Genome-Wide Identification and Expression Analysis of Carotenoid Cleavage Dioxygenase Genes in Salvia miltiorrhiza
Abstract
1. Introduction
2. Results
2.1. Identification of CCD Genes Family Members
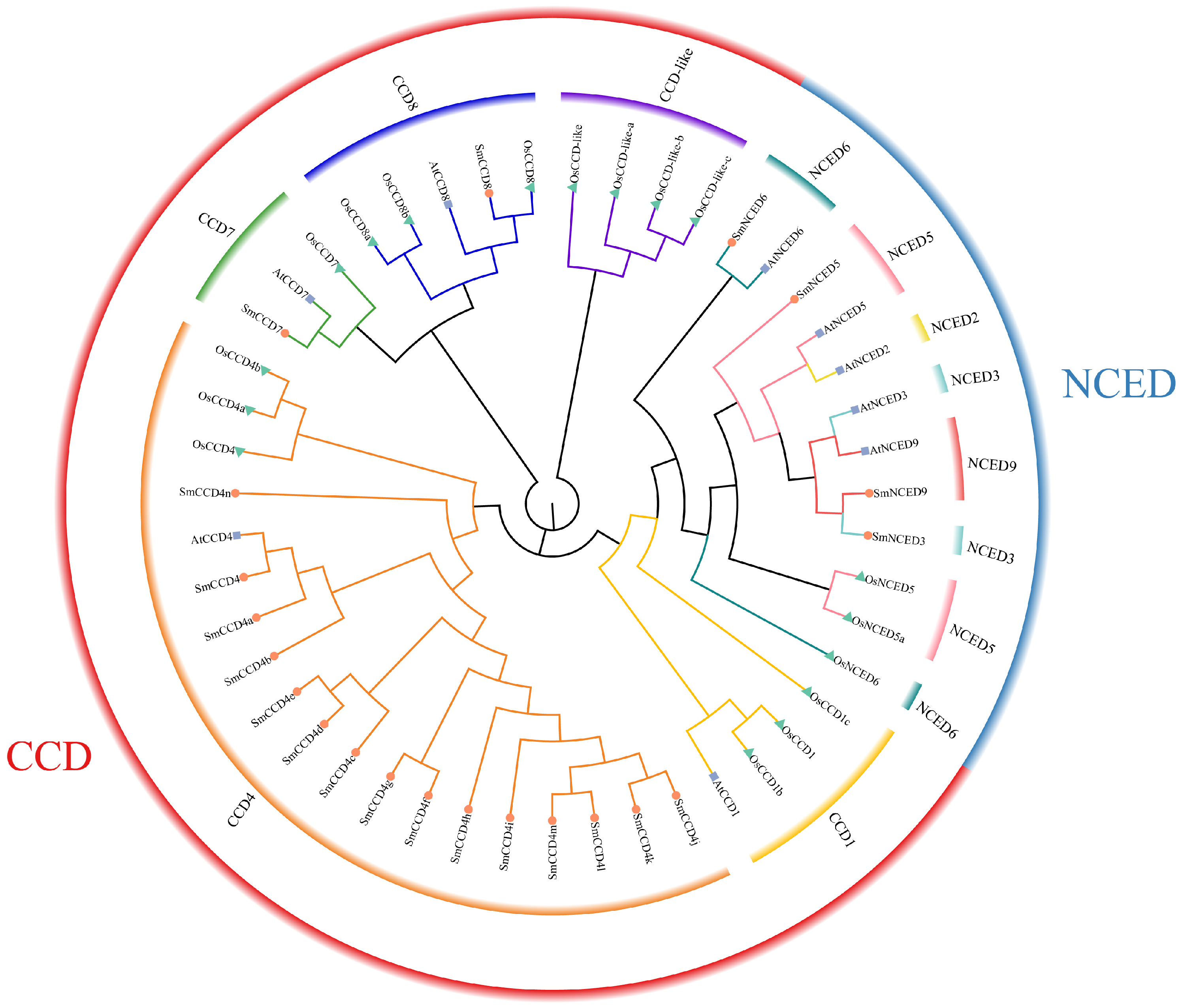
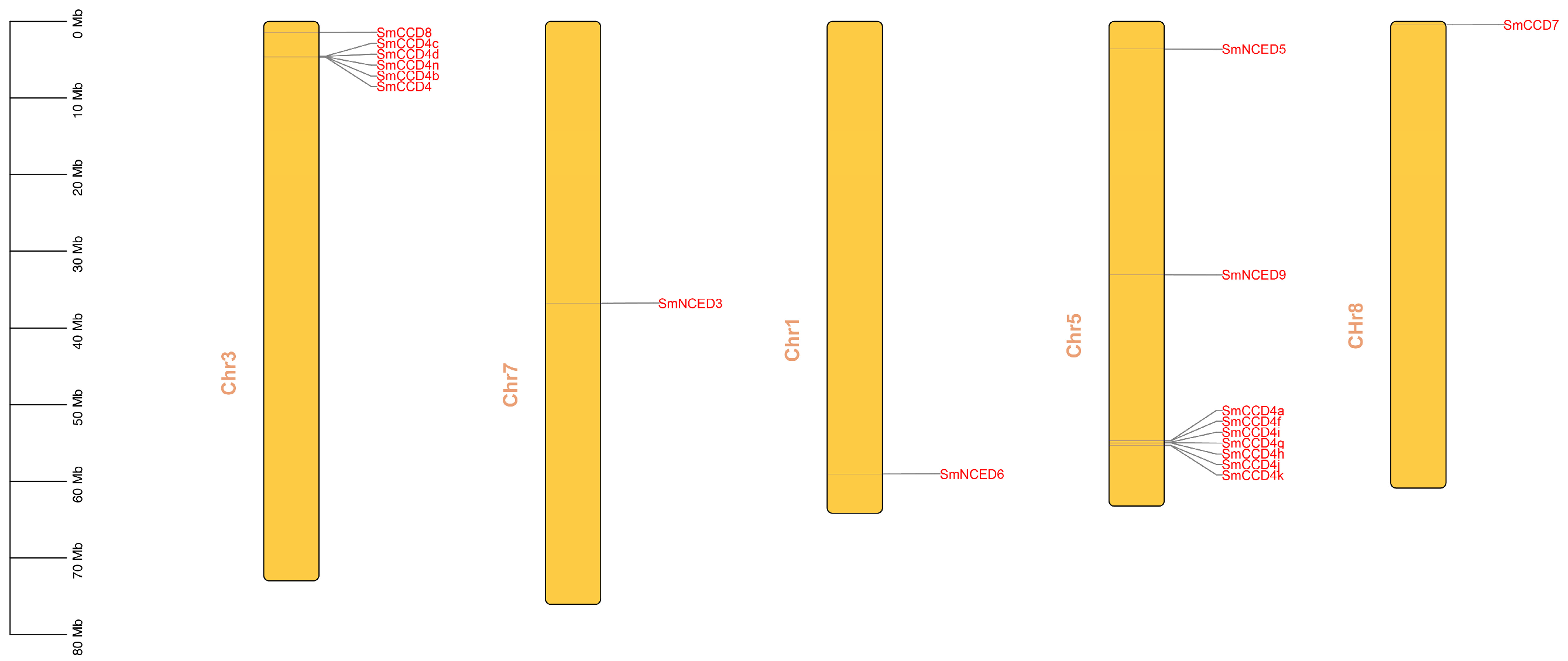
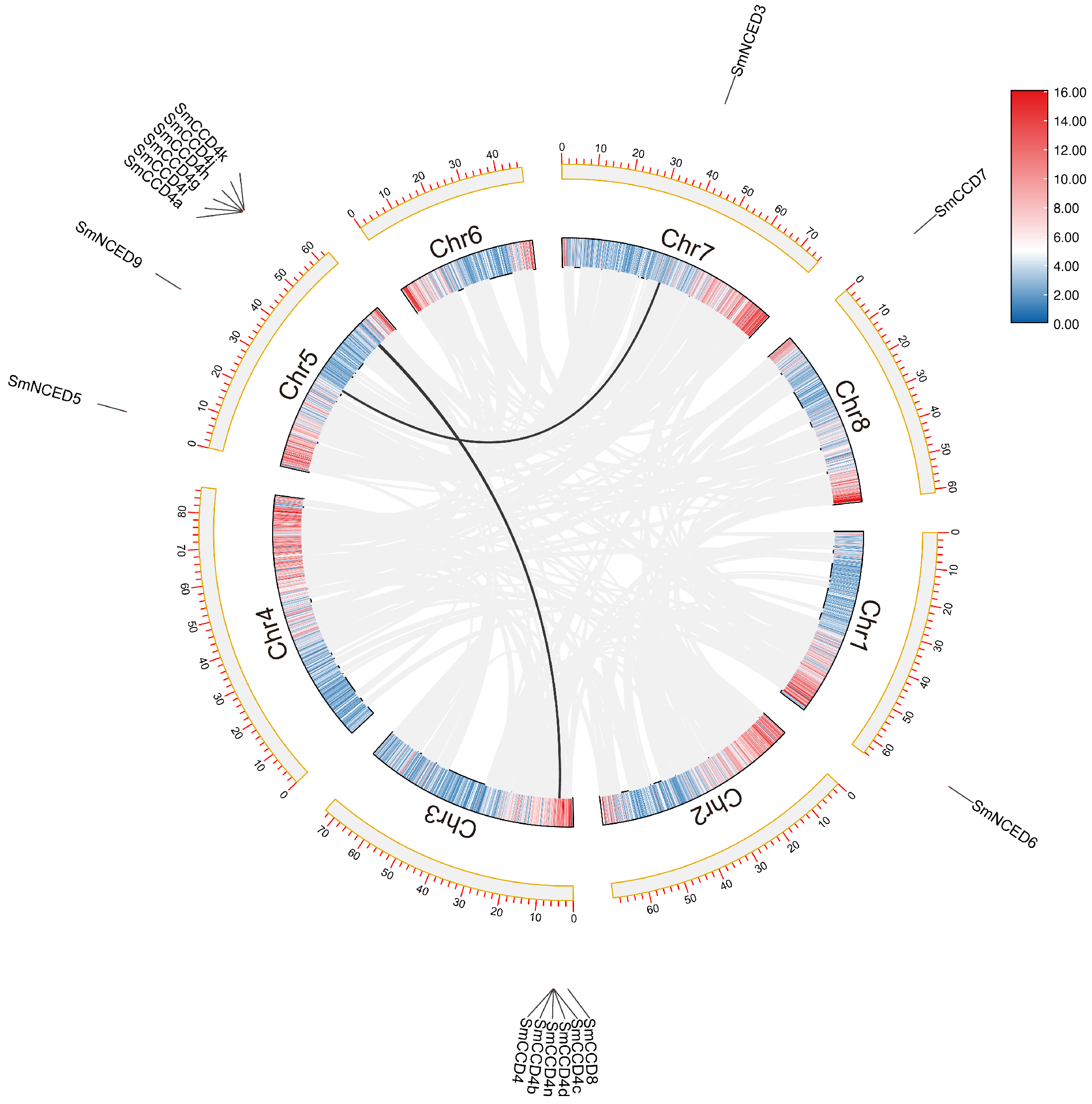
2.2. Phylogenetic Tree Analysis, Chromosome Locations, and Collinearity Analysis
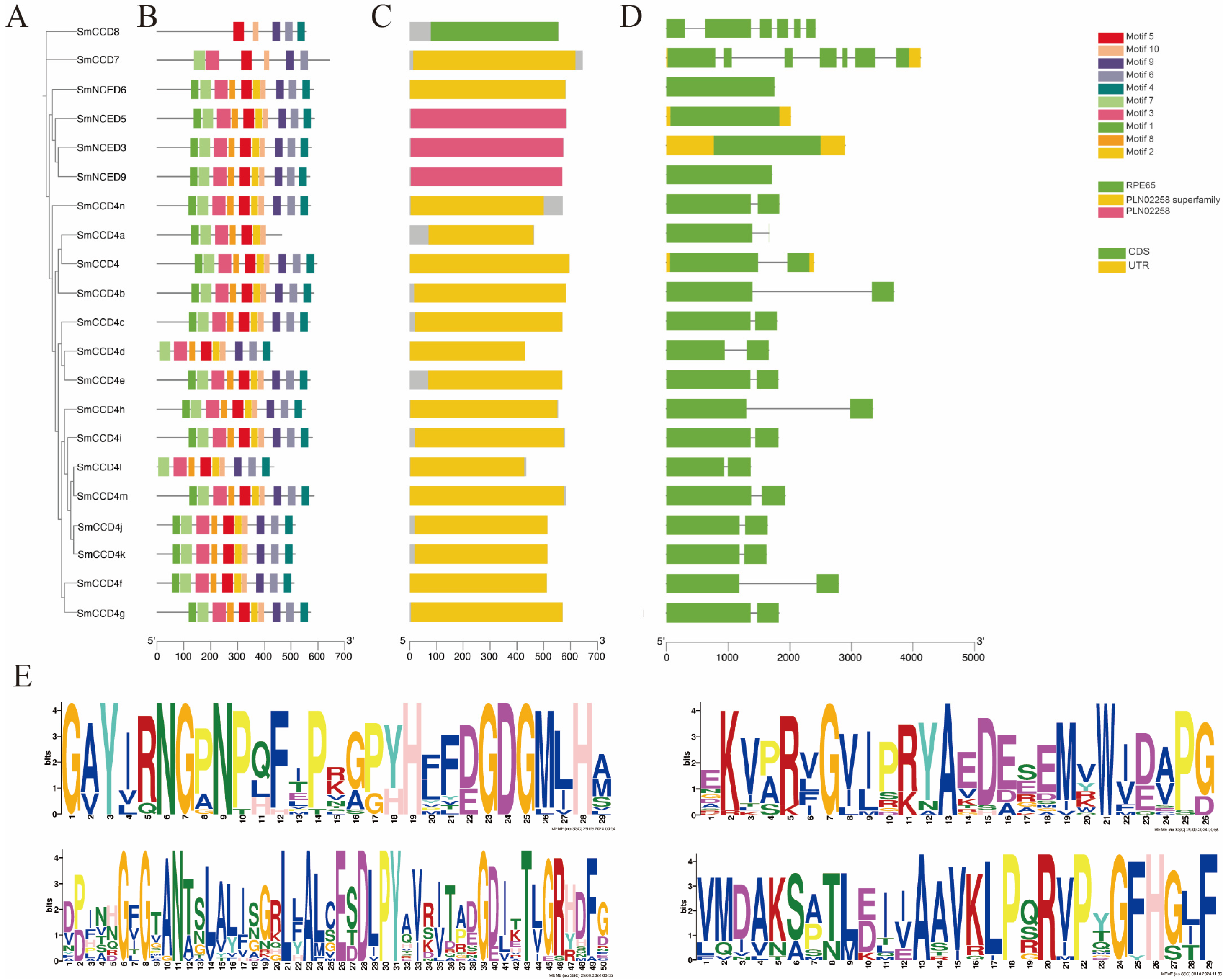
2.3. Motif Analysis and Cis-Element Analysis
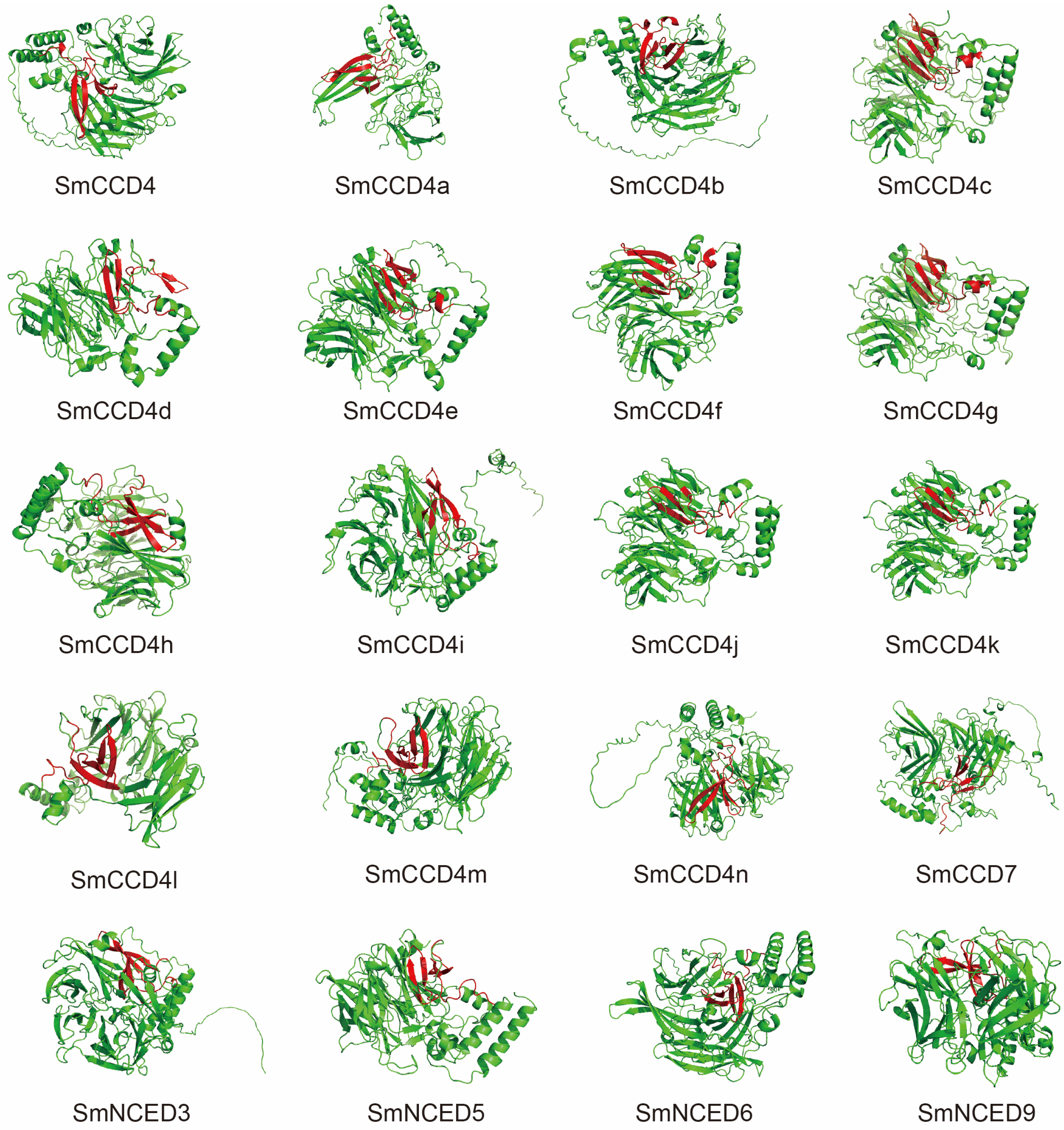
2.4. Prediction of Three-Dimensional (3D) Structure of SmCCDs
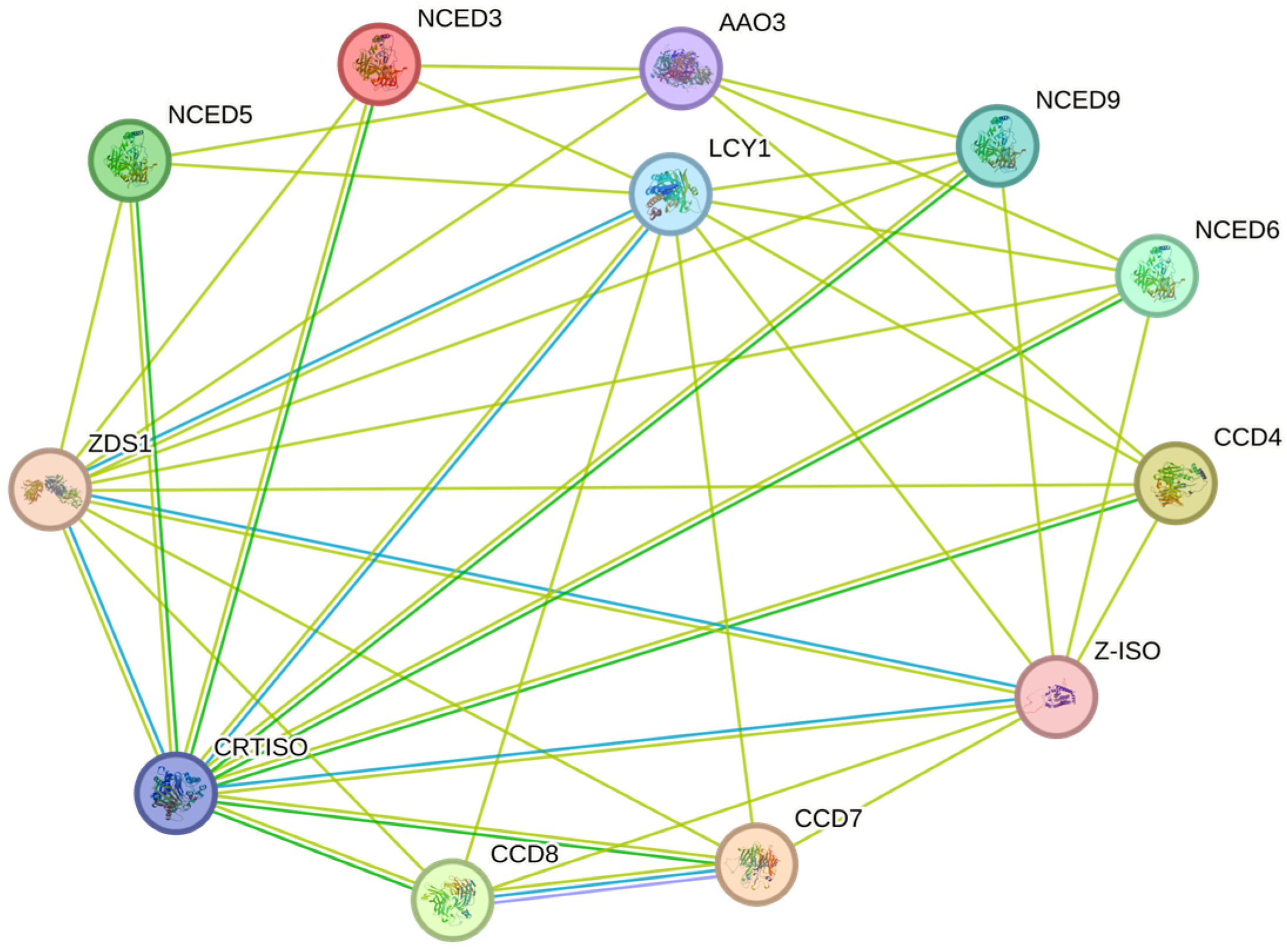
2.5. Protein Interaction Network Analysis
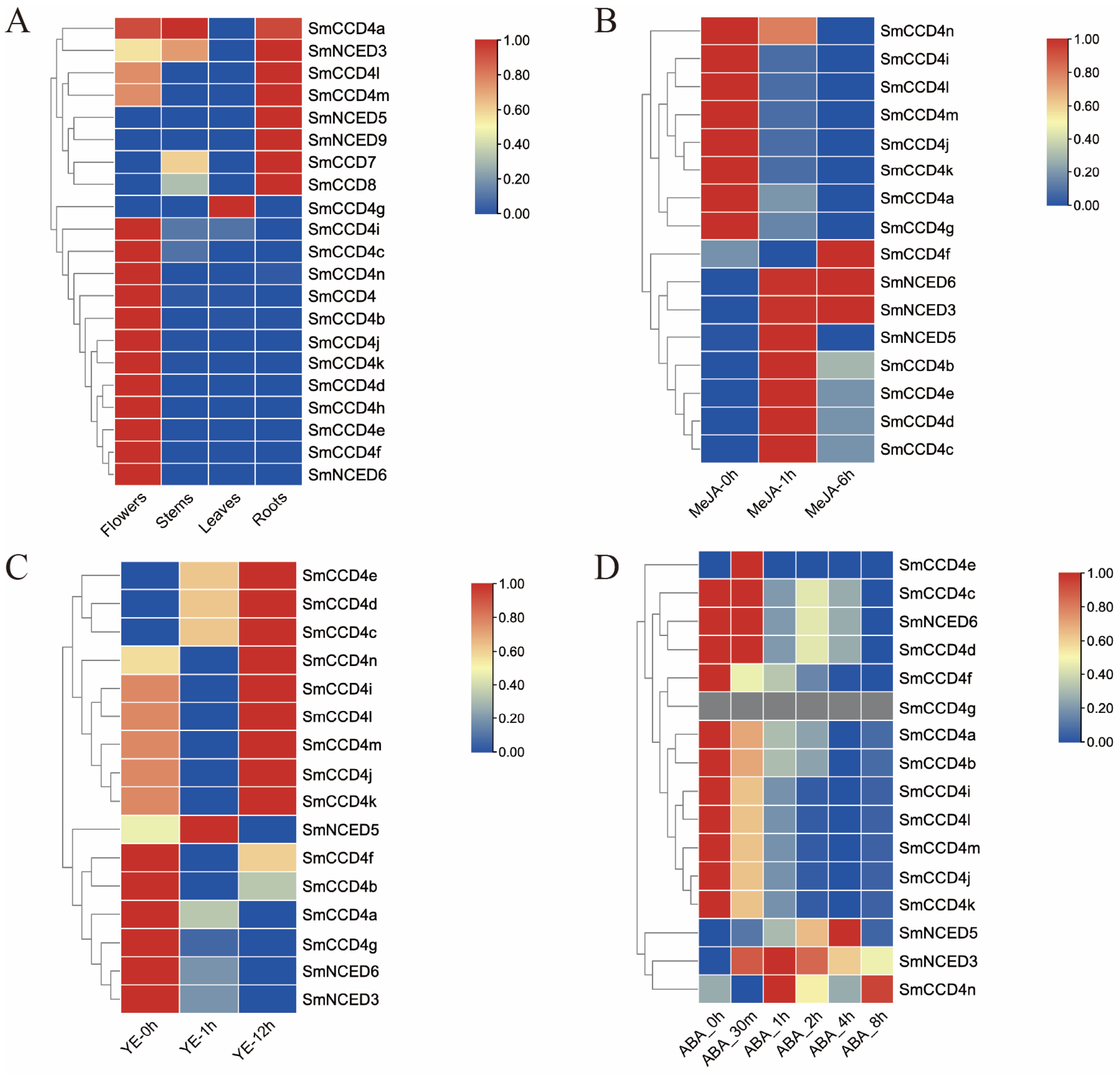
2.6. Expression Patterns of SmCCDs in Different Tissues, Stress Condition, and the Screening of Candidate Functional SmCCD Genes
3. Discussion
4. Materials and Methods
4.1. Identification of CCD Genes Family Members in S. miltiorrhiza
4.2. Phylogenetic Tree Analysis, Chromosome Locations, and Collinearity Analysis of SmCCDs
4.3. Motif Analysis and Cis-Element Analysis
4.4. Prediction of Secondary and Three-Dimensional (3D) Structure of SmCCDs
4.5. Protein Interaction Network of SmCCDs
4.6. Expression Patterns of SmCCDs in Different Tissues and Elicitation
4.7. Pre-Experiment for CCD Screening, Transcriptome Sequencing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maoka, T. Recent progress in structural studies of carotenoids in animals and plants. Arch. Biochem. Biophys. 2009, 483, 191–195. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.A.; Rodriguez-Concepcion, M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arab. Book 2012, 10, e0158. [Google Scholar] [CrossRef] [PubMed]
- Gamon, J.A.; Huemmrich, K.F.; Wong, C.Y.; Ensminger, I.; Garrity, S.; Hollinger, D.Y.; Noormets, A.; Peñuelas, J. A remotely sensed pigment index reveals photosynthetic phenology in evergreen conifers. Proc. Natl. Acad. Sci. USA 2016, 113, 13087–13092. [Google Scholar] [CrossRef]
