M.; Pérez, A.; Quesada, M.D.; Gordillo, S.; Vilaplana, C.; Pallarés, M.A.; Molinos, S.; Sánchez, M.D.; et al. Comparative Evaluation of the New Version of the INNO-LiPA Mycobacteria and GenoType Mycobacterium Assays for Identification of Mycobacterium Species from MB/BacT Liquid Cultures Artificially Inoculated with Mycobacterial Strains. J. Clin. Microbiol. 2004, 42, 3083–3088. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, K.; Singh, S. Multiplex PCR assay for simultaneous detection and differentiation of Mycobacterium tuberculosis, Mycobacterium avium complexes and other Mycobacterial species directly from clinical specimens. J. Appl. Microbiol. 2009, 107, 425–435. [Google Scholar] [CrossRef]
- Smirnova, T.; Ustinova, V.; Andreevskaya, S.; Larionova, E.; Kiseleva, E.; Chernousova, L.; Varlamov, D.; Sochivko, D.; Ergeshov, A. Evaluation of a new assay for nontuberculous mycobacteria species identification in diagnostic material and cultures. Tuberculosis 2021, 130, 102124. [Google Scholar] [CrossRef]
- Shrestha, N.K.; Tuohy, M.J.; Hall, G.S.; Reischl, U.; Gordon, S.M.; Procop, G.W. Detection and Differentiation of Mycobacterium tuberculosis and Nontuberculous Mycobacterial Isolates by Real-Time PCR. J. Clin. Microbiol. 2003, 41, 5121–5126. [Google Scholar] [CrossRef]
- Zelazny, A.M.; Calhoun, L.B.; Li, L.; Shea, Y.R.; Fischer, S.H. Identification of Mycobacterium Species by secA1 Sequences. J. Clin. Microbiol. 2005, 43, 1051–1058. [Google Scholar] [CrossRef]
- Zhu, L.; Jiang, G.; Wang, S.; Wang, C.; Li, Q.; Yu, H.; Zhou, Y.; Zhao, B.; Huang, H.; Xing, W.; et al. Biochip System for Rapid and Accurate Identification of Mycobacterial Species from Isolates and Sputum. J. Clin. Microbiol. 2010, 48, 3654–3660. [Google Scholar] [CrossRef]
- Gingeras, T.R.; Ghandour, G.; Wang, E.; Berno, A.; Small, P.M.; Drobniewski, F.; Alland, D.; Desmond, E.; Holodniy, M.; Drenkow, J. Simultaneous Genotyping and Species Identification Using Hybridization Pattern Recognition Analysis of Generic Mycobacterium DNA Arrays. Genome Res. 1998, 8, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Bang, H.-E.; Bai, G.-H.; Cho, S.-N. Novel Polymorphic Region of the rpoB Gene Containing Mycobacterium Species-Specific Sequences and Its Use in Identification of Mycobacteria. J. Clin. Microbiol. 2003, 41, 2213–2218. [Google Scholar] [CrossRef] [PubMed]
