Molecular Pathway and Immune Profile Analysis of IPMN-Derived Versus PanIN-Derived Pancreatic Ductal Adenocarcinomas
Abstract
1. Introduction
2. Results
3. Discussion
4. Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tonini, V.; Zanni, M. Pancreatic cancer in 2021: What you need to know to win. World J. Gastroenterol. 2021, 27, 5851–5889. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2023-cancer-facts-figures.html (accessed on 10 June 2024).
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.D.; Mazzilli, S.A.; Reid, M.E.; Dhillon, S.S.; Platero, S.; Beane, J.; Spira, A.E. The Case for a Pre-Cancer Genome Atlas (PCGA). Cancer Prev. Res. 2016, 9, 119–124. [Google Scholar] [CrossRef]
- Srivastava, S.; Wagner, P.D.; Hughes, S.K.; Ghosh, S. PreCancer Atlas: Present and Future. Cancer Prev. Res. 2023, 16, 379–384. [Google Scholar] [CrossRef]
- Moris, M.; Bridges, M.D.; Pooley, R.A.; Raimondo, M.; Woodward, T.A.; Stauffer, J.A.; Asbun, H.J.; Wallace, M.B. Association Between Advances in High-Resolution Cross-Section Imaging Technologies and Increase in Prevalence of Pancreatic Cysts From 2005 to 2014. Clin. Gastroenterol. Hepatol. 2016, 14, 585–593.e583. [Google Scholar] [CrossRef]
- Laffan, T.A.; Horton, K.M.; Klein, A.P.; Berlanstein, B.; Siegelman, S.S.; Kawamoto, S.; Johnson, P.T.; Fishman, E.K.; Hruban, R.H. Prevalence of unsuspected pancreatic cysts on MDCT. AJR Am. J. Roentgenol. 2008, 191, 802–807. [Google Scholar] [CrossRef]
- Ricci, C.; Migliori, M.; Imbrogno, A.; Mazzotta, E.; Felicani, C.; Serra, C.; Bergonzoni, B.; Calculli, L.; Casadei, R. Prevalence of Asymptomatic Intraductal Papillary Mucinous Neoplasms in Healthy and Ill Populations Detected by Ultrasonography: A Single-Center Study of 6353 Outpatients. Pancreas 2019, 48, 113–120. [Google Scholar] [CrossRef]
- Latenstein, A.E.J.; Mackay, T.M.; Beane, J.D.; Busch, O.R.; van Dieren, S.; Gleeson, E.M.; Koerkamp, B.G.; van Santvoort, H.C.; Wellner, U.F.; Williamsson, C.; et al. The use and clinical outcome of total pancreatectomy in the United States, Germany, the Netherlands, and Sweden. Surgery 2021, 170, 563–570. [Google Scholar] [CrossRef]
- Hosoda, W.; Sasaki, E.; Murakami, Y.; Yamao, K.; Shimizu, Y.; Yatabe, Y. GNAS mutation is a frequent event in pancreatic intraductal papillary mucinous neoplasms and associated adenocarcinomas. Virchows Arch. 2015, 466, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Distler, M.; Aust, D.; Weitz, J.; Pilarsky, C.; Grutzmann, R. Precursor lesions for sporadic pancreatic cancer: PanIN, IPMN, and MCN. BioMed Res. Int. 2014, 2014, 474905. [Google Scholar] [CrossRef] [PubMed]
- Grutzmann, R.; Niedergethmann, M.; Pilarsky, C.; Kloppel, G.; Saeger, H.D. Intraductal papillary mucinous tumors of the pancreas: Biology, diagnosis, and treatment. Oncologist 2010, 15, 1294–1309. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.C.; Basturk, O.; Brannon, A.R.; Bhanot, U.; Scott, S.N.; Bouvier, N.; LaFemina, J.; Jarnagin, W.R.; Berger, M.F.; Klimstra, D.; et al. GNAS and KRAS Mutations Define Separate Progression Pathways in Intraductal Papillary Mucinous Neoplasm-Associated Carcinoma. J. Am. Coll. Surg. 2015, 220, 845–854.e841. [Google Scholar] [CrossRef] [PubMed]