- Hou, X.; Rivers, J.; León, P.; McQuinn, R.P.; Pogson, B.J. Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; Block, A.; Vogel, J.T.; Dabney-Smith, C.; Mila, I.; Bouzayen, M.; Magallanes-Lundback, M.; DellaPenna, D.; McCarty, D.R.; Klee, H.J. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.Q.; Zhang, Y.; Yang, C.K.; Li, J.G.; Rui, X.; Ding, F.; Hu, F.C.; Wang, X.H.; Ma, W.Q.; Zhou, K.B. Genome-wide identification and expression analysis of carotenoid cleavage oxygenase genes in Litchi (Litchi chinensis Sonn.). BMC Plant Biol. 2022, 22, 394. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef] [PubMed]
- Floss, D.S.; Walter, M.H. Role of carotenoid cleavage dioxygenase 1 (CCD1) in apocarotenoid biogenesis revisited. Plant Signaling Behav. 2009, 4, 172–175. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Qin, X.; Loewen, M.C. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J. Biol. Chem. 2004, 279, 46940–46945. [Google Scholar] [CrossRef]
- Bruno, M.; Koschmieder, J.; Wuest, F.; Schaub, P.; Fehling-Kaschek, M.; Timmer, J.; Beyer, P.; Al-Babili, S. Enzymatic study on AtCCD4 and AtCCD7 and their potential to form acyclic regulatory metabolites. J. Exp. Bot. 2016, 67, 5993–6005. [Google Scholar] [CrossRef]
- Ahrazem, O.; Trapero, A.; Gómez, M.D.; Rubio-Moraga, A.; Gómez-Gómez, L. Genomic analysis and gene structure of the plant carotenoid dioxygenase 4 family: A deeper study in Crocus sativus and its allies. Genomics 2010, 96, 239–250. [Google Scholar] [CrossRef]
- Adami, M.; De Franceschi, P.; Brandi, F.; Liverani, A.; Giovannini, D.; Rosati, C.; Dondini, L.; Tartarini, S. Identifying a carotenoid cleavage dioxygenase (ccd4) gene controlling yellow/white fruit flesh color of peach. Plant Mol. Biol. Rep. 2013, 31, 1166–1175. [Google Scholar] [CrossRef]
- Campbell, R.; Ducreux, L.J.M.; Morris, W.L.; Morris, J.A.; Suttle, J.C.; Ramsay, G.; Bryan, G.J.; Hedley, P.E.; Taylor, M.A. The metabolic and developmental roles of carotenoid cleavage dioxygenase4 from potato. Plant Physiol. 2010, 154, 656–664. [Google Scholar] [CrossRef]
