Genome-Wide Identification and Expression Profiles of Nuclear Factor Y A Transcription Factors in Blueberry Under Abiotic Stress
Abstract
1. Introduction
2. Results
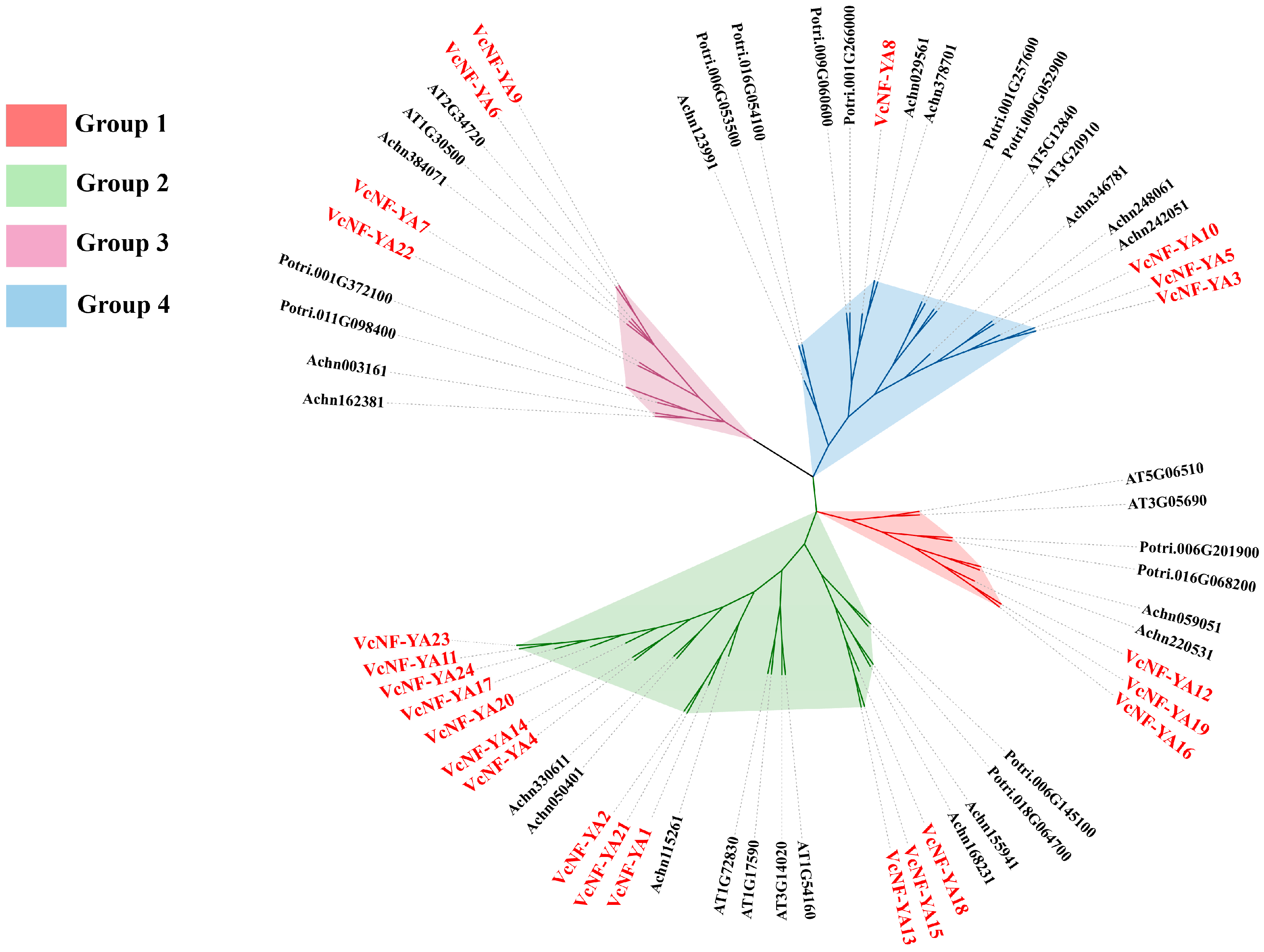
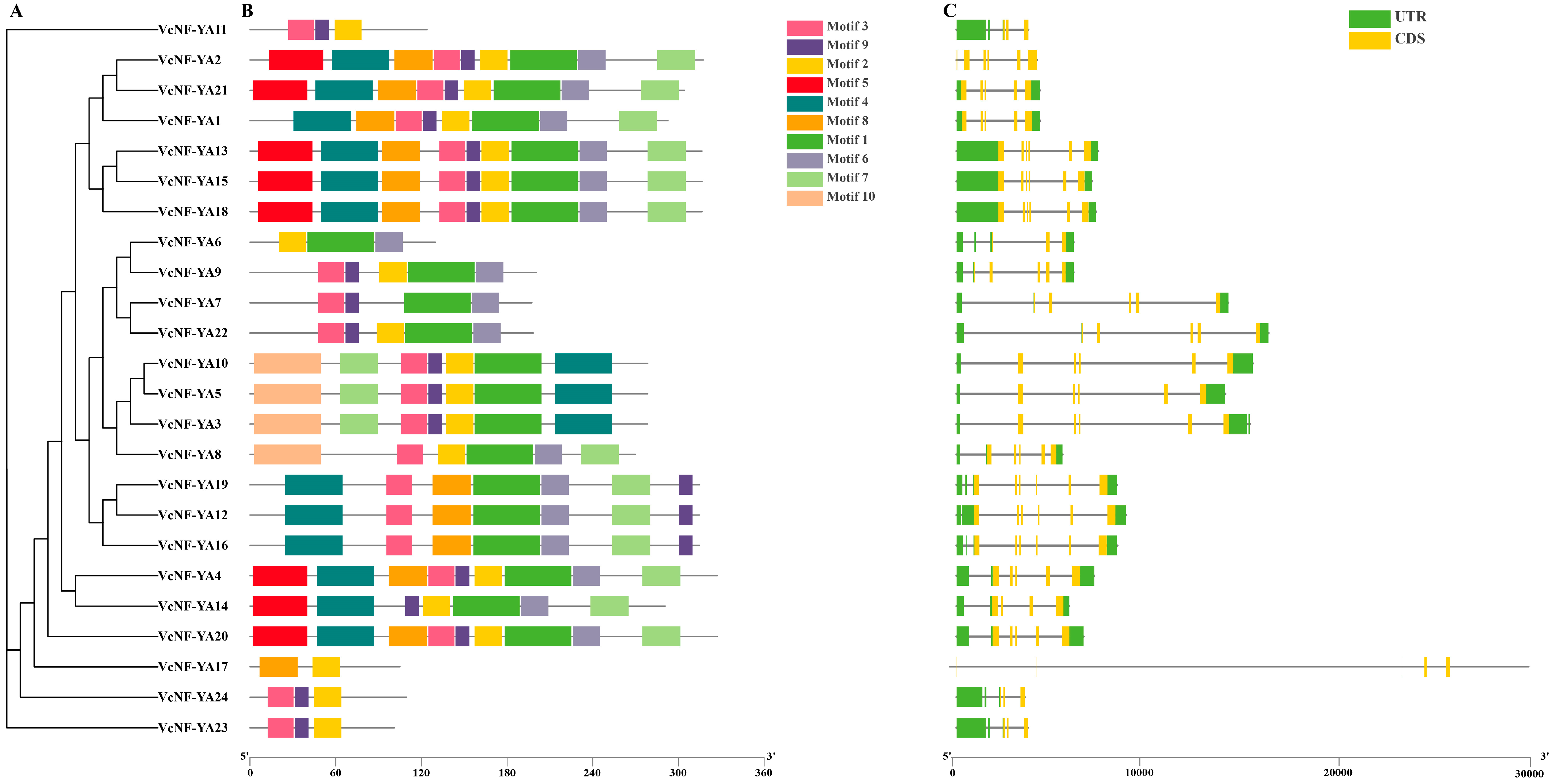
2.1. Identification and Sequence Analysis of NF-YA Transcription Factors in Blueberry
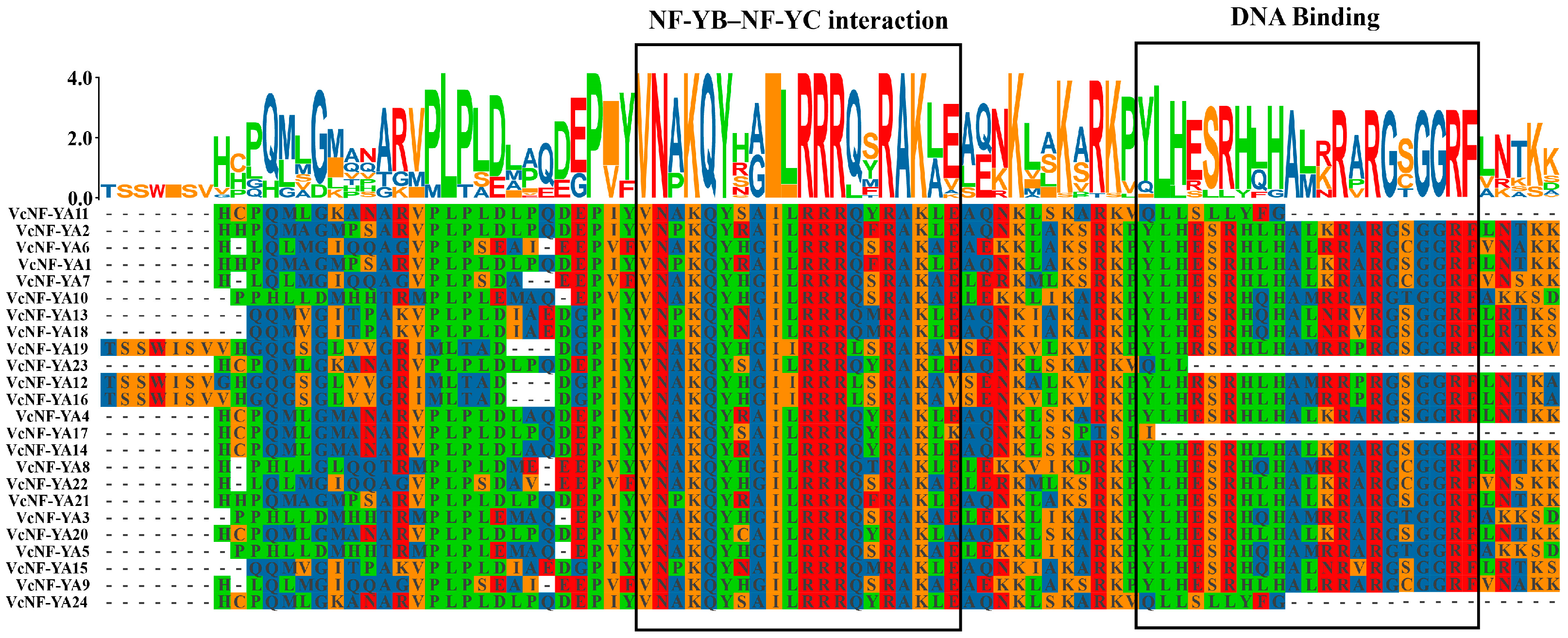
2.2. Multiple Sequence Alignment Analysis of the 24 VcNF-YAs
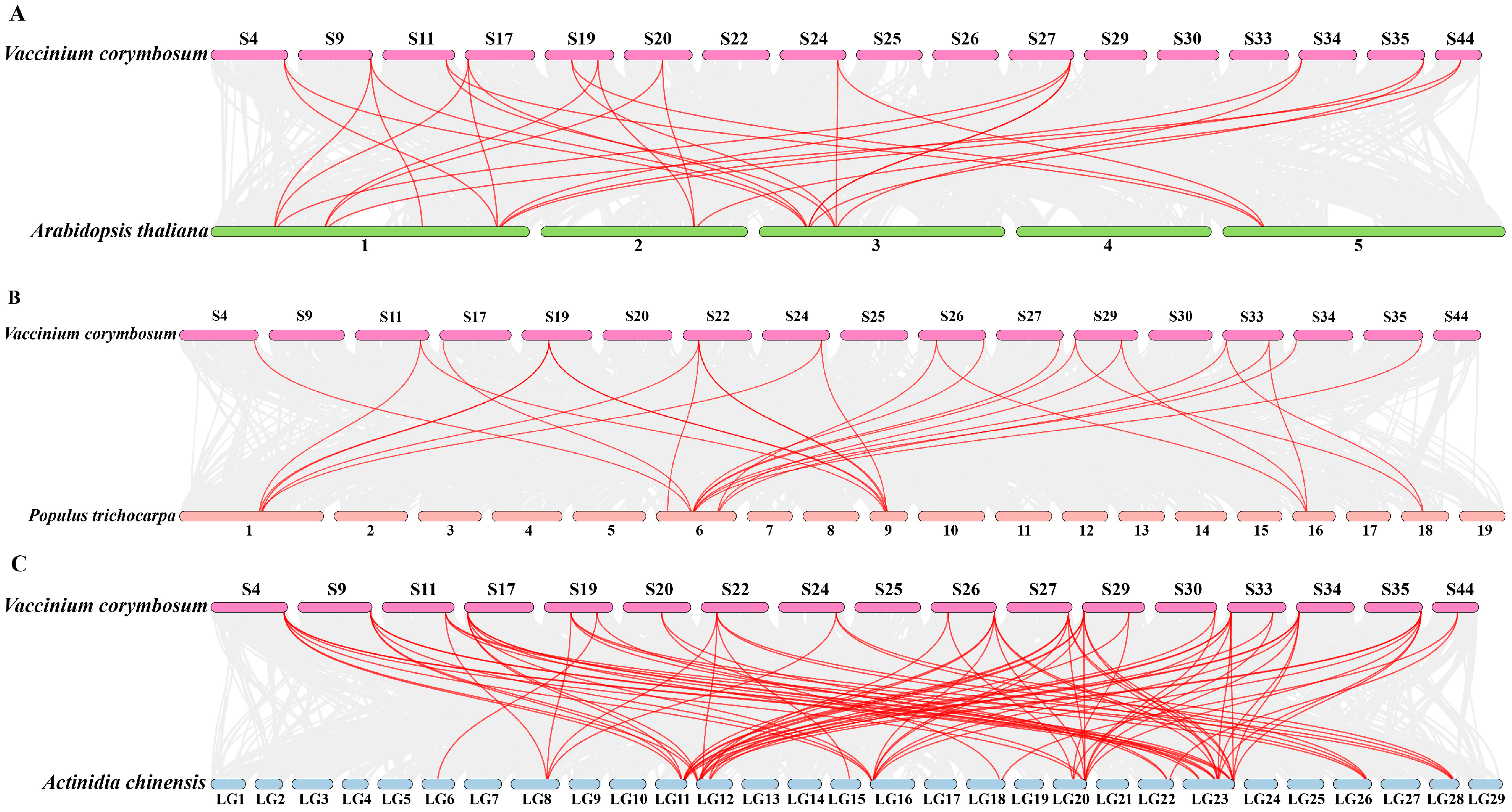
2.3. Chromosome Location and Intraspecies Collinearity of VcNF-YAs
2.4. Interspecies Collinearity of VcNF-YAs
2.5. Analysis of cis-Acting Elements in the Promoter Regions of VcNF-YAs
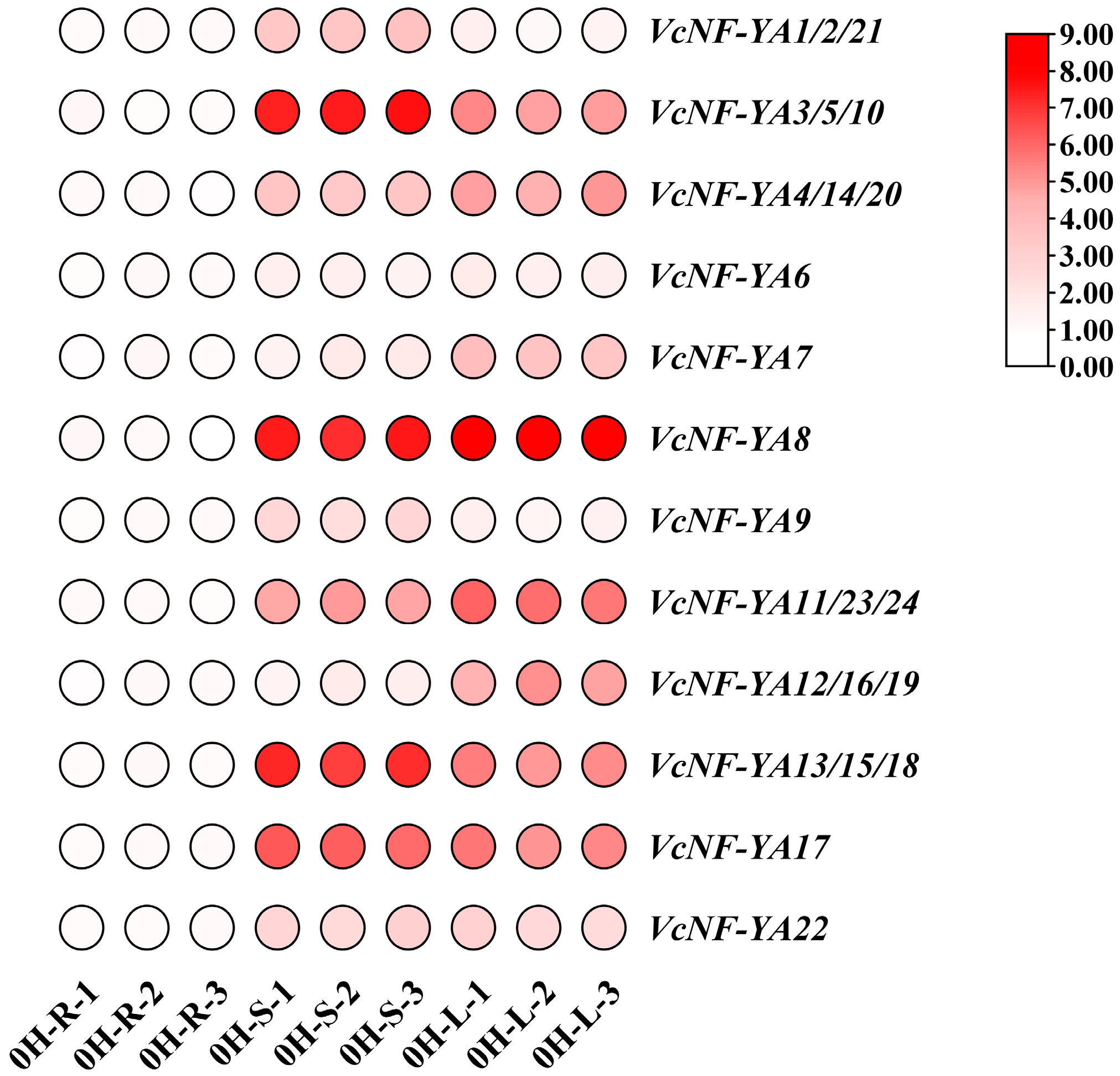
2.6. Tissue-Specific Expression Analysis of VcNF-YAs
2.7. Expression Patterns of VcNF-YAs Under ABA Treatment
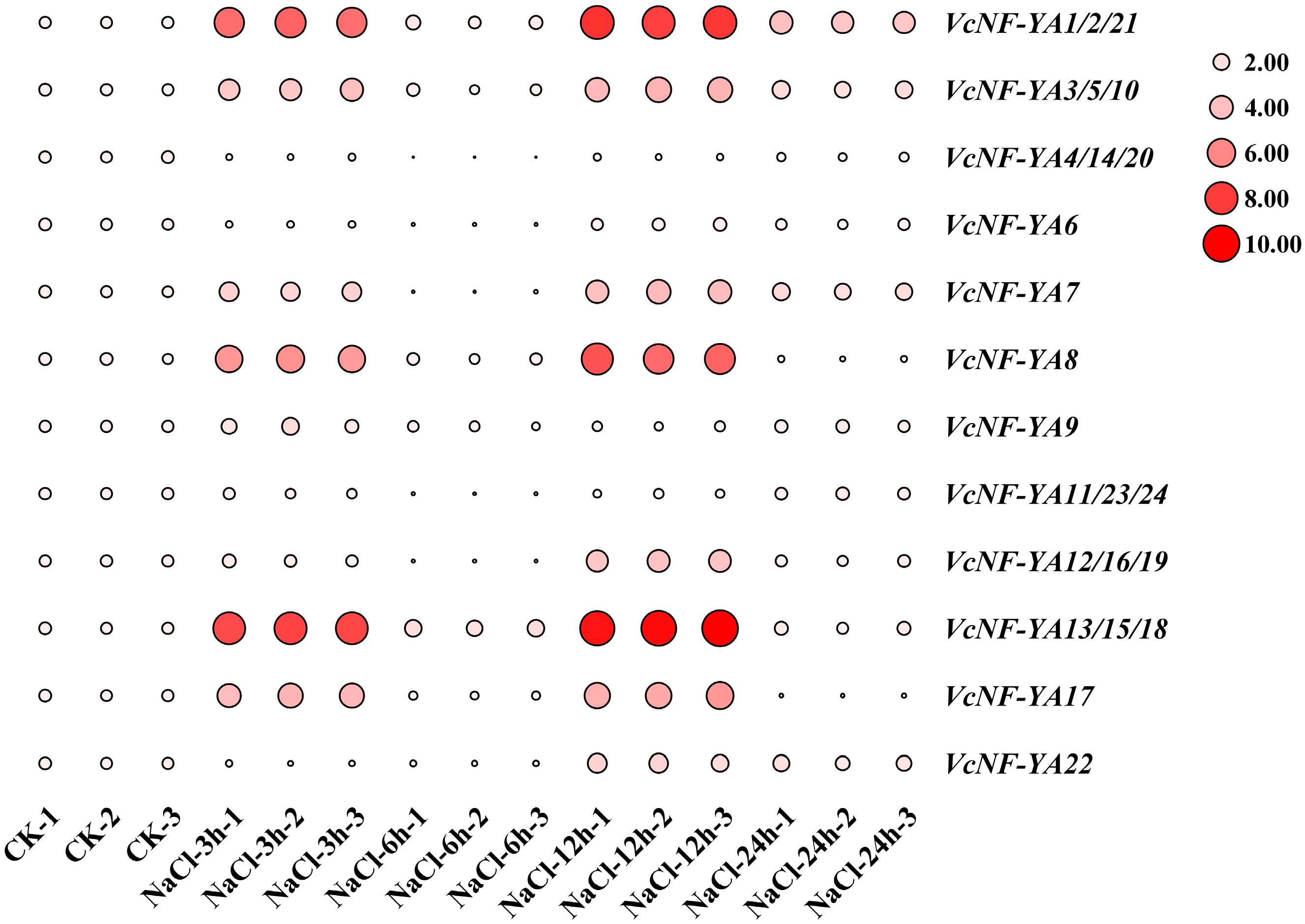
2.8. Expression Patterns of VcNF-YAs Under Salt Stress
2.9. Expression Patterns of VcNF-YAs Under Cold Stress
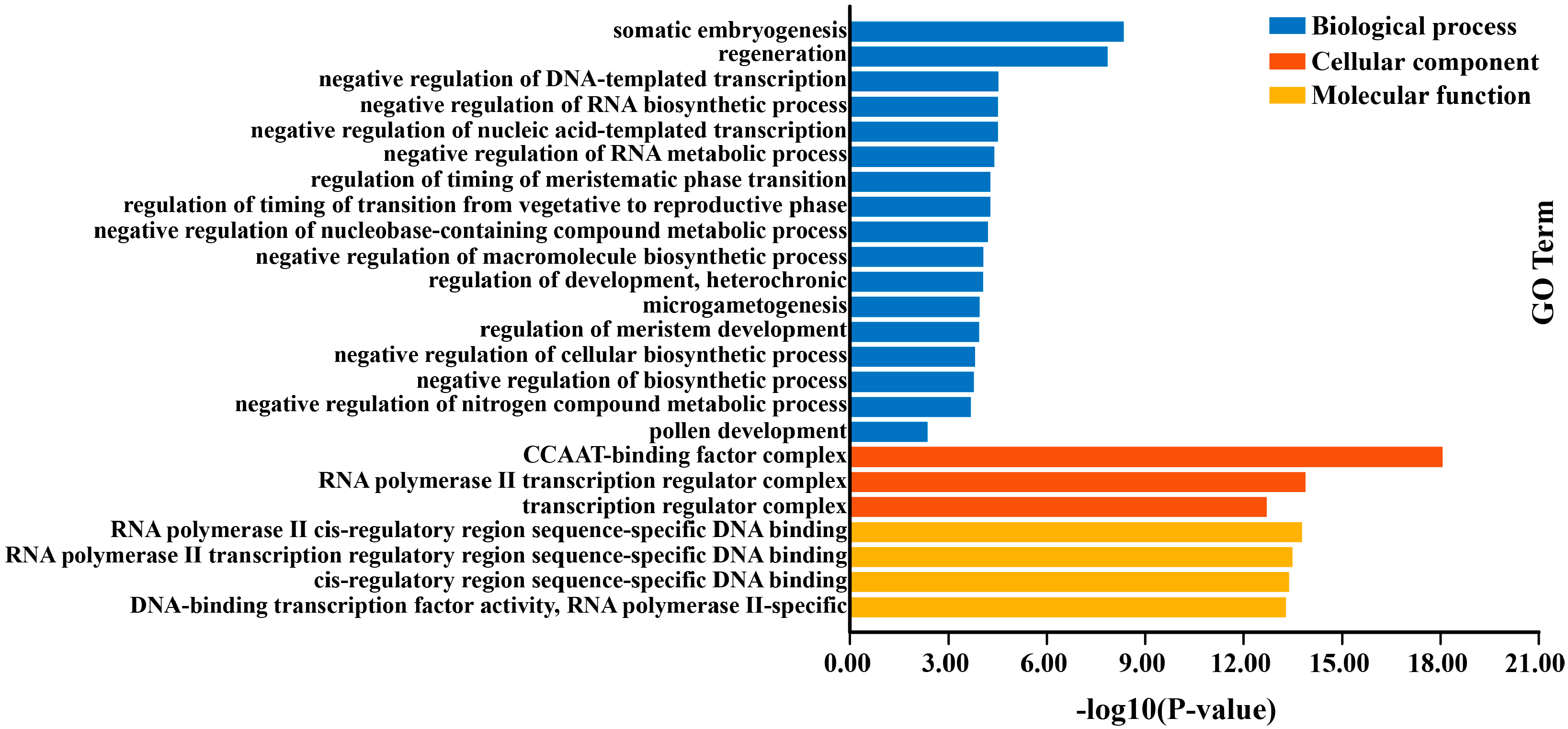
2.10. GO Enrichmentof VcNF-YA Proteins
2.11. Transcription-Activating Activity of VcNF-YAs
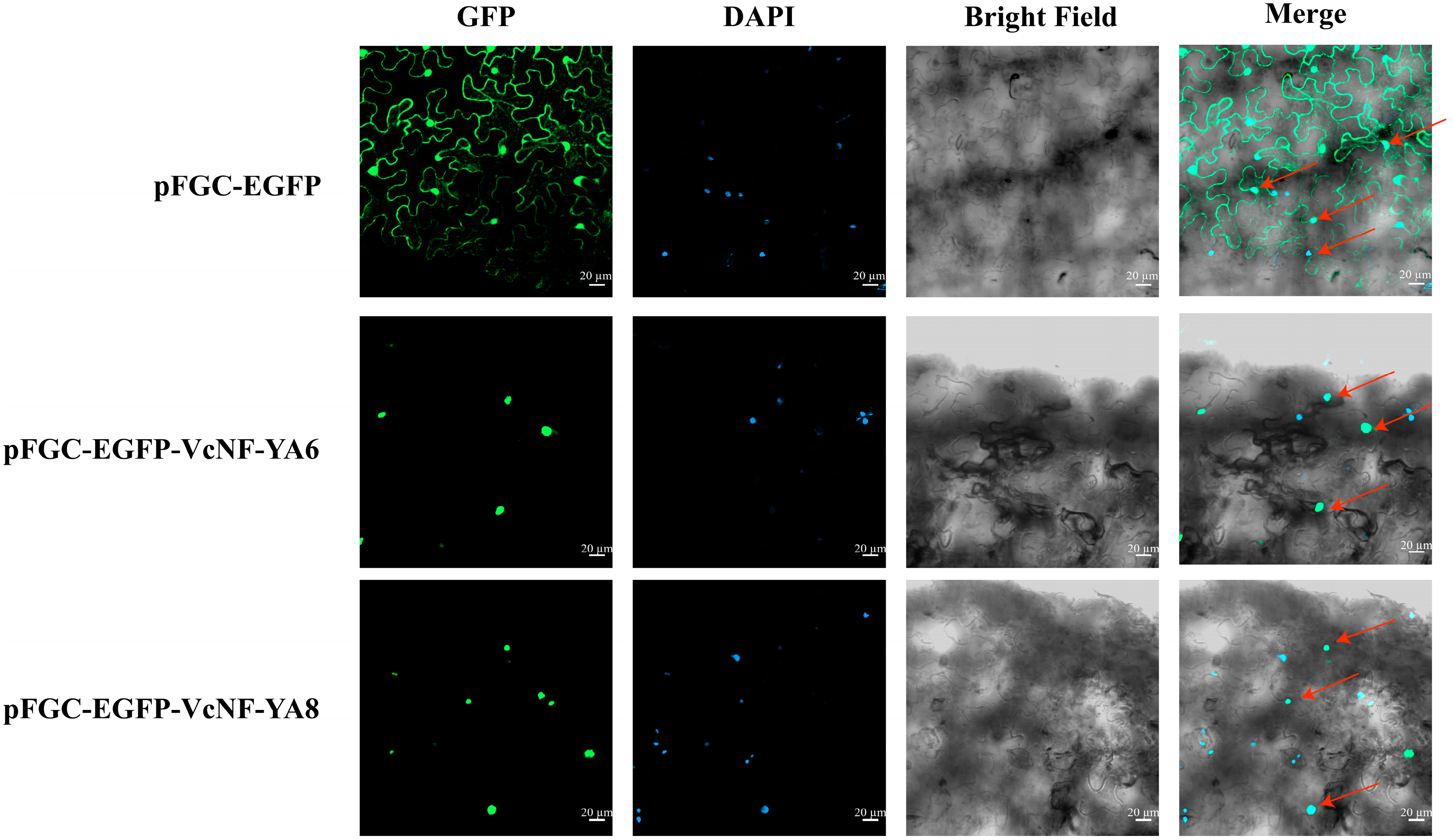
2.12. Subcellular Localization of VcNF-YA6 and VcNF-YA8 Proteins
3. Discussion
4. Materials and Methods
4.1. Identification and Analysis of VcNF-YA Transcription Factor Family in Vaccinium corymbosum
4.2. Analysis of Gene Structure and Conserved Motifs
4.3. Multiple Sequence Alignment and Phylogenetic Analysis of the 24 VcNF-YAs
4.4. Chromosome Distribution, Intraspecies and Interspecies Collinearity Analysis, and Ka/Ks Calculation
4.5. Analysis of cis-Acting Elements
4.6. Plant Materials, Growth Conditions, Treatments, and Sampling
4.7. RNA Extraction and Expression Analysis