- Eshleman, J.R.; Norris, A.L.; Sadakari, Y.; Debeljak, M.; Borges, M.; Harrington, C.; Lin, E.; Brant, A.; Barkley, T.; Almario, J.A.; et al. KRAS and guanine nucleotide-binding protein mutations in pancreatic juice collected from the duodenum of patients at high risk for neoplasia undergoing endoscopic ultrasound. Clin. Gastroenterol. Hepatol. 2015, 13, 963–969.e964. [Google Scholar] [CrossRef]
- Huang, X.; Feng, Y.; Ma, D.; Ding, H.; Dong, G.; Chen, Y.; Huang, X.; Zhang, J.; Xu, X.; Chen, C. The molecular, immune features, and risk score construction of intraductal papillary mucinous neoplasm patients. Front. Mol. Biosci. 2022, 9, 887887. [Google Scholar] [CrossRef]
- Zhang, C.L.; Shen, Q.; Liu, F.D.; Yang, F.; Gao, M.Q.; Jiang, X.C.; Li, Y.; Zhang, X.Y.; En, G.E.; Pan, X.; et al. SDC1 and ITGA2 as novel prognostic biomarkers for PDAC related to IPMN. Sci. Rep. 2023, 13, 18727. [Google Scholar] [CrossRef]
- Fischer, C.G.; Beleva Guthrie, V.; Braxton, A.M.; Zheng, L.; Wang, P.; Song, Q.; Griffin, J.F.; Chianchiano, P.E.; Hosoda, W.; Niknafs, N.; et al. Intraductal Papillary Mucinous Neoplasms Arise From Multiple Independent Clones, Each With Distinct Mutations. Gastroenterology 2019, 157, 1123–1137.e1122. [Google Scholar] [CrossRef]
- Hosoda, W.; Chianchiano, P.; Griffin, J.F.; Pittman, M.E.; Brosens, L.A.; Noe, M.; Yu, J.; Shindo, K.; Suenaga, M.; Rezaee, N.; et al. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J. Pathol. 2017, 242, 16–23. [Google Scholar] [CrossRef]
- Yoon, J.G.; Smith, D.; Ojili, V.; Paspulati, R.M.; Ramaiya, N.H.; Tirumani, S.H. Pancreatic cystic neoplasms: A review of current recommendations for surveillance and management. Abdom. Radiol. 2021, 46, 3946–3962. [Google Scholar] [CrossRef]
- Reid, M.D.; Bhattarai, S.; Graham, R.P.; Pehlivanoglu, B.; Sigel, C.S.; Shi, J.; Saqi, A.; Shirazi, M.; Xue, Y.; Basturk, O.; et al. Pancreatoblastoma: Cytologic and histologic analysis of 12 adult cases reveals helpful criteria in their diagnosis and distinction from common mimics. Cancer Cytopathol. 2019, 127, 708–719. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Sharma, A.; Chhoda, A.; Ruzgar, N.; Hasan, N.; Kwak, R.; Wolfgang, C.L.; Wang, T.H.; Kunstman, J.W.; Salem, R.R.; et al. Methylation-based Cell-free DNA Signature for Early Detection of Pancreatic Cancer. Pancreas 2021, 50, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Fujikura, K.; Hosoda, W.; Felsenstein, M.; Song, Q.; Reiter, J.G.; Zheng, L.; Beleva Guthrie, V.; Rincon, N.; Dal Molin, M.; Dudley, J.; et al. Multiregion whole-exome sequencing of intraductal papillary mucinous neoplasms reveals frequent somatic KLF4 mutations predominantly in low-grade regions. Gut 2021, 70, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Lee, K.; Lockhart, J.H.; Cukras, S.D.; Carvajal, R.; Beg, A.A.; Flores, E.R.; Teng, M.; Chung, C.H.; Tan, A.C. TIMEx: Tumor-immune microenvironment deconvolution web-portal for bulk transcriptomics using pan-cancer scRNA-seq signatures. Bioinformatics 2021, 37, 3681–3683. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wang, J.; Han, Y.; Dong, X.; Ge, J.; Zheng, R.; Shi, X.; Wang, B.; Li, Z.; Ren, P.; et al. TISCH: A comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021, 49, D1420–D1430. [Google Scholar] [CrossRef] [PubMed]