- Daruwalla, A.; Kiser, P.D. Structural and mechanistic aspects of carotenoid cleavage dioxygenases (CCDs). BBA Mol. Cell Biol. Lipids 2020, 1865, 158590. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Jia, L.; Cheng, Y.; Ruan, M.; Ye, Q.; Wang, R.; Yao, Z.; Zhou, G.; Liu, J.; Yu, J.; et al. Evolutionary origin of the carotenoid cleavage oxygenase family in plants and expression of pepper genes in response to abiotic stresses. Front. Plant Sci. 2022, 12, 792832. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.R.; Li, J.Y.; Zhang, J.; Chen, D.; Zhang, H.P.; Liu, C.Y.; Qin, G.H. Genome-wide identification and expression analysis of the carotenoid cleavage oxygenase gene family in five Rosaceae species. Plant Mol. Biol. Rep. 2021, 39, 739–751. [Google Scholar] [CrossRef]
- Zhang, X.H.; Liu, H.Q.; Guo, Q.W.; Zheng, C.F.; Li, C.S.; Xiang, X.M.; Zhao, D.F.; Liu, J.; Luo, J.; Zhao, D.K.; et al. Genome-wide identification, phylogenetic relationships, and expression analysis of the carotenoid cleavage oxygenase gene family in pepper. Genet. Mol. Res. 2016, 15, 15048695. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.H.; Masuda, J.; Miyajima, I.; Thien, N.Q.; Mojtahedi, N.; Hiramatsu, M.; Kim, J.H.; Okubo, H. Involvement of carotenoid cleavage dioxygenase 4 gene in tepal color change in Lilium brownii var. colchesteri. J. Jpn. Soc. Hortic. Sci. 2012, 81, 366–373. [Google Scholar]
- Li, T.; Deng, Y.J.; Liu, J.X.; Duan, A.Q.; Liu, H.; Xiong, A.S. DcCCD4 catalyzes the degradation of α-carotene and β-carotene to affect carotenoid accumulation and taproot color in carrot. Plant J. 2021, 108, 1116–1130. [Google Scholar] [CrossRef]
- Ilg, A.; Yu, Q.J.; Schaub, P.; Beyer, P.; Al-Babili, S. Overexpression of the rice carotenoid cleavage dioxygenase 1 gene in Golden Rice endosperm suggests apocarotenoids as substrates in planta. Planta 2010, 232, 691–699. [Google Scholar] [CrossRef]