4.8. Identification of Transcriptional Activation Activity of VcNF-YAs
4.9. GO Enrichment Analysis of VcNF-YA Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular Events of Rice AP2/ERF Transcription Factors. Int. J. Mol. Sci. 2022, 19, 12031. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana Nuclear Factor Y Transcription Factors. Front. Plant Sci. 2017, 7, 2045. [Google Scholar] [CrossRef]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-Specific Transcription Factor NF-Y Displays Histone-Like DNA Binding and H2B-Like Ubiquitination. Cell 2013, 152, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F., III; Mantovani, R. The Promiscuous Life of Plant Nuclear Factor Y Transcription Factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef] [PubMed]
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-Box Binding Transcription Factors in Plants: Y So Many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Calvenzani, V.; Testoni, G.; Gusmaroli, B.; Gusmaroli, M.; Lorenzo, N.; Gnesutta, K.; Petroni, R.; Mantovani, C.; Tonelli, C. Interactions and CCAAT-Binding of Arabidopsis thaliana NF-Y Subunits. PLoS ONE 2012, 7, e42902. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Mantovani, R.; Hooft van Huijsduijnen, R.; Andre, I.; Benoist, C.; Mathis, D. Evolutionary Variation of the CCAAT-Binding Transcription Factor NF-Y. Nucleic Acids Res. 1992, 20, 1087–1091. [Google Scholar] [CrossRef]
- Albani, D.; Robert, L.S. Cloning and Characterization of a Brassica Napus Gene Encoding a Homologue of the B Subunit of a Heteromeric CCAAT-Binding Factor. Gene 1995, 167, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.; Murray, J.A.; Smith, A.G. Multiple Genes Encoding the Conserved CCAAT-Box Transcription Factor Complex Are Expressed in Arabidopsis. Plant Physiol. 1998, 117, 1015–1022. [Google Scholar] [CrossRef]