- Griss, J.; Viteri, G.; Sidiropoulos, K.; Nguyen, V.; Fabregat, A.; Hermjakob, H. ReactomeGSA—Efficient Multi-Omics Comparative Pathway Analysis. Mol. Cell. Proteom. 2020, 19, 2115–2125. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Marin-Garcia, P.; Ping, P.; Stein, L.; D’Eustachio, P.; Hermjakob, H. Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics 2018, 34, 1208–1214. [Google Scholar] [CrossRef]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Huang, B.; Trujillo, M.A.; Fujikura, K.; Qiu, M.; Chen, F.; Felsenstein, M.; Zhou, C.; Skaro, M.; Gauthier, C.; Macgregor-Das, A.; et al. Molecular characterization of organoids derived from pancreatic intraductal papillary mucinous neoplasms. J. Pathol. 2020, 252, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Fukushima, N.; Maitra, A.; Iacobuzio-Donahue, C.A.; van Heek, N.T.; Cameron, J.L.; Yeo, C.J.; Hruban, R.H.; Goggins, M. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am. J. Pathol. 2004, 164, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Gaiser, R.A.; Pessia, A.; Ateeb, Z.; Davanian, H.; Fernandez Moro, C.; Alkharaan, H.; Healy, K.; Ghazi, S.; Arnelo, U.; Valente, R.; et al. Integrated targeted metabolomic and lipidomic analysis: A novel approach to classifying early cystic precursors to invasive pancreatic cancer. Sci. Rep. 2019, 9, 10208. [Google Scholar] [CrossRef]
- Tang, Y.; Tian, W.; Xie, J.; Zou, Y.; Wang, Z.; Li, N.; Zeng, Y.; Wu, L.; Zhang, Y.; Wu, S.; et al. Prognosis and Dissection of Immunosuppressive Microenvironment in Breast Cancer Based on Fatty Acid Metabolism-Related Signature. Front. Immunol. 2022, 13, 843515. [Google Scholar] [CrossRef]
- Swierczynski, J.; Hebanowska, A.; Sledzinski, T. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World J. Gastroenterol. 2014, 20, 2279–2303. [Google Scholar] [CrossRef]
- Ose, J.; Gigic, B.; Brezina, S.; Lin, T.; Baierl, A.; Geijsen, A.; van Roekel, E.; Robinot, N.; Gicquiau, A.; Achaintre, D.; et al. Targeted Plasma Metabolic Profiles and Risk of Recurrence in Stage II and III Colorectal Cancer Patients: Results from an International Cohort Consortium. Metabolites 2021, 11, 129. [Google Scholar] [CrossRef]
- Gong, Y.; Wu, S.; Dong, S.; Chen, S.; Cai, G.; Bao, K.; Yang, H.; Jiao, Y. Development of a prognostic metabolic signature in stomach adenocarcinoma. Clin. Transl. Oncol. 2022, 24, 1615–1630. [Google Scholar] [CrossRef]
- Chakraborty, P.; George, J.T.; Woodward, W.A.; Levine, H.; Jolly, M.K. Gene expression profiles of inflammatory breast cancer reveal high heterogeneity across the epithelial-hybrid-mesenchymal spectrum. Transl. Oncol. 2021, 14, 101026. [Google Scholar] [CrossRef]
- El Rayes, T.; Catena, R.; Lee, S.; Stawowczyk, M.; Joshi, N.; Fischbach, C.; Powell, C.A.; Dannenberg, A.J.; Altorki, N.K.; Gao, D.; et al. Lung inflammation promotes metastasis through neutrophil protease-mediated degradation of Tsp-1. Proc. Natl. Acad. Sci. USA 2015, 112, 16000–16005. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Cai, W.; Yang, J.; Fu, X.; Putha, L.; Xia, Q.; Windsor, J.A.; Phillips, A.R.; Tyndall, J.D.A.; Du, D.; et al. Targeting Macrophage Migration Inhibitory Factor in Acute Pancreatitis and Pancreatic Cancer. Front. Pharmacol. 2021, 12, 638950. [Google Scholar] [CrossRef] [PubMed]
- Moschovis, D.; Bamias, G.; Delladetsima, I. Mucins in neoplasms of pancreas, ampulla of Vater and biliary system. World J. Gastrointest. Oncol. 2016, 8, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.S.; Ciprani, D.; O’Shea, A.; Liss, A.S.; Yang, R.; Fletcher-Mercaldo, S.; Mino-Kenudson, M.; Fernandez-Del Castillo, C.; Weissleder, R. Extracellular Vesicle Analysis Allows for Identification of Invasive IPMN. Gastroenterology 2021, 160, 1345–1358.e11. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, J.; Zheng, S.; Lu, Z.; Che, Y.; Mao, S.; Lei, Y.; Zang, R.; Liu, C.; Wang, X.; et al. Tumor microenvironment characterization identifies two lung adenocarcinoma subtypes with specific immune and metabolic state. Cancer Sci. 2020, 111, 1876–1886. [Google Scholar] [CrossRef]