- Wang, J.M.; Wu, B.; Zhang, N.; Zhao, M.Y.; Jing, T.T.; Wu, Y.; Hu, Y.Q.; Yu, F.; Wan, X.C.; Schwab, W.; et al. Dehydration-induced carotenoid cleavage dioxygenase 1 reveals a novel route for β-ionone formation during tea (Camellia sinensis) withering. J. Agric. Food Chem. 2020, 68, 10815–10821. [Google Scholar] [CrossRef] [PubMed]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Fei, R.W.; Guan, S.X.; Duan, S.Y.; Ge, J.Y.; Sun, T.Y.; Sun, X.M. Elucidating biological functions of 9-cis-epoxycarotenoid dioxygenase genes involved in seed dormancy in Paeonia lactiflora. Plants 2023, 12, 710. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Zhao, C.L.; Du, T.F.; Li, A.X.; Qin, Z.; Zhang, L.M.; Dong, S.X.; Wang, Q.M.; Hou, F.Y. Overexpression of 9-cis-epoxycarotenoid dioxygenase gene, IbNCED1, negatively regulates plant height in transgenic sweet potato. Int. J. Mol. Sci. 2023, 24, 10421. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef]
- Timmins, J.J.B.; Kroukamp, H.; Walker, R.S.K.; Pretorius, I.S.; Paulsen, I.T. Comparative evaluation of secreted plant carotenoid cleavage dioxygenase 1 (CCD1) enzymes in Saccharomyces cerevisiae. Fermentation 2022, 8, 395. [Google Scholar] [CrossRef]
- Lee, H.G.; Lee, K.; Seo, P.J. The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 2015, 87, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liang, G.; Yu, D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant. 2012, 5, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Hong, X.; Zhang, L.; Yang, W.; Zeng, Y.; Hou, Z. Transcriptomic analysis provides insight into the regulation mechanism of silver ions (Ag+) and jasmonic acid methyl ester (MeJA) on secondary metabolism in the hairy roots of Salvia miltiorrhiza Bunge (Lamiaceae). Med. Plant Biol. 2023, 2, 3. [Google Scholar] [CrossRef]
- Xu, Z.; Peters, R.J.; Weirather, J.; Luo, H.; Liao, B.; Zhang, X.; Zhu, Y.; Ji, A.; Zhang, B.; Hu, S.; et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015, 82, 951–961. [Google Scholar] [CrossRef]
- Timmins, J.J.B.; Kroukamp, H.; Paulsen, I.T.; Pretorius, I.S. The sensory significance of apocarotenoids in wine: Importance of carotenoid cleavage dioxygenase 1 (CCD1) in the production of β-ionone. Molecules 2020, 25, 2779. [Google Scholar] [CrossRef]