- Rubina, A.; Pan’kov, S.; Dementieva, E.; Pen’kov, D.; Butygin, A.; Vasiliskov, V.; Chudinov, A.; Mikheikin, A.; Mikhailovich, V.; Mirzabekov, A. Hydrogel drop microchips with immobilized DNA: Properties and methods for large-scale production. Anal. Biochem. 2003, 325, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Frederiksen, W.; Garrity, G.M.; Grimont, P.A.D.; Kämpfer, P.; Maiden, M.C.J.; Nesme, X.; Rosselló-Mora, R.; Swings, J.; Trüper, H.G.; et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 2002, 52, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Beye, M.; Fahsi, N.; Raoult, D.; Fournier, P.-E. Careful use of 16S rRNA gene sequence similarity values for the identification of Mycobacterium species. New Microbes New Infect. 2017, 22, 24–29. [Google Scholar] [CrossRef]
- Won, S.; Cho, S.; Kim, H. rRNA operon improves species-level classification of bacteria and microbial community analysis compared to 16S rRNA. Microbiol. Spectr. 2024, 12, e0093124. [Google Scholar] [CrossRef]
- Park, H.; Jang, H.; Song, E.; Chang, C.L.; Lee, M.; Jeong, S.; Park, J.; Kang, B.; Kim, C. Detection and Genotyping of Mycobacterium Species from Clinical Isolates and Specimens by Oligonucleotide Array. J. Clin. Microbiol. 2005, 43, 1782–1788. [Google Scholar] [CrossRef]
- Tortoli, E.; Nanetti, A.; Piersimoni, C.; Cichero, P.; Farina, C.; Mucignat, G.; Scarparo, C.; Bartolini, L.; Valentini, R.; Nista, D.; et al. Performance Assessment of New Multiplex Probe Assay for Identification of Mycobacteria. J. Clin. Microbiol. 2001, 39, 1079–1084. [Google Scholar] [CrossRef]
- Häfner, B.; Haag, H.; Geiss, H.-K.; Nolte, O. Different molecular methods for the identification of rarely isolated non-tuberculous mycobacteria and description of new hsp65 restriction fragment length polymorphism patterns. Mol. Cell. Probes 2004, 18, 59–65. [Google Scholar] [CrossRef]
- Cooksey, R.C.; Limor, J.; Morlock, G.P.; Crawford, J.T. Identifying Mycobacterium species and strain typing using a microfluidic labchip instrument. BioTechniques 2003, 35, 786–794. [Google Scholar] [CrossRef]