- Stewart, P.A.; Welsh, E.A.; Slebos, R.J.C.; Fang, B.; Izumi, V.; Chambers, M.; Zhang, G.; Cen, L.; Pettersson, F.; Zhang, Y.; et al. Proteogenomic landscape of squamous cell lung cancer. Nat. Commun. 2019, 10, 3578. [Google Scholar] [CrossRef]
- Yang, C.; Huang, X.; Liu, Z.; Qin, W.; Wang, C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol. Oncol. 2020, 14, 896–913. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, D.; Chen, Y.; Li, H.; Wang, J.; Yuan, Z.; Sun, L.; Zhou, Z.; Chen, M.; Zhang, Y.; et al. Development and validation of robust metabolism-related gene signature in the prognostic prediction of hepatocellular carcinoma. J. Cell. Mol. Med. 2023, 27, 1006–1020. [Google Scholar] [CrossRef]
- Pihlak, R.; Weaver, J.M.J.; Valle, J.W.; McNamara, M.G. Advances in Molecular Profiling and Categorisation of Pancreatic Adenocarcinoma and the Implications for Therapy. Cancers 2018, 10, 17. [Google Scholar] [CrossRef]
- Smith, H.; Arbe-Barnes, E.; Shah, E.A.; Sivakumar, S. Manipulating regulatory T cells: Is it the key to unlocking effective immunotherapy for pancreatic ductal adenocarcinoma? Front. Immunol. 2024, 15, 1406250. [Google Scholar] [CrossRef]
- Sarfraz, Z.; Sarfraz, A.; Farooq, M.D.; Khalid, M.; Cheema, K.; Javad, F.; Khan, T.; Pervaiz, Z.; Sarfraz, M.; Jaan, A.; et al. The Current Landscape of Clinical Trials for Immunotherapy in Pancreatic Cancer: A State-of-the-Art Review. J. Gastrointest. Cancer 2024, 55, 1026–1057. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; Koay, E.J.; Chari, S.T.; Maitra, A. Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology 2019, 156, 2024–2040. [Google Scholar] [CrossRef] [PubMed]
- Oyama, H.; Hamada, T.; Nakai, Y.; Tanaka, M.; Takagi, K.; Fukuda, R.; Hakuta, R.; Ishigaki, K.; Kanai, S.; Kawaguchi, Y.; et al. Intraductal Papillary Mucinous Neoplasm Surveillance Leads to Early Diagnosis and Better Outcomes of Concomitant Cancer. Ann. Surg. 2024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xue, H.; Dong, Y.; Zhang, J.; Pan, Y.; Shi, L.; Xiong, P.; Zhu, J.; Li, W.; Zheng, W.; et al. GKN2 promotes oxidative stress-induced gastric cancer cell apoptosis via the Hsc70 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 338. [Google Scholar] [CrossRef]
- Steiner, S.; Seleznik, G.M.; Reding, T.; Stopic, M.; Lenggenhager, D.; Ten Buren, E.; Eshmuminov, D.; Endhardt, K.; Hagedorn, C.; Heidenblut, A.M.; et al. De novo expression of gastrokines in pancreatic precursor lesions impede the development of pancreatic cancer. Oncogene 2022, 41, 1507–1517. [Google Scholar] [CrossRef]
- Ouyang, J.; Pan, X.; Lin, H.; Hu, Z.; Xiao, P.; Hu, H. GKN2 increases apoptosis, reduces the proliferation and invasion ability of gastric cancer cells through down-regulating the JAK/STAT signaling pathway. Am. J. Transl. Res. 2017, 9, 803–811. [Google Scholar]
- Tarhan, Y.E.; Kato, T.; Jang, M.; Haga, Y.; Ueda, K.; Nakamura, Y.; Park, J.H. Morphological Changes, Cadherin Switching, and Growth Suppression in Pancreatic Cancer by GALNT6 Knockdown. Neoplasia 2016, 18, 265–272. [Google Scholar] [CrossRef]