- Dhar, M.K.; Mishra, S.; Bhat, A.; Chib, S.; Kaul, S. Plant carotenoid cleavage oxygenases: Structure-function relationships and role in development and metabolism. Brief. Funct. Genomics 2020, 19, 1–9. [Google Scholar] [CrossRef]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Zhou, X.T.; Jia, L.D.; Duan, M.Z.; Chen, X.; Qiao, C.L.; Ma, J.Q.; Zhang, C.; Jing, F.Y.; Zhang, S.S.; Yang, B.; et al. Genome-wide identification and expression profiling of the carotenoid cleavage dioxygenase (CCD) gene family in Brassica napus L. PLoS ONE 2020, 15, e0238179. [Google Scholar] [CrossRef]
- Akram, J.; Siddique, R.; Shafiq, M.; Tabassum, B.; Manzoor, M.T.; Javed, M.A.; Anwar, S.; Nisa, B.U.; Saleem, M.H.; Javed, B.; et al. Genome-wide identification of CCO gene family in cucumber (Cucumis sativus) and its comparative analysis with A. thaliana. BMC Plant Biol. 2023, 23, 640. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Liu, G.; Li, Y.; Liu, S.; Yu, C.; Chen, Y.; Zhong, F.; Zhang, J. Comprehensive analysis of carotenoid cleavage dioxygenases gene family and its expression in response to abiotic stress in poplar. Int. J. Mol. Sci. 2022, 23, 1418. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, G.; Gu, T.; Ding, J.; Li, Y. Bioinformatic and expression analyses on carotenoid dioxygenase genes in fruit development and abiotic stress responses in Fragaria vesca. Mol. Genet. Genom. 2017, 292, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yu, X.; Chen, L.; Zhao, G.; Li, S.; Zhou, H.; Dai, Y.; Sun, N.; Xie, Y.; Gao, J.; et al. Genome-wide identification and expression analysis of the NCED family in cotton (Gossypium hirsutum L.). PLoS ONE 2021, 16, e0246021. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Kishimoto, S.; Yamamizo, C.; Fukuta, N.; Ohmiya, A. Carotenoid accumulations and carotenogenic gene expressions in the petals of Eustoma grandiflorum. Plant Breed. 2013, 132, 417–422. [Google Scholar] [CrossRef]
- Cheng, D.; Wang, Z.; Li, S.; Zhao, J.; Wei, C.; Zhang, Y. Genome-wide identification of CCD gene family in six cucurbitaceae species and its expression profiles in melon. Genes 2022, 13, 262. [Google Scholar] [CrossRef] [PubMed]