- Fukushima, M.; Kakinuma, K.; Hayashi, H.; Nagai, H.; Ito, K.; Kawaguchi, R. Detection and Identification of Mycobacterium Species Isolates by DNA Microarray. J. Clin. Microbiol. 2003, 41, 2605–2615. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kasai, H.; Ezaki, T.; Harayama, S. Differentiation of Phylogenetically Related Slowly Growing Mycobacteria by Their gyrB Sequences. J. Clin. Microbiol. 2000, 38, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Dauendorffer, J.-N.; Guillemin, I.; Aubry, A.; Truffot-Pernot, C.; Sougakoff, W.; Jarlier, V.; Cambau, E. Identification of Mycobacterial Species by PCR Sequencing of Quinolone Resistance-Determining Regions of DNA Gyrase Genes. J. Clin. Microbiol. 2003, 41, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Zimenkov, D.V.; Kulagina, E.V.; Antonova, O.V.; Krasnova, M.A.; Chernyaeva, E.N.; Zhuravlev, V.Y.; Kuz’Min, A.V.; Popov, S.A.; Zasedatelev, A.S.; Gryadunov, D.A. Evaluation of a Low-Density Hydrogel Microarray Technique for Mycobacterial Species Identification. J. Clin. Microbiol. 2015, 53, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Zimenkov, D.; Atanasova, Y.; Ushtanit, A.; Yordanova, S.; Baykova, A.; Filippova, M.; Semenova, U.; Mokrousov, I.; Bachiyska, E. The Intriguing Pattern of Nontuberculous Mycobacteria in Bulgaria and Description of Mycobacterium bulgaricum sp. nov. Int. J. Mol. Sci. 2024, 25, 10434. [Google Scholar] [CrossRef]
- Bannantine, J.P.; Conde, C.; Bayles, D.O.; Branger, M.; Biet, F. Genetic Diversity Among Mycobacterium avium Subspecies Revealed by Analysis of Complete Genome Sequences. Front. Microbiol. 2020, 11, 1701. [Google Scholar] [CrossRef]
- Turenne, C.Y.; Collins, D.M.; Alexander, D.C.; Behr, M.A. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium Are Independently Evolved Pathogenic Clones of a Much Broader Group of M. avium Organisms. J. Bacteriol. 2008, 190, 2479–2487. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef]
- Boyle, D.P.; Zembower, T.R.; Qi, C. Relapse versus Reinfection of Mycobacterium avium Complex Pulmonary Disease. Patient Characteristics and Macrolide Susceptibility. Ann. Am. Thorac. Soc. 2016, 13, 1956–1961. [Google Scholar] [CrossRef]