- Mantovani, R. The Molecular Biology of the CCAAT-Binding Factor NF-Y. Gene 1999, 239, 15–27. [Google Scholar] [CrossRef]
- Gusmaroli, G.; Tonelli, C.; Mantovani, R. Regulation of the CCAAT-Binding NF-Y Subunits in Arabidopsis thaliana. Gene 2001, 264, 173–185. [Google Scholar] [CrossRef]
- Baskin, J.M.; Baskin, C.C. A Classification System for Seed Dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Finch, W.E.; Leubner-Metzger, G. Seed Dormancy and the Control of Germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Zhang, H.; Forde, B.G. An Arabidopsis Mads Box Gene That Controls Nutrient-Induced Changes in Root Architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Declerck, M.; Christ, A.; Blein, T.; Ma, L.; Lelandais-Brière, C.; Njo, M.F.; Beeckman, T.; Crespi, M.; Hartmann, C. A MiR169 Isoform Regulates Specific NF-YA Targets and Root Architecture in Arabidopsis. New Phytol. 2014, 202, 1197–1211. [Google Scholar] [CrossRef] [PubMed]
- Dolfini, D.; Imbriano, C.; Mantovani, R. The Role(S) of NF-Y in Development and Differentiation. Cell Death Differ. 2024, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; Alonso-Blanco, C.; Vreugdenhil, D. Naturally Occurring Genetic Variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004, 55, 141–172. [Google Scholar] [CrossRef] [PubMed]
- Mouradov, A.; Fau-Coupland, G.C.F.; Coupland, G. Control of Flowering Time: Interacting Pathways as a Basis for Diversity. Plant Cell 2002, 14, S111–S130. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G.; Dean, C. Arabidopsis, the Rosetta Stone of Flowering Time? Science 2002, 296, 285–289. [Google Scholar] [CrossRef]