- Pin, F.; Novinger, L.J.; Huot, J.R.; Harris, R.A.; Couch, M.E.; O’Connell, T.M.; Bonetto, A. PDK4 drives metabolic alterations and muscle atrophy in cancer cachexia. FASEB J. 2019, 33, 7778–7790. [Google Scholar] [CrossRef]
- Yang, X.; Jiang, S.; Yuan, Z.; Jiang, J.; Yang, M.; Luo, J.; Ye, T. SPINK4 modulates inhibition of glycolysis against colorectal cancer progression. Biomol. Biomed. 2024, 24, 1571–1585. [Google Scholar] [CrossRef]
- Pu, N.; Gao, S.; Xu, Y.; Zhao, G.; Lv, Y.; Nuerxiati, A.; Li, J.A.; Wang, D.; Xu, X.; Kuang, T.; et al. Alkaline Phosphatase-To-Albumin Ratio as a Prognostic Indicator in Pancreatic Ductal Adenocarcinoma after Curative Resection. J. Cancer 2017, 8, 3362–3370. [Google Scholar] [CrossRef]
- Chen, D.T.; Davis-Yadley, A.H.; Huang, P.Y.; Husain, K.; Centeno, B.A.; Permuth-Wey, J.; Pimiento, J.M.; Malafa, M. Prognostic Fifteen-Gene Signature for Early Stage Pancreatic Ductal Adenocarcinoma. PLoS ONE 2015, 10, e0133562. [Google Scholar] [CrossRef] [PubMed]
- Helm, O.; Held-Feindt, J.; Grage-Griebenow, E.; Reiling, N.; Ungefroren, H.; Vogel, I.; Kruger, U.; Becker, T.; Ebsen, M.; Rocken, C.; et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer 2014, 135, 843–861. [Google Scholar] [CrossRef] [PubMed]
- Karakhanova, S.; Link, J.; Heinrich, M.; Shevchenko, I.; Yang, Y.; Hassenpflug, M.; Bunge, H.; von Ahn, K.; Brecht, R.; Mathes, A.; et al. Characterization of myeloid leukocytes and soluble mediators in pancreatic cancer: Importance of myeloid-derived suppressor cells. Oncoimmunology 2015, 4, e998519. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Morine, Y.; Ikemoto, T.; Imura, S.; Iwahashi, S.; Saito, Y.; Shimada, M. Nab-paclitaxel interrupts cancer-stromal interaction through C-X-C motif chemokine 10-mediated interleukin-6 downregulation in vitro. Cancer Sci. 2018, 109, 2509–2519. [Google Scholar] [CrossRef] [PubMed]
- McAllister, F.; Bailey, J.M.; Alsina, J.; Nirschl, C.J.; Sharma, R.; Fan, H.; Rattigan, Y.; Roeser, J.C.; Lankapalli, R.H.; Zhang, H.; et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell 2014, 25, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Fan, W.; Xu, Z.; Chen, H.; He, Y.; Yang, G.; Yang, G.; Hu, H.; Tang, S.; Wang, P.; et al. Inhibiting tumor necrosis factor-alpha diminishes desmoplasia and inflammation to overcome chemoresistance in pancreatic ductal adenocarcinoma. Oncotarget 2016, 7, 81110–81122. [Google Scholar] [CrossRef]
- Senturk, Z.N.; Akdag, I.; Deniz, B.; Sayi-Yazgan, A. Pancreatic cancer: Emerging field of regulatory B-cell-targeted immunotherapies. Front. Immunol. 2023, 14, 1152551. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Pontious, C.; Kovalenko, I.; Lapienyte, L.; Dreyer, S.; Lee, H.J.; Thurston, G.; Zhang, Y.; Lazarus, J.; Sajjakulnukit, P.; et al. Macrophage-Released Pyrimidines Inhibit Gemcitabine Therapy in Pancreatic Cancer. Cell Metab. 2019, 29, 1390–1399.e6. [Google Scholar] [CrossRef]
- Shi, J.; Yi, Z.; Jin, L.; Zhao, L.; Raskind, A.; Yeomans, L.; Nwosu, Z.C.; Simeone, D.M.; Lyssiotis, C.A.; Stringer, K.A.; et al. Cyst fluid metabolites distinguish malignant from benign pancreatic cysts. Neoplasia 2021, 23, 1078–1088. [Google Scholar] [CrossRef]
- Seo, J.; Jeong, D.W.; Park, J.W.; Lee, K.W.; Fukuda, J.; Chun, Y.S. Fatty-acid-induced FABP5/HIF-1 reprograms lipid metabolism and enhances the proliferation of liver cancer cells. Commun. Biol. 2020, 3, 638. [Google Scholar] [CrossRef]