- Daley, C.L.; Iaccarino, J.M.; Lange, C.; Cambau, E.; Wallace, R.J., Jr.; Andrejak, C.; Böttger, E.C.; Brozek, J.; Griffith, D.E.; Guglielmetti, L.; et al. Treatment of Nontuberculous Mycobacterial Pulmonary Disease: An Official ATS/ERS/ESCMID/IDSA Clinical Practice Guideline. Clin. Infect. Dis. 2020, 71, 905–913. [Google Scholar] [CrossRef]
- Jiang, S.; Redelman-Sidi, G. BCG in Bladder Cancer Immunotherapy. Cancers 2022, 14, 3073. [Google Scholar] [CrossRef] [PubMed]
- Central Tuberculosis Research Institute; Ustinova, V.V.; Smirnova, T.G.; Andreevskaya, S.N.; Andrievskaya, I.Y.; Larionova, E.E.; Chernousova, L.N. Sequencing of six genomes of clinical strains of non-tuberculous mycobacteria. Tuberc. Lung Dis. 2019, 96, 70–71. [Google Scholar] [CrossRef]
- Bouam, A.; Armstrong, N.; Levasseur, A.; Drancourt, M. Mycobacterium terramassiliense, Mycobacterium rhizamassiliense and Mycobacterium numidiamassiliense sp. nov., three new Mycobacterium simiae complex species cultured from plant roots. Sci. Rep. 2018, 8, 9309. [Google Scholar] [CrossRef]
- Deng, Y.; Mou, T.; Wang, J.; Su, J.; Yan, Y.; Zhang, Y.-Q. Characterization of three rapidly growing novel Mycobacterium species with significant polycyclic aromatic hydrocarbon bioremediation potential. Front. Microbiol. 2023, 14, 1225746. [Google Scholar] [CrossRef] [PubMed]
- Hitch, T.C.; Riedel, T.; Oren, A.; Overmann, J.; Lawley, T.D.; Clavel, T. Automated analysis of genomic sequences facilitates high-throughput and comprehensive description of bacteria. ISME Commun. 2021, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Nouioui, I.; Carro, L.; Teramoto, K.; Igual, J.M.; Jando, M.; Montero-Calasanz, M.D.C.; Sutcliffe, I.; Sangal, V.; Goodfellow, M.; Klenk, H.-P. Mycobacterium eburneum sp. nov., a non-chromogenic, fast-growing strain isolated from sputum. Int. J. Syst. Evol. Microbiol. 2017, 67, 3174–3181. [Google Scholar] [CrossRef]
- Davidson, R.M.; DeGroote, M.A.; Marola, J.L.; Buss, S.; Jones, V.; McNeil, M.R.; Freifeld, A.G.; Epperson, L.E.; Hasan, N.A.; Jackson, M.; et al. Mycobacterium talmoniae sp. nov., a slowly growing mycobacterium isolated from human respiratory samples. Int. J. Syst. Evol. Microbiol. 2017, 67, 2640–2645. [Google Scholar] [CrossRef]