- Blázquez, M.A.; Ahn, J.H.; Weigel, D. A Thermosensory Pathway Controlling Flowering Time in Arabidopsis thaliana. Nat. Genet. 2003, 33, 168–171. [Google Scholar] [CrossRef]
- Yanovsky, M.J.; Kay, S.A. Molecular Basis of Seasonal Time Measurement in Arabidopsis. Nature 2002, 419, 308–312. [Google Scholar] [CrossRef]
- Lee, D.-K.; Kim, H.I.; Jang, G.; Chung, P.J.; Jeong, J.S.; Kim, Y.S.; Bang, S.W.; Jung, H.; Choi, Y.D.; Kim, J.-K. The NF-YA Transcription Factor OsNF-YA7 Confers Drought Stress Tolerance of Rice in an Abscisic Acid Independent Manner. Plant Sci. 2015, 241, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding Plant Responses to Drought-from Genes to the Whole Plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Muhammad, M.A.; Waseem, M.; Jakada, B.; Okal, E.J.; Lei, Z.; Saqib, H.S.A.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of Abscisic Acid-Mediated Drought Stress Responses in Plants. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-Specific NF-Y Family Transcription Factor, PdNF-Yb21, Positively Regulates Root Growth and Drought Resistance by Abscisic Acid-Mediated Indoylacetic Acid Transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Quach, T.N.; Nguyen, H.T.; Valliyodan, B.; Joshi, T.; Xu, D.; Nguyen, H.T. Genome-Wide Expression Analysis of Soybean NF-Y Genes Reveals Potential Function in Development and Drought Response. Mol. Genet. Genom. 2015, 290, 1095–1115. [Google Scholar] [CrossRef]