- Pita-Grisanti, V.; Velez-Bonet, E.; Chasser, K.; Hurst, Z.; Liette, A.; Vulic, G.; Dubay, K.; Lahooti, A.; Badi, N.; Ueltschi, O.; et al. Physical Activity Decreases Inflammation and Delays the Development of Obesity-Associated Pancreatic Ductal Adenocarcinoma. Cancer Res. 2024, 84, 3058–3071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wei, Y.; Wang, C. Impacts of an Exercise Intervention on the Health of Pancreatic Beta-Cells: A Review. Int. J. Environ. Res. Public. Health 2022, 19, 7229. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.C.; Westermark, A.M.; Zhang, Y.; Yuan, C.; Li, Z.; Lau, A.N.; Sapp, K.M.; Wolpin, B.M.; Vander Heiden, M.G. Low glycaemic diets alter lipid metabolism to influence tumour growth. Nature 2021, 599, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, N.D.; Wijma, A.G.; Bongers, B.C.; Rensen, S.S.; den Dulk, M.; Klaase, J.M.; Olde Damink, S.W.M. Supervised Home-Based Exercise Prehabilitation in Unfit Patients Scheduled for Pancreatic Surgery: Protocol for a Multicenter Feasibility Study. JMIR Res. Protoc. 2023, 12, e46526. [Google Scholar] [CrossRef] [PubMed]
- Nimmakayala, R.K.; Leon, F.; Rachagani, S.; Rauth, S.; Nallasamy, P.; Marimuthu, S.; Shailendra, G.K.; Chhonker, Y.S.; Chugh, S.; Chirravuri, R.; et al. Metabolic programming of distinct cancer stem cells promotes metastasis of pancreatic ductal adenocarcinoma. Oncogene 2021, 40, 215–231. [Google Scholar] [CrossRef]
- Nimmakayala, R.K.; Rauth, S.; Chirravuri Venkata, R.; Marimuthu, S.; Nallasamy, P.; Vengoji, R.; Lele, S.M.; Rachagani, S.; Mallya, K.; Malafa, M.P.; et al. PGC1alpha-Mediated Metabolic Reprogramming Drives the Stemness of Pancreatic Precursor Lesions. Clin. Cancer Res. 2021, 27, 5415–5429. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, L.; Ding, G.; Huang, H.; Cao, G.; Sun, X.; Lou, N.; Wei, Q.; Shen, T.; Xu, X.; et al. Interferon gamma inhibits CXCL8-CXCR2 axis mediated tumor-associated macrophages tumor trafficking and enhances anti-PD1 efficacy in pancreatic cancer. J. Immunother. Cancer 2020, 8, e000308. [Google Scholar] [CrossRef]
- Detjen, K.M.; Farwig, K.; Welzel, M.; Wiedenmann, B.; Rosewicz, S. Interferon gamma inhibits growth of human pancreatic carcinoma cells via caspase-1 dependent induction of apoptosis. Gut 2001, 49, 251–262. [Google Scholar] [CrossRef]
- Imai, D.; Yoshizumi, T.; Okano, S.; Itoh, S.; Ikegami, T.; Harada, N.; Aishima, S.; Oda, Y.; Maehara, Y. IFN-gamma Promotes Epithelial-Mesenchymal Transition and the Expression of PD-L1 in Pancreatic Cancer. J. Surg. Res. 2019, 240, 115–123. [Google Scholar] [CrossRef]
- Gupta, A.; Zhang, D.; Braithwaite, D.; Karanth, S.D.; Tailor, T.D.; Clarke, J.M.; Akinyemiju, T. Racial Differences in Survival Among Advanced-stage Non-small-Cell Lung Cancer Patients Who Received Immunotherapy: An Analysis of the US National Cancer Database (NCDB). J. Immunother. 2022, 45, 132–137. [Google Scholar] [CrossRef]
- Olateju, O.A.; Zeng, Z.; Adenaiye, O.O.; Varisco, T.J.; Zakeri, M.; Sujit, S.S. Investigation of racial differences in survival from non-small cell lung cancer with immunotherapy use: A Texas study. Front. Oncol. 2022, 12, 1092355. [Google Scholar] [CrossRef] [PubMed]