- Tortoli, E.; Meehan, C.J.; Grottola, A.; Serpini, G.F.; Fabio, A.; Trovato, A.; Pecorari, M.; Cirillo, D.M. Genome-based taxonomic revision detects a number of synonymous taxa in the genus Mycobacterium. Infect. Genet. Evol. 2019, 75, 103983. [Google Scholar] [CrossRef]
- Stanford, J.L.; Gunthorpe, W.J. A study of some fast-growing scotochromogenic mycobacteria including species descriptions of Mycobacterium gilvum (new species) and Mycobacterium duvalii (new species). Br. J. Exp. Pathol. 1971, 52, 627–637. [Google Scholar]
- Bouam, A.; Heidarieh, P.; Shahraki, A.H.; Pourahmad, F.; Mirsaeidi, M.; Hashemzadeh, M.; Baptiste, E.; Armstrong, N.; Levasseur, A.; Robert, C.; et al. Mycobacterium ahvazicum sp. nov., the nineteenth species of the Mycobacterium simiae complex. Sci. Rep. 2018, 8, 4138. [Google Scholar] [CrossRef] [PubMed]
- De Beenhouwer, H.; Liang, Z.; De Rijk, P.; Van Eekeren, C.; Portaels, F. Detection and identification of mycobacteria by DNA amplification and oligonucleotide-specific capture plate hybridization. J. Clin. Microbiol. 1995, 33, 2994–2998. [Google Scholar] [CrossRef] [PubMed]
- Andenmatten, S.; Opota, O.; Mazza-Stalder, J.; Nicod, L.; Greub, G.; Jaton, K. Added diagnostic value of 16S rRNA gene pan-mycobacterial PCR for nontuberculous mycobacterial infections: A 10-year retrospective study. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1873–1881. [Google Scholar] [CrossRef] [PubMed]
- Springer, B.; Stockman, L.; Teschner, K.; Roberts, G.D.; Böttger, E.C. Two-laboratory collaborative study on identification of mycobacteria: Molecular versus phenotypic methods. J. Clin. Microbiol. 1996, 34, 296–303. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Uh, Y.; Kim, S.; Lee, H. Performance of the Quantamatrix Multiplexed Assay Platform system for the differentiation and identification of Mycobacterium species. J. Med. Microbiol. 2017, 66, 777–787. [Google Scholar] [CrossRef]
- Wang, H.; Kim, H.; Kim, S.; Bang, H.; Kim, D.; Lee, H. Evaluation of PCR-reverse blot hybridization assay for the differentiation and identification of Mycobacterium species in liquid cultures. J. Appl. Microbiol. 2014, 118, 142–151. [Google Scholar] [CrossRef]
- Sun, Q.; Yan, J.; Liao, X.; Wang, C.; Wang, C.; Jiang, G.; Dong, L.; Wang, F.; Huang, H.; Wang, G.; et al. Trends and Species Diversity of Non-tuberculous Mycobacteria Isolated From Respiratory Samples in Northern China, 2014–2021. Front. Public Health 2022, 10, 923968. [Google Scholar] [CrossRef]