- Basso, M.F.; Contaldi, F.; Celso, F.L.; Karalija, E.; Paz-Carrasco, L.C.; Barone, G.; Ferrante, A.; Martinelli, F. Expression profile of the NCED/CCD genes in chickpea and lentil during abiotic stress reveals a positive correlation with increased plant tolerance. Plant Sci. 2023, 336, 111817. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; He, L.; Dong, J.; Zhao, C.; Tang, R.; Jia, X. Overexpression of sweet potato carotenoid cleavage dioxygenase 4 (IbCCD4) decreased salt tolerance in Arabidopsis thaliana. Int. J. Mol Sci. 2022, 23, 9963. [Google Scholar] [CrossRef]
- Zhou, W.; Huang, Q.; Wu, X.; Zhou, Z.; Ding, M.; Shi, M.; Huang, F.; Li, S.; Wang, Y.; Kai, G. Comprehensive transcriptome profiling of Salvia miltiorrhiza for discovery of genes associated with the biosynthesis of tanshinones and phenolic acids. Sci. Rep. 2017, 7, 10554. [Google Scholar] [CrossRef]
- Kalladan, R.; Lasky, J.R.; Sharma, S.; Kumar, M.N.; Juenger, T.E.; Des Marais, D.L.; Verslues, P.E. Natural variation in 9-cis-epoxycartenoid dioxygenase 3 and ABA accumulation. Plant Physiol. 2019, 179, 1620–1631. [Google Scholar] [CrossRef]
- Zhu, K.; Sun, Q.; Chen, H.; Mei, X.; Lu, S.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. Ethylene activation of carotenoid biosynthesis by a novel transcription factor CsERF061. J. Exp. Bot. 2021, 72, 3137–3154. [Google Scholar] [CrossRef]
- Wang, R.K.; Wang, C.E.; Fei, Y.Y.; Gai, J.Y.; Zhao, T.J. Genome-wide identification and transcription analysis of soybean carotenoid oxygenase genes during abiotic stress treatments. Mol. Biol. Rep. 2013, 40, 4737–4745. [Google Scholar] [CrossRef]
- Liu, Y.; Dong, B.; Zhang, C.; Yang, L.; Wang, Y.; Zhao, H. Effects of exogenous abscisic acid (ABA) on carotenoids and petal color in Osmanthus fragrans ‘Yanhonggui’. Plants 2020, 9, 454. [Google Scholar] [CrossRef]