- Wang, T.; Yu, L.; Li, Q.; Zhang, Q.; Yu, Z.; Ding, X.; Yang, S. Overexpression of GmNF-YA14 Produced Multiple Phenotypes in Soybean. Environ. Exp. Bot. 2023, 210, 105316. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, S.; Ren, T.; Niu, M.; Liu, X.; Liu, C.; Wang, H.; Yin, W.; Xia, X. Crucial Abiotic Stress Regulatory Network of NF-Y Transcription Factor in Plants. Int. J. Mol. Sci. 2023, 5, 4426. [Google Scholar] [CrossRef]
- Zhang, T.; Zhou, Y. Chapter 14—Plant Transcription Factors and Temperature Stress. In Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2023; pp. 287–300. [Google Scholar]
- Kavi Kishor, P.B.; Ganie, S.A.; Wani, S.H.; Guddimalli, R.; Karumanchi, A.R.; Edupuganti, S.; Naravula, J.; Kumar, V.; Polavarapu, R.; Suravajhala, P.; et al. Nuclear Factor-Y (NF-Y): Developmental and Stress-Responsive Roles in the Plant Lineage. J. Plant Growth Regul. 2023, 42, 2711–2735. [Google Scholar] [CrossRef]
- Li, S.; Li, K.; Ju, Z.; Cao, D.; Fu, D.; Zhu, H.; Zhu, B.; Luo, Y. Genome-Wide Analysis of Tomato NF-Y Factors and Their Role in Fruit Ripening. BMC Genom. 2016, 17, 36. [Google Scholar] [CrossRef]
- Delarue, M.; Benhamed, M.; Fragkostefanakis, S. The Role of Epigenetics in Tomato Stress Adaptation. New Crops 2025, 2, 100044. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Dong, K.; Liu, S.; Li, Y.; Xu, G.; Sun, H. Genome-Wide Identification of the WRKY Gene Family in Blueberry (Vaccinium spp.) and Expression Analysis under Abiotic Stress. Front. Plant Sci. 2024, 15, 1447749. [Google Scholar] [CrossRef]
- Chen, W.; Shao, J.; Ye, M.; Yu, K.; Bednarek, S.Y.; Duan, X.; Guo, W. Blueberry VcLON 2, a Peroxisomal LON Protease, Is Involved in Abiotic Stress Tolerance. Environ. Exp. Bot. 2017, 134, 1–11. [Google Scholar] [CrossRef]