- Liu, X.; Lin, Z.; Li, Y.; Zhong, Z.; Wu, A.; Jiang, Y. Analysis of Clinical Isolation Characteristics of Nontuberculous Mycobacteria and Drug Sensitivity of Rapidly Growing Mycobacteria in the General Hospital of Guangzhou, China. Infect. Drug Resist. 2024, 17, 4079–4088. [Google Scholar] [CrossRef]
- Turenne, C.Y. Nontuberculous mycobacteria: Insights on taxonomy and evolution. Infect. Genet. Evol. 2019, 72, 159–168. [Google Scholar] [CrossRef]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front. Microbiol. 2018, 9, 67. [Google Scholar] [CrossRef]
- Cao, X.-J.; Li, Y.-P.; Wang, J.-Y.; Zhou, J.; Guo, X.-G. MPT64 assays for the rapid detection of Mycobacterium tuberculosis. BMC Infect. Dis. 2021, 21, 336. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, S.-J.; Kim, S.; Na Lee, H.; Sung, H.; Shim, T.S.; Jo, K.-W. Isolation of genetically distinct strains within the same species during treatment of MAC pulmonary disease. Chest 2024. [Google Scholar] [CrossRef] [PubMed]
- Wetzstein, N.; Kohl, T.A.; Andres, S.; Schultze, T.G.; Geil, A.; Kim, E.; Biciusca, T.; Hügel, C.; Hogardt, M.; Lehn, A.; et al. Comparative analysis of phenotypic and genotypic antibiotic susceptibility patterns in Mycobacterium avium complex. Int. J. Infect. Dis. 2020, 93, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoon, S.H.; Kim, T.S.; Seong, M.-W.; Han, S.K.; Yim, J.-J. Implication of species change of Nontuberculous Mycobacteria during or after treatment. BMC Pulm. Med. 2017, 17, 213. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.-J.; Park, C.M.; Park, Y.S.; Lee, J.; Lee, S.-M.; Yang, S.-C.; Yoo, C.-G.; Kim, Y.W.; Han, S.K.; Yim, J.-J. Isolation of multiple nontuberculous mycobacteria species in the same patients. Int. J. Infect. Dis. 2011, 15, e795–e798. [Google Scholar] [CrossRef]
- Rawlings, J.O.; Pantula, S.G.; Dickey, D.A. Applied Regression Analysis: A Research Tool, 2nd ed.; Springer Texts in Statistics; Springer: New York, NY, USA; Berlin/Heidelberg, Germany, 1998; ISBN 978-0-387-98454-4. [Google Scholar]