- Nazha, B.; Mishra, M.; Pentz, R.; Owonikoko, T.K. Enrollment of Racial Minorities in Clinical Trials: Old Problem Assumes New Urgency in the Age of Immunotherapy. Am. Soc. Clin. Oncol. Educ. Book. 2019, 39, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Temple, J.R.; Breitkopf, C.R.; Berenson, A.B. Racial differences in body fat distribution among reproductive-aged women. Metabolism 2009, 58, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Hunter, G.R.; Weinsier, R.L.; Darnell, B.E.; Zuckerman, P.A.; Goran, M.I. Racial differences in energy expenditure and aerobic fitness in premenopausal women. Am. J. Clin. Nutr. 2000, 71, 500–506. [Google Scholar] [CrossRef]
- Kimm, S.Y.; Glynn, N.W.; Aston, C.E.; Damcott, C.M.; Poehlman, E.T.; Daniels, S.R.; Ferrell, R.E. Racial differences in the relation between uncoupling protein genes and resting energy expenditure. Am. J. Clin. Nutr. 2002, 75, 714–719. [Google Scholar] [CrossRef]
- Berman, D.M.; Rodrigues, L.M.; Nicklas, B.J.; Ryan, A.S.; Dennis, K.E.; Goldberg, A.P. Racial disparities in metabolism, central obesity, and sex hormone-binding globulin in postmenopausal women. J. Clin. Endocrinol. Metab. 2001, 86, 97–103. [Google Scholar] [CrossRef]
- Fenstermacher, D.A.; Wenham, R.M.; Rollison, D.E.; Dalton, W.S. Implementing personalized medicine in a cancer center. Cancer J. 2011, 17, 528–536. [Google Scholar] [CrossRef]
- Tanaka, M.; Fernandez-Del Castillo, C.; Kamisawa, T.; Jang, J.Y.; Levy, P.; Ohtsuka, T.; Salvia, R.; Shimizu, Y.; Tada, M.; Wolfgang, C.L. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 2017, 17, 738–753. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Results Based upon Data Generated by the TCGA Research Network. Available online: https://www.cancer.gov/tcga (accessed on 29 September 2021).
- Gu, Z. Complex heatmap visualization. Imeta 2022, 1, e43. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://github.com/friendly/ggbiplot (accessed on 8 April 2023).
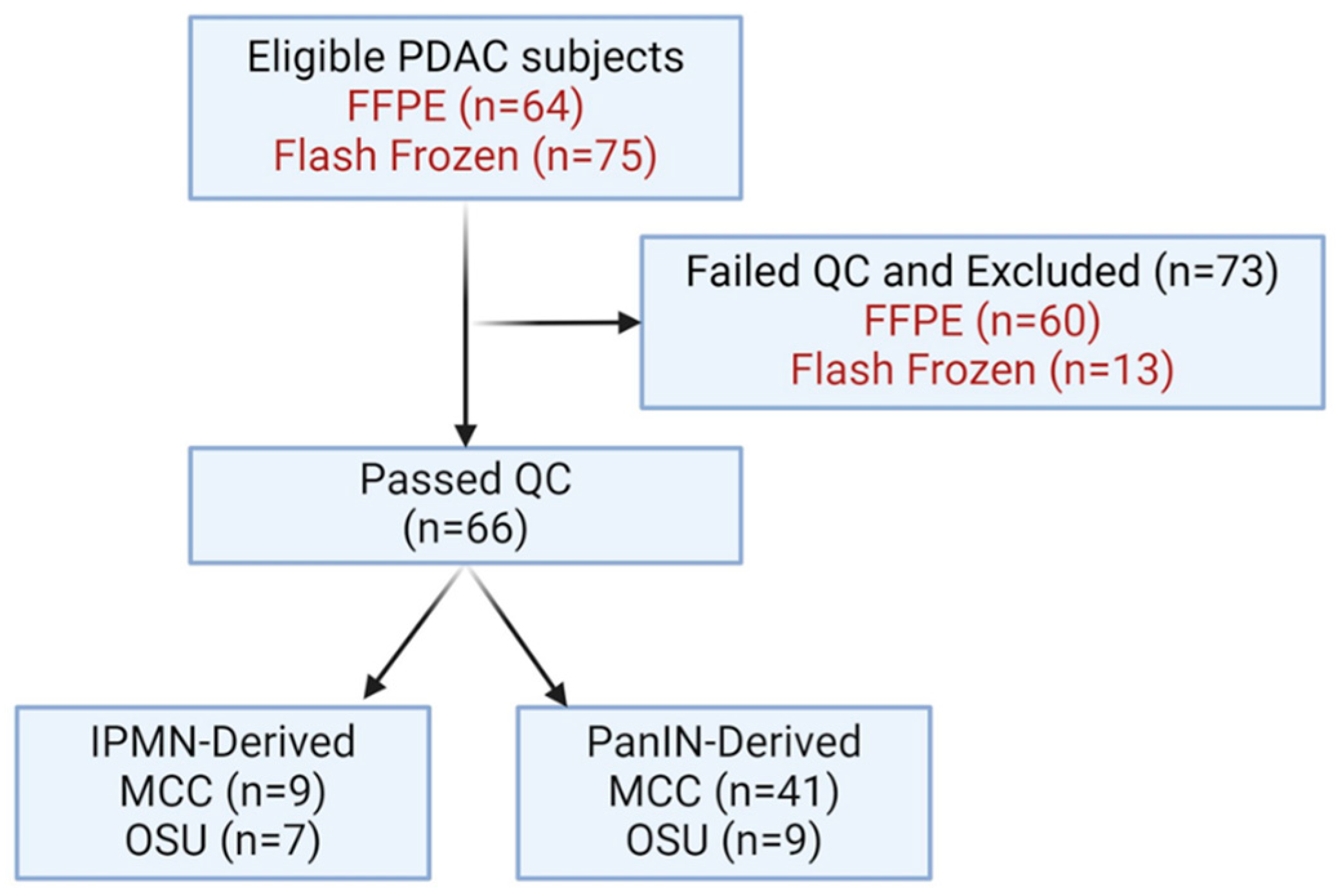
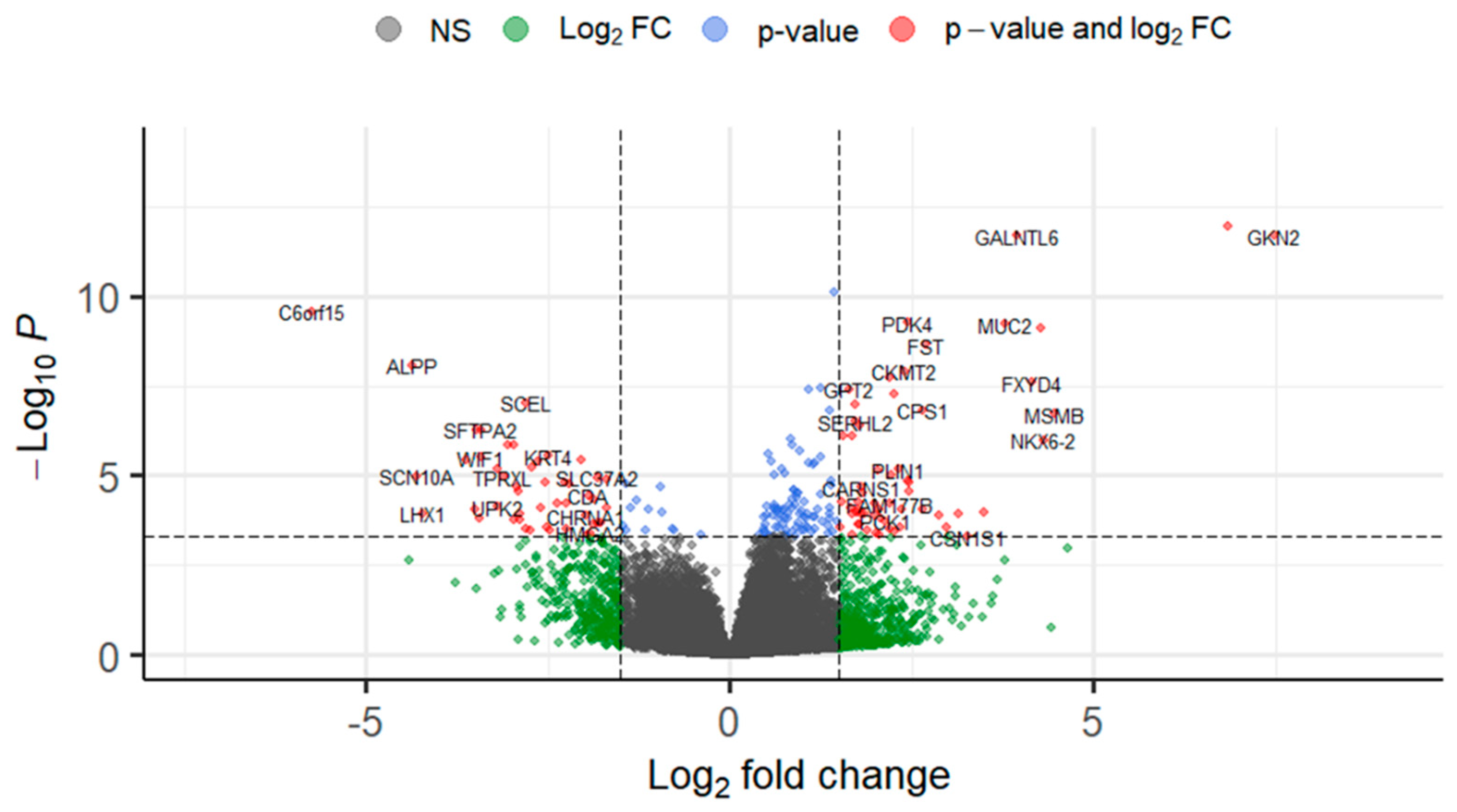
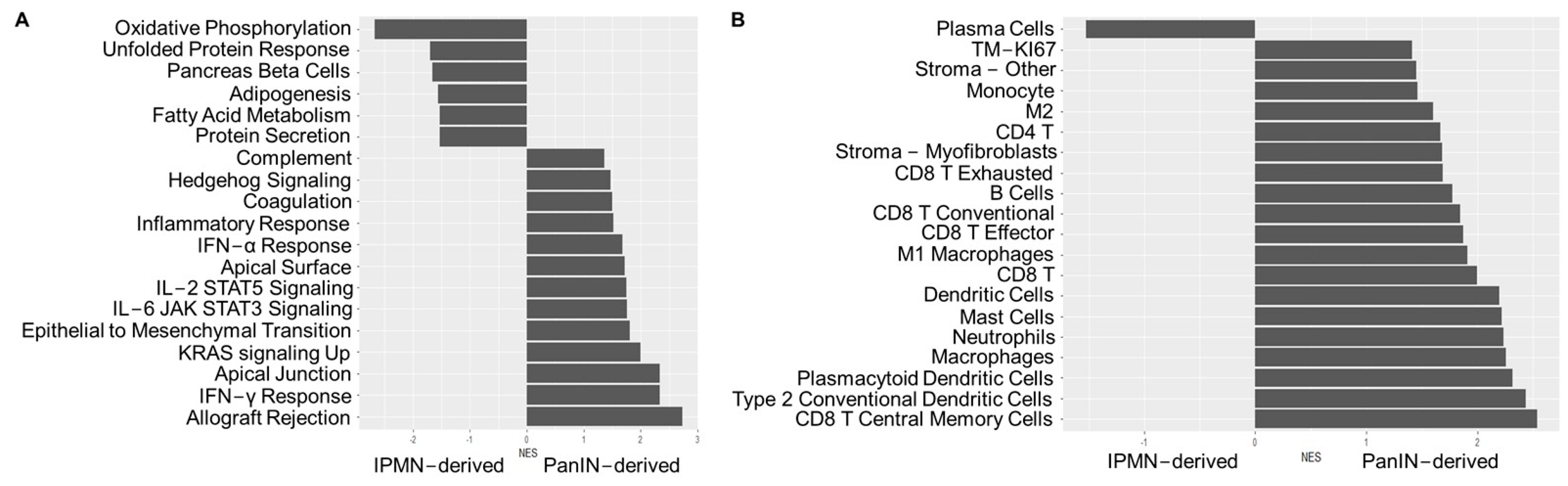
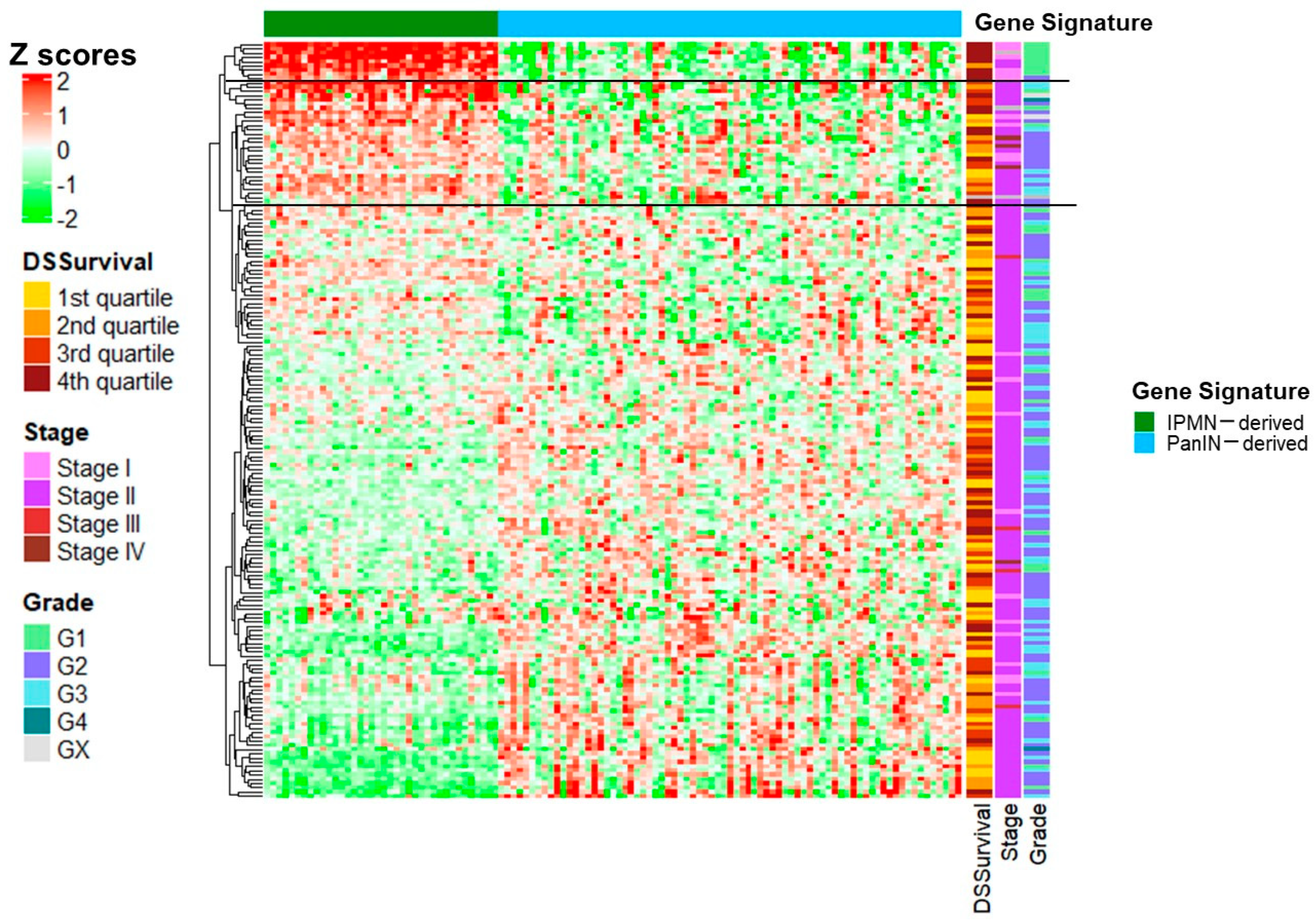
| Characteristics (n = 66) | |
|---|---|
| Age at Diagnosis, Mean (Range) | 68.3 (Min 48; Max 87) |
| Sex, N (%) | |
| Male | 37 (56.1%) |
| Female | 29 (43.9%) |
| Race &Ethnicity, N (%) | |
| Non-Hispanic White | 58 (87.9%) |
| Non-Hispanic Black | 3 (4.5%) |
| Hispanic | 3 (4.5%) |
| Other/missing | 2 (3.0%) |
| Histology, N (%) | |
| Adenocarcinoma, NOS | 33 (50.0%) |
| IPMN, non-invasive | 2 (3.0%) |
| Invasive carcinoma of no special type | 28 (42.4%) |
| Adenocarcinoma in situ | 1 (1.5%) |
| Mucinous adenocarcinoma | 1 (1.5%) |
| Adenocarcinoma with mixed subtypes | 1 (1.5%) |
| Grade, N % | |
| Grade 1–2 | 33 (50.0%) |
| PanIN-derived | 26 (39.4%) |
| IPMN-derived | 7 (10.6%) |
| Grade 3 | 17 (25.8%) |
| PanIN-derived | 14 (21.1%) |
| IPMN-derived | 3 (4.5%) |
| Stage, N % | |
| Early (stage I/II) | 56 (84.8%) |
| PanIN-derived | 43 (65.2%) |
| IPMN-derived | 13 (19.7%) |
| Late (stage III/IV) | 5 (7.6%) |
| PanIN-derived | 5 (7.6%) |
| IPMN-derived | 0 (0%) |
| Sample Treatment Status | |
| Treatment Naïve | 41 (31 PanIN-derived) |
| Neoadjuvant chemotherapy | 9 (9 PanIN-derived) |
| Group Description | |
| IPMN-derived | 16 (24.2%) |
| PanIN-derived | 50 (75.8%) |
| Survival (in months, median (LCL;UCL)) | |
| PanIN-derived | 27 (19; 37) |
| IPMN-derived | 36 (25; NA) |
| Gene Name | Log2 FC | lfc SE | p-Value | Padj |
|---|---|---|---|---|
| GKN2 | 7.489 | 1.065 | <0.001 | <0.001 |
| INSL4 | 6.838 | 0.961 | <0.001 | <0.001 |
| C6orf15 | −5.736 | 0.908 | <0.001 | <0.001 |