- Chen, H.; Zuo, X.; Shao, H.; Fan, S.; Ma, J.; Zhang, D.; Zhao, C.; Yan, X.; Liu, X.; Han, M. Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica). Plant Physiol. Biochem. 2018, 123, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Hua, Q.; Kai, G.Y. Comprehensive transcriptomic analysis in response to abscisic acid in Salvia miltiorrhiza. Plant Cell Tissue Organ Cult. 2021, 147, 389–404. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, C.; Tang, X.; Wang, L.; Guo, R. Transcriptomic and metabolomic insights into ABA-related genes in Cerasus humilis under drought stress. Int. J. Mol. Sci. 2024, 25, 7635. [Google Scholar] [CrossRef]
- Ma, Y.; Cui, G.; Chen, T.; Ma, X.; Wang, R.; Jin, B.; Yang, J.; Kang, L.; Tang, J.; Lai, C.; et al. Expansion within the CYP71D subfamily drives the heterocyclization of tanshinones synthesis in Salvia miltiorrhiza. Nat. Commun. 2021, 12, 685. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Wang, C.; Li, J.; Gao, X.; Huang, Q.; Gong, Y.; Hao, X.; Maoz, I.; Kai, G.; Zhou, W. Genome-wide analysis of U-box E3 ubiquitin ligase family in response to ABA treatment in Salvia miltiorrhiza. Front. Plant Sci. 2022, 13, 829447. [Google Scholar] [CrossRef]
- Wang, H.B.; Wei, T.; Wang, X.; Zhang, L.P.; Yang, M.L.; Chen, L.; Song, W.Q.; Wang, C.G.; Chen, C.B. Transcriptome analyses from mutant Salvia miltiorrhiza reveals important roles for SmGASA4 during plant development. Int. J. Mol. Sci. 2018, 19, 2088. [Google Scholar] [CrossRef]

| Name | Number of Amino Acids | Molecular Weight/kDa | Theoretical pI | Instability Index | Aliphatic Index | Subcellular Localization |
|---|---|---|---|---|---|---|
| SmCCD4 | 596 | 65.44 | 6.64 | 36.34 | 77.42 | Chloroplast, Cytoplasm |
| SmCCD4a | 464 | 49.99 | 7.31 | 36.06 | 90.41 | Chloroplast, Cytoplasm |
| SmCCD4b | 584 | 64.06 | 6.36 | 41.02 | 85.36 | Chloroplast, Cytoplasm |
| SmCCD4c | 571 | 62.16 | 5.95 | 37.01 | 84.20 | Chloroplast, Cytoplasm |
| SmCCD4d | 432 | 48.07 | 5.59 | 29.09 | 93.63 | Cytoplasm |
| SmCCD4e | 570 | 62.25 | 6.79 | 34.29 | 88.14 | Chloroplast, Cytoplasm |
| SmCCD4f | 511 | 55.63 | 5.91 | 34.45 | 83.78 | Cytoplasm |
| SmCCD4g | 572 | 62.12 | 6.29 | 36.13 | 87.19 | Chloroplast, Cytoplasm |
| SmCCD4h | 554 | 61.04 | 6.03 | 37.18 | 80.61 | Mitochondria, Chloroplast, Cytoplasm |
| SmCCD4i | 579 | 64.18 | 6.85 | 36.23 | 90.6 | Cytoplasm |