- Forbes, B.A.; Hall, G.S.; Miller, M.B.; Novak, S.M.; Rowlinson, M.-C.; Salfinger, M.; Somoskövi, A.; Warshauer, D.M.; Wilson, M.L. Practical Guidance for Clinical Microbiology Laboratories: Mycobacteria. Clin. Microbiol. Rev. 2018, 31, 10–1128. [Google Scholar] [CrossRef]
- Griffith, D.E.; Aksamit, T.; Brown-Elliott, B.A.; Catanzaro, A.; Daley, C.; Gordin, F.; Holland, S.M.; Horsburgh, R.; Huitt, G.; Iademarco, M.F.; et al. An Official ATS/IDSA Statement: Diagnosis, Treatment, and Prevention of Nontuberculous Mycobacterial Diseases. Am. J. Respir. Crit. Care Med. 2007, 175, 367–416. [Google Scholar] [CrossRef]
- Kim, D.; Park, S.; Chun, J. Introducing EzAAI: A pipeline for high throughput calculations of prokaryotic average amino acid identity. J. Microbiol. 2021, 59, 476–480. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Arahal, D.R.; Göker, M.; Moore, E.R.B.; Rossello-Mora, R.; Sutcliffe, I.C. International Code of Nomenclature of Prokaryotes. Prokaryotic Code (2022 Revision). Int. J. Syst. Evol. Microbiol. 2023, 73, 005585. [Google Scholar] [CrossRef] [PubMed]

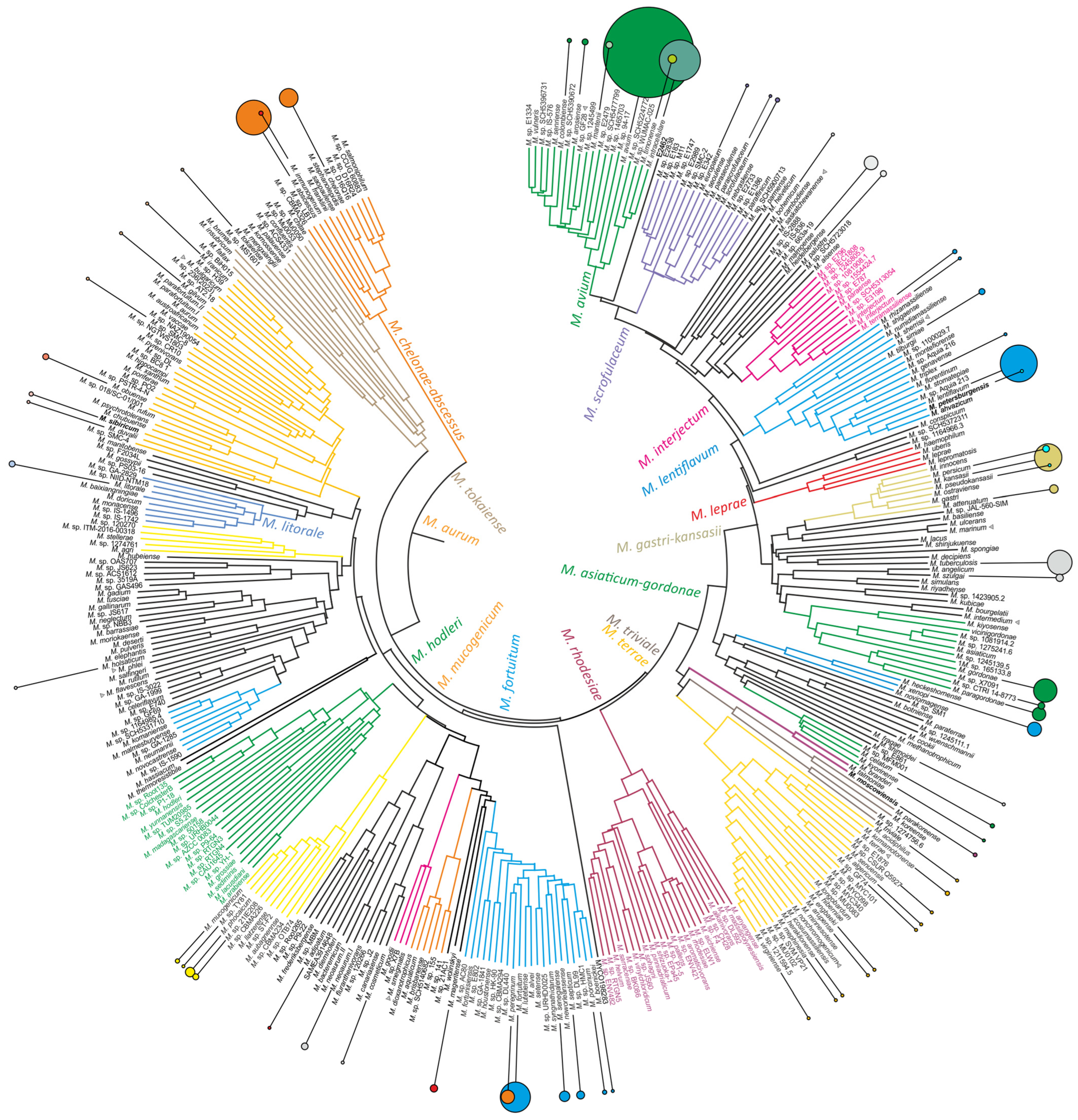
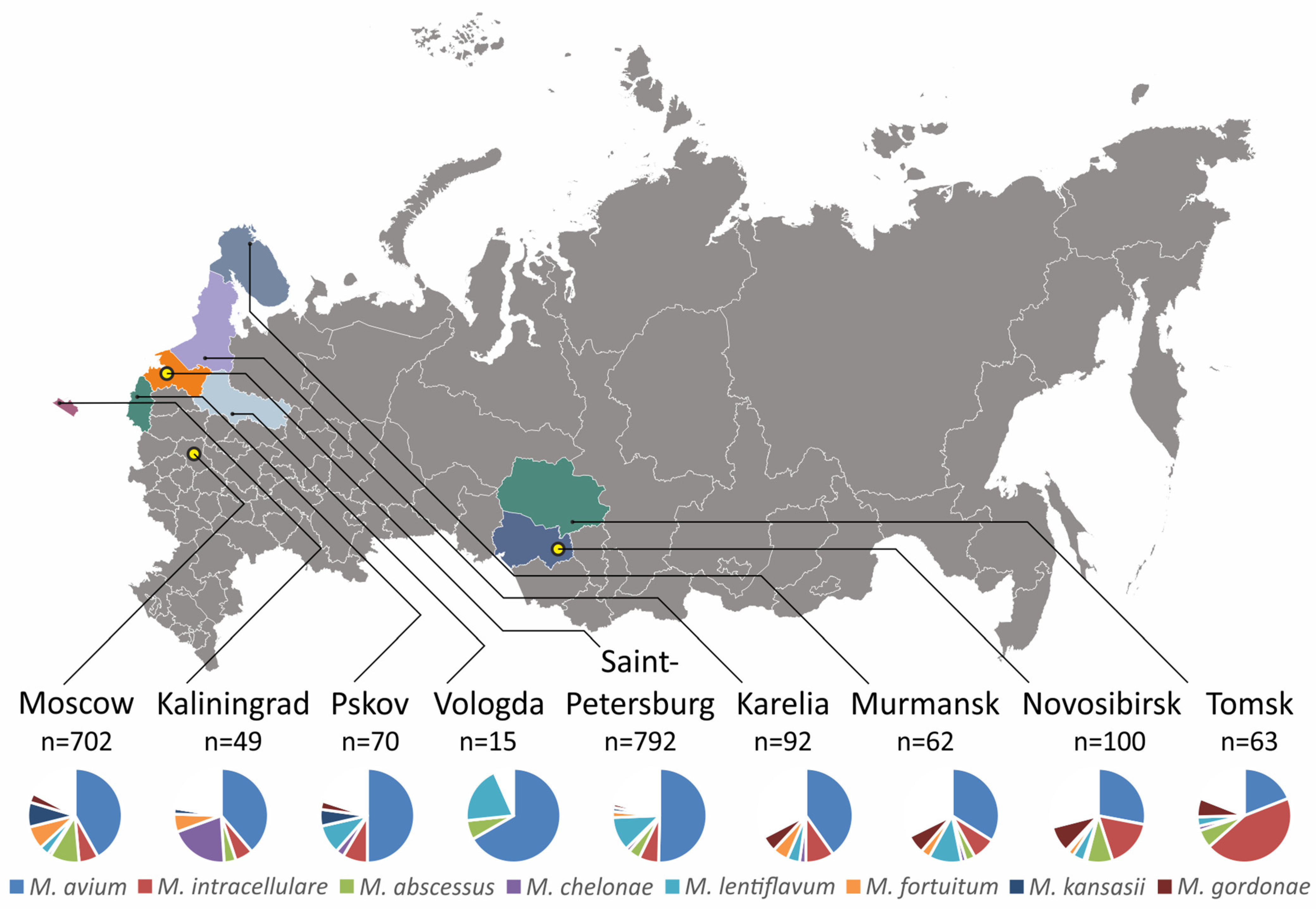
| Cluster or Complex | Growth Rate | Species in Cluster | Total Cases | Cases with Repeated Isolation |
|---|---|---|---|---|
| M. avium | slow | 6 | 1135 | 306 |
| M. chelonae-abscessus | rapid | 3 | 177 | 38 |
| M. lentiflavum | slow | 4 | 159 | 17 |
| M. fortuitum | rapid | 6 | 141 | 15 |
| M. gastri-kansasii | slow | 4 | 98 | 45 |
| M. asiaticum-gordonae | slow | 3 | 86 | 6 |
| M. tuberculosis | slow | 1 | 68 | - |
| M. malmoense | slow | 2 | 25 | 6 |
| M. xenopi | slow | 1 | 22 | 5 |
| M. mucogenicum | rapid | 4 | 17 | 4 |
| M. terrae | slow | 9 | 8 | 0 |
| M. neoaurum | rapid | 3 | 7 | 0 |
| M. szulgai | slow | 1 | 6 | 3 |
| M. aurum | rapid | 3 | 5 | 3 |
| M. chubuense | rapid | 1 | 4 | 1 |
| M. shimodei | slow | 3 | 4 | 1 |
| M. scrofulaceum | slow | 3 | 4 | 2 |
| M. litorale | rapid | 1 | 4 | 1 |
| M. duvalii | rapid | 2 | 3 | 0 |
| M. interjectum | slow | 2 | 3 | 0 |
| M. talmoniae | slow | 1 | 2 | 0 |
| M. triviale | slow | 1 | 1 | 0 |
| M. bohemicum | slow | 1 | 1 | 0 |
| M. tokaiense | rapid | 1 | 1 | 0 |
| M. elephantis | rapid | 1 | 1 | 1 |
| (M. mageritense) | rapid | 1 | 6 | 1 |
| Cluster | Isolate | Sample | Reference | Total Cases | Cases with Repeated Isolation |