- Yocca, A.E.; Platts, A.; Alger, E.; Teresi, S.; Mengist, M.F.; Benevenuto, J.; Ferrão, L.F.V.; Jacobs, M.; Babinski, M.; Magallanes-Lundback, M.; et al. Blueberry and Cranberry Pangenomes as a Resource for Future Genetic Studies and Breeding Efforts. Hortic. Res. 2023, 10, 202. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. Hmmer Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Allen, J.; Amode, R.M.; Azov, A.G.; Barba, M.; Becerra, A.; Bhai, J.; Campbell, L.I.; Martinez, M.C.; Chakiachvili, M.; et al. Ensembl Genomes 2022: An Expanding Genome Resource for Non-Vertebrates. Nucleic Acids Res. 2022, 50, D996–D1003. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. Plantcare, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ryynänen, L. Effect of Abscisic Acid, Cold Hardening, and Photoperiod on Recovery of Cryopreservedin Vitroshoot Tips of Silver Birch. Cryobiology 1998, 36, 32–39. [Google Scholar] [CrossRef]
- Hu, P.; Zhang, K.; Yang, C. Functional Roles of the Birch Bprav1 Transcription Factor in Salt and Osmotic Stress Response. Plant Sci. 2022, 315, 111131. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, F.; Wang, B.; Wu, H.; Wu, J.; Liu, J.; Sun, Y.; Cheng, C.; Qiu, D. Identification, Characterization and Expression Analysis of Anthocyanin Biosynthesis-Related bHLH Genes in Blueberry (Vaccinium corymbosum L.). Int. J. Mol. Sci. 2021, 22, 13274. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N. Statistical Analysis of Real-Time Pcr Data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef] [PubMed]





| Gene Name | Locus Name | Amino Acid No. | Molecular Weight (Da) | Isoelectric Point | Aromaticity | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| VcNF-YA1 | Vc_DUK_00005681-RA | 307 | 33,660.66 | 9.61 | 0.0912 | 61.59 | 61.73 | −0.58 |