| SmCCD4j | 515 | 56.79 | 5.63 | 36.54 | 88.04 | Chloroplast, Cytoplasm |
| SmCCD4k | 515 | 56.78 | 5.84 | 38.38 | 89.01 | Cytoplasm |
| SmCCD4l | 435 | 47.87 | 4.88 | 34.43 | 90.32 | Mitochondria, Chloroplast, Cytoplasm |
| SmCCD4m | 585 | 64.24 | 5.47 | 44.66 | 87.50 | Chloroplast, Cytoplasm |
| SmCCD4n | 572 | 62.74 | 6.41 | 34.71 | 84.90 | Chloroplast, Cytoplasm |
| SmCCD7 | 645 | 72.15 | 8.67 | 42.39 | 78.19 | Mitochondria, Chloroplast, Cytoplasm |
| SmCCD8 | 556 | 62.21 | 6.56 | 39.95 | 78.38 | Chloroplast, Cytoplasm |
| SmNCED3 | 574 | 63.87 | 6.24 | 41.97 | 78.01 | Cytoplasm |
| SmNCED5 | 586 | 65.03 | 6.15 | 43.65 | 77.20 | Cytoplasm |
| SmNCED6 | 583 | 64.33 | 5.89 | 42.28 | 88.27 | Chloroplast, Cytoplasm |
| SmNCED9 | 569 | 63.02 | 7.23 | 44.97 | 84.85 | Chloroplast, Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shou, M.; Lin, Q.; Peng, L.; Wang, Z.; Xu, Y.; Qi, J.; Zhao, D.; Shi, M.; Kai, G. Genome-Wide Identification and Expression Analysis of Carotenoid Cleavage Dioxygenase Genes in Salvia miltiorrhiza. Int. J. Mol. Sci. 2024, 25, 13138. https://doi.org/10.3390/ijms252313138
Shou M, Lin Q, Peng L, Wang Z, Xu Y, Qi J, Zhao D, Shi M, Kai G. Genome-Wide Identification and Expression Analysis of Carotenoid Cleavage Dioxygenase Genes in Salvia miltiorrhiza. International Journal of Molecular Sciences. 2024; 25(23):13138. https://doi.org/10.3390/ijms252313138
Chicago/Turabian StyleShou, Minyu, Qinzhe Lin, Lulu Peng, Zijie Wang, Ying Xu, Jiaochen Qi, Degang Zhao, Min Shi, and Guoyin Kai. 2024. "Genome-Wide Identification and Expression Analysis of Carotenoid Cleavage Dioxygenase Genes in Salvia miltiorrhiza" International Journal of Molecular Sciences 25, no. 23: 13138. https://doi.org/10.3390/ijms252313138
APA StyleShou, M., Lin, Q., Peng, L., Wang, Z., Xu, Y., Qi, J., Zhao, D., Shi, M., & Kai, G. (2024). Genome-Wide Identification and Expression Analysis of Carotenoid Cleavage Dioxygenase Genes in Salvia miltiorrhiza. International Journal of Molecular Sciences, 25(23), 13138. https://doi.org/10.3390/ijms252313138