|---|---|---|---|---|---|
| M. aurum | SMC-8 | SAMN20167551 | - | 2 | 1 |
| M. mucogenicum | 21IE208 | SAMN31811490 | - | 4 | 3 |
| M. mucogenicum | TY81 | SAMD00239442 | - | 1 | |
| M. terrae | CSUR_Q5927 * | SAMN32979486 | - | 1 | |
| M. asiaticum | CTRI_14-8773 | SAMN04123344 | [52] | 5 | 1 |
| Strain Name | pU-009 | pV-006 | pR-1184 |
|---|---|---|---|
| Proposed name | M. moscowiensis | M. sibiricum | M. petersburgensis |
| Genome accession | GCA_041287095.1 | GCA_041287115.1 | GCA_041287235.1 |
| Assembly (bp) | 5,730,899 | 5,120,508 | 5,944,740 |
| GC (%) | 68.44 | 68.01 | 66.1 |
| Genes | 5538 | 4963 | 5632 |
| CDSs | 5486 | 4906 | 5578 |
| Pseudo Genes | 109 | 94 | 83 |
| RNA | 52 | 57 | 54 |
| Closest species | M. talmoniae | M. duvalii | M. lentiflavum |
| ANI (%) | 92.11 | 84.20 | 91.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimenkov, D.; Zhuravlev, V.; Ushtanit, A.; Filippova, M.; Semenova, U.; Solovieva, N.; Sviridenko, M.; Khakhalina, A.; Safonova, S.; Makarova, M.; et al. Biochip-Based Identification of Mycobacterial Species in Russia. Int. J. Mol. Sci. 2024, 25, 13200. https://doi.org/10.3390/ijms252313200
Zimenkov D, Zhuravlev V, Ushtanit A, Filippova M, Semenova U, Solovieva N, Sviridenko M, Khakhalina A, Safonova S, Makarova M, et al. Biochip-Based Identification of Mycobacterial Species in Russia. International Journal of Molecular Sciences. 2024; 25(23):13200. https://doi.org/10.3390/ijms252313200
Chicago/Turabian StyleZimenkov, Danila, Vyacheslav Zhuravlev, Anastasia Ushtanit, Marina Filippova, Uliana Semenova, Natalia Solovieva, Maria Sviridenko, Anastasia Khakhalina, Svetlana Safonova, Marina Makarova, and et al. 2024. "Biochip-Based Identification of Mycobacterial Species in Russia" International Journal of Molecular Sciences 25, no. 23: 13200. https://doi.org/10.3390/ijms252313200
APA StyleZimenkov, D., Zhuravlev, V., Ushtanit, A., Filippova, M., Semenova, U., Solovieva, N., Sviridenko, M., Khakhalina, A., Safonova, S., Makarova, M., Gordeeva, E., Guselnikova, E., Schwartz, Y., Stavitskaya, N., & Yablonsky, P. (2024). Biochip-Based Identification of Mycobacterial Species in Russia. International Journal of Molecular Sciences, 25(23), 13200. https://doi.org/10.3390/ijms252313200