| VcNF-YA2 | Vc_DUK_00049900-RA | 333 | 36,417.54 | 8.73 | 0.0871 | 59.22 | 60.75 | −0.61 |
| VcNF-YA3 | Vc_DUK_00060112-RA | 292 | 32,545.71 | 6.96 | 0.0616 | 66.5 | 51.64 | −1.06 |
| VcNF-YA4 | Vc_DUK_00005992-RA | 343 | 37,833.08 | 7.96 | 0.0787 | 50.93 | 61.22 | −0.68 |
| VcNF-YA5 | Vc_DUK_00024170-RA | 292 | 32,502.69 | 6.96 | 0.0616 | 67.86 | 51.99 | −1.04 |
| VcNF-YA6 | Vc_DUK_00024941-RA | 136 | 14,961.01 | 10.57 | 0.0368 | 74.77 | 69.71 | −0.74 |
| VcNF-YA7 | Vc_DUK_00022255-RA | 207 | 23,013.43 | 6.41 | 0.0676 | 64.11 | 52.42 | −1.01 |
| VcNF-YA8 | Vc_DUK_00052734-RA | 283 | 31,469.02 | 9.06 | 0.0459 | 57.26 | 55.51 | −1.02 |
| VcNF-YA9 | Vc_DUK_00014736-RA | 210 | 23,137.95 | 9.33 | 0.0667 | 74.77 | 57.76 | −0.81 |
| VcNF-YA10 | Vc_DUK_00015745-RA | 292 | 32,589.83 | 6.96 | 0.0616 | 65.48 | 51.64 | −1.05 |
| VcNF-YA11 | Vc_DUK_00044442-RA | 130 | 14,694.45 | 9.75 | 0.1077 | 39.08 | 105.85 | 0.06 |
| VcNF-YA12 | Vc_DUK_00011698-RA | 330 | 35,606 | 10.2 | 0.0727 | 58.77 | 60 | −0.59 |
| VcNF-YA13 | Vc_DUK_00013377-RA | 332 | 36,651.14 | 8.9 | 0.0602 | 61.1 | 60.54 | −0.73 |
| VcNF-YA14 | Vc_DUK_00034638-RA | 305 | 33,701.71 | 9.43 | 0.0721 | 48.39 | 63.05 | −0.67 |
| VcNF-YA15 | Vc_DUK_00025714-RA | 332 | 36,651.14 | 8.9 | 0.0602 | 61.1 | 60.54 | −0.73 |
| VcNF-YA16 | Vc_DUK_00027345-RA | 330 | 35,814.3 | 10.26 | 0.0727 | 59.37 | 60.27 | −0.61 |
| VcNF-YA17 | Vc_DUK_00036894-RA | 110 | 12,382.36 | 9 | 0.0636 | 64.42 | 86.09 | −0.36 |
| VcNF-YA18 | Vc_DUK_00060457-RA | 332 | 36,557.11 | 9.27 | 0.0633 | 59.98 | 59.1 | −0.73 |
| VcNF-YA19 | Vc_DUK_00061817-RA | 330 | 35,773.25 | 10.2 | 0.0727 | 57.96 | 60.85 | −0.59 |