| MSMB | 4.464 | 0.857 | <0.001 | <0.001 |
| ALPP | −4.376 | 0.759 | <0.001 | <0.001 |
| NKX6-2 | 4.316 | 0.883 | <0.001 | 0.001 |
| SCN10A | −4.314 | 0.978 | <0.001 | 0.004 |
| SPINK4 | 4.280 | 0.695 | <0.001 | <0.001 |
| FXYD4 | 4.161 | 0.747 | <0.001 | <0.001 |
| GALNTL6 | 3.950 | 0.562 | <0.001 | <0.001 |
| MUC2 | 3.775 | 0.610 | <0.001 | <0.001 |
| ALPPL2 | −3.623 | 0.783 | <0.001 | <0.001 |
| DSG3 | −3.490 | 0.697 | <0.001 | <0.001 |
| SFTPA2 | −3.431 | 0.683 | <0.001 | <0.001 |
| WIF1 | −3.417 | 0.733 | <0.001 | 0.002 |
| CLDN6 | −3.187 | 0.708 | <0.001 | 0.003 |
| TPRXL | −3.117 | 0.708 | <0.001 | 0.004 |
| NCCRP1 | −3.046 | 0.632 | <0.001 | 0.001 |
| TNNT1 | −2.966 | 0.616 | <0.001 | 0.001 |
| PRSS33 | −2.930 | 0.688 | <0.001 | 0.006 |
| PADI3 | −2.901 | 0.693 | <0.001 | 0.008 |
| SCEL | −2.809 | 0.525 | <0.001 | 0.000 |
| CALB1 | −2.717 | 0.600 | <0.001 | 0.002 |
| FST | 2.686 | 0.449 | <0.001 | <0.001 |
| CPS1 | 2.654 | 0.505 | <0.001 | <0.001 |
| CHIT1 | −2.639 | 0.572 | <0.001 | 0.002 |
| MUC21 | −2.539 | 0.587 | <0.001 | 0.005 |
| KRT4 | −2.493 | 0.532 | <0.001 | 0.001 |
| SCARA5 | 2.466 | 0.588 | <0.001 | 0.007 |
| ORM1 | 2.457 | 0.569 | <0.001 | 0.005 |
| PDK4 | 2.448 | 0.394 | <0.001 | <0.001 |
| CA9 | 2.433 | 0.562 | <0.001 | <0.001 |
| CKMT2 | 2.398 | 0.421 | <0.001 | <0.001 |
| PLIN1 | 2.318 | 0.515 | <0.001 | 0.003 |
| KCNS1 | −2.275 | 0.527 | <0.001 | 0.005 |
| PPARGC1A | 2.260 | 0.415 | <0.001 | <0.001 |
| TRPA1 | 2.221 | 0.502 | <0.001 | 0.004 |
| IL20RB | −2.212 | 0.514 | <0.001 | 0.005 |
| MAMDC4 | 2.187 | 0.389 | <0.001 | <0.001 |
| PPP1R14D | −2.043 | 0.442 | <0.001 | 0.002 |
| LINGO4 | 2.027 | 0.452 | <0.001 | 0.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, M.A.; Gumpper-Fedus, K.; Krishna, S.G.; Genilo-Delgado, M.C.; Brantley, S.; Hart, P.A.; Dillhoff, M.E.; Gomez, M.F.; Basinski, T.L.; Mok, S.R.; et al. Molecular Pathway and Immune Profile Analysis of IPMN-Derived Versus PanIN-Derived Pancreatic Ductal Adenocarcinomas. Int. J. Mol. Sci. 2024, 25, 13164. https://doi.org/10.3390/ijms252313164
Park MA, Gumpper-Fedus K, Krishna SG, Genilo-Delgado MC, Brantley S, Hart PA, Dillhoff ME, Gomez MF, Basinski TL, Mok SR, et al. Molecular Pathway and Immune Profile Analysis of IPMN-Derived Versus PanIN-Derived Pancreatic Ductal Adenocarcinomas. International Journal of Molecular Sciences. 2024; 25(23):13164. https://doi.org/10.3390/ijms252313164
Chicago/Turabian StylePark, Margaret A., Kristyn Gumpper-Fedus, Somashekar G. Krishna, Maria C. Genilo-Delgado, Stephen Brantley, Phil A. Hart, Mary E. Dillhoff, Maria F. Gomez, Toni L. Basinski, Shaffer R. Mok, and et al. 2024. "Molecular Pathway and Immune Profile Analysis of IPMN-Derived Versus PanIN-Derived Pancreatic Ductal Adenocarcinomas" International Journal of Molecular Sciences 25, no. 23: 13164. https://doi.org/10.3390/ijms252313164
APA StylePark, M. A., Gumpper-Fedus, K., Krishna, S. G., Genilo-Delgado, M. C., Brantley, S., Hart, P. A., Dillhoff, M. E., Gomez, M. F., Basinski, T. L., Mok, S. R., Luthra, A. K., Fleming, J. B., Mohammadi, A., Centeno, B. A., Jiang, K., Karolak, A., Jeong, D., Chen, D.-T., Stewart, P. A., ... Permuth, J. B. (2024). Molecular Pathway and Immune Profile Analysis of IPMN-Derived Versus PanIN-Derived Pancreatic Ductal Adenocarcinomas. International Journal of Molecular Sciences, 25(23), 13164. https://doi.org/10.3390/ijms252313164