| VcNF-YA20 | Vc_DUK_00070132-RA | 343 | 37,763 | 7.91 | 0.0787 | 52.89 | 60.09 | −0.67 |
| VcNF-YA21 | Vc_DUK_00039037-RA | 319 | 35,103.18 | 9.52 | 0.0878 | 62.31 | 60.34 | −0.65 |
| VcNF-YA22 | Vc_DUK_00094119-RA | 208 | 22,994.42 | 6.41 | 0.0625 | 65.96 | 55.91 | −0.94 |
| VcNF-YA23 | Vc_DUK_00099783-RA | 106 | 12,033.23 | 9.76 | 0.0755 | 65.2 | 95.85 | −0.3 |
| VcNF-YA24 | Vc_DUK_00101891-RA | 115 | 12,983.33 | 9.77 | 0.0957 | 66.15 | 90.87 | −0.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Su, H.; Sun, S.; Sun, J.; Zhang, X.; Yu, J. Genome-Wide Identification and Expression Profiles of Nuclear Factor Y A Transcription Factors in Blueberry Under Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 12832. https://doi.org/10.3390/ijms252312832
Xu X, Su H, Sun S, Sun J, Zhang X, Yu J. Genome-Wide Identification and Expression Profiles of Nuclear Factor Y A Transcription Factors in Blueberry Under Abiotic Stress. International Journal of Molecular Sciences. 2024; 25(23):12832. https://doi.org/10.3390/ijms252312832
Chicago/Turabian StyleXu, Xiuyue, Hong Su, Shuwei Sun, Jing Sun, Xiang Zhang, and Jiajie Yu. 2024. "Genome-Wide Identification and Expression Profiles of Nuclear Factor Y A Transcription Factors in Blueberry Under Abiotic Stress" International Journal of Molecular Sciences 25, no. 23: 12832. https://doi.org/10.3390/ijms252312832
APA StyleXu, X., Su, H., Sun, S., Sun, J., Zhang, X., & Yu, J. (2024). Genome-Wide Identification and Expression Profiles of Nuclear Factor Y A Transcription Factors in Blueberry Under Abiotic Stress. International Journal of Molecular Sciences, 25(23), 12832. https://doi.org/10.3390/ijms252312832




