Genome-Wide Analysis of Nuclear factor-YC Genes in the Tea Plant (Camellia sinensis) and Functional Identification of CsNF-YC6
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of CsNF-YC Subunits in the Tea Plant Genome
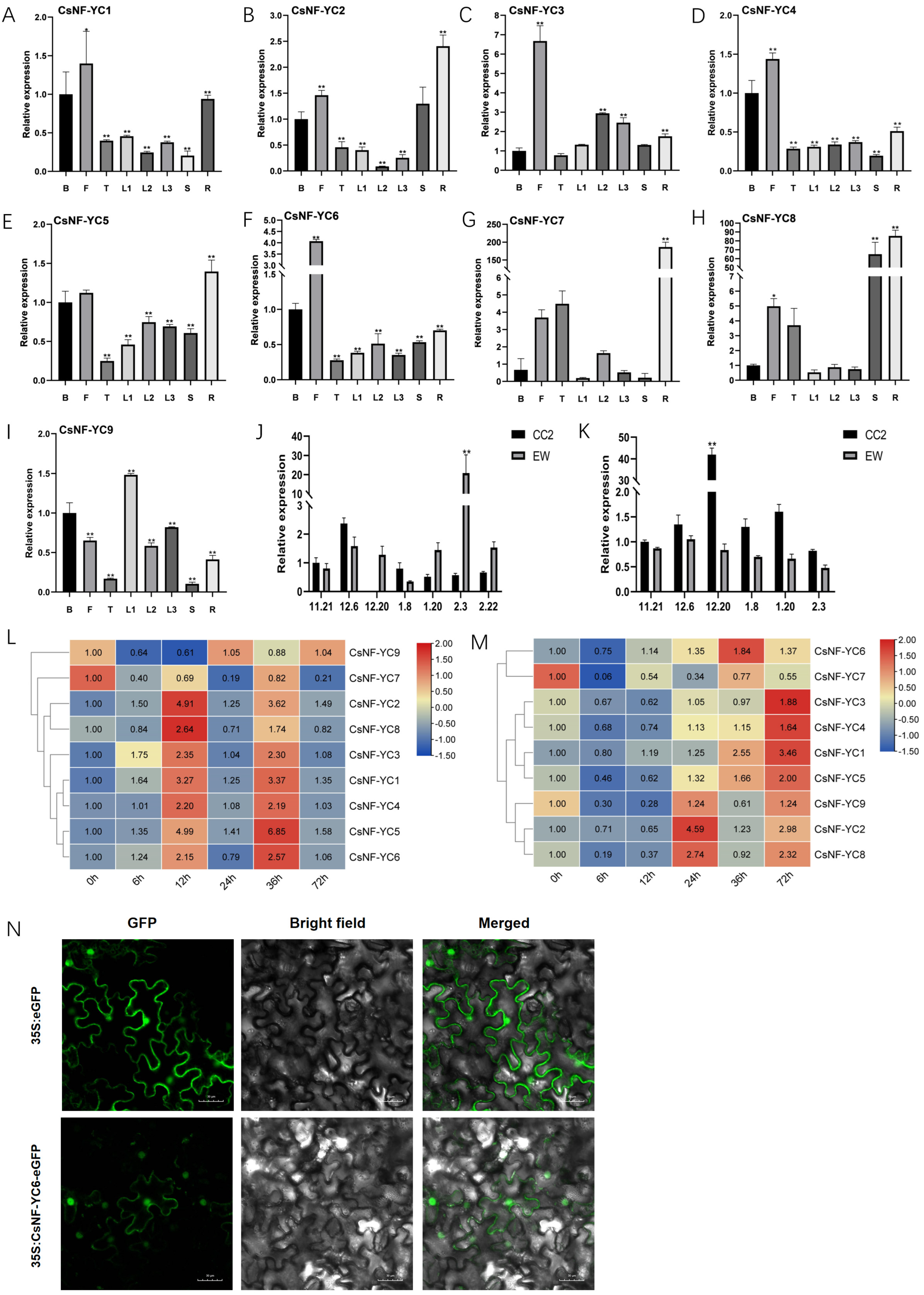
2.2. Expression Patterns of CsNF-YC Genes in Different Tissues and Their Responses to Abiotic Stresses
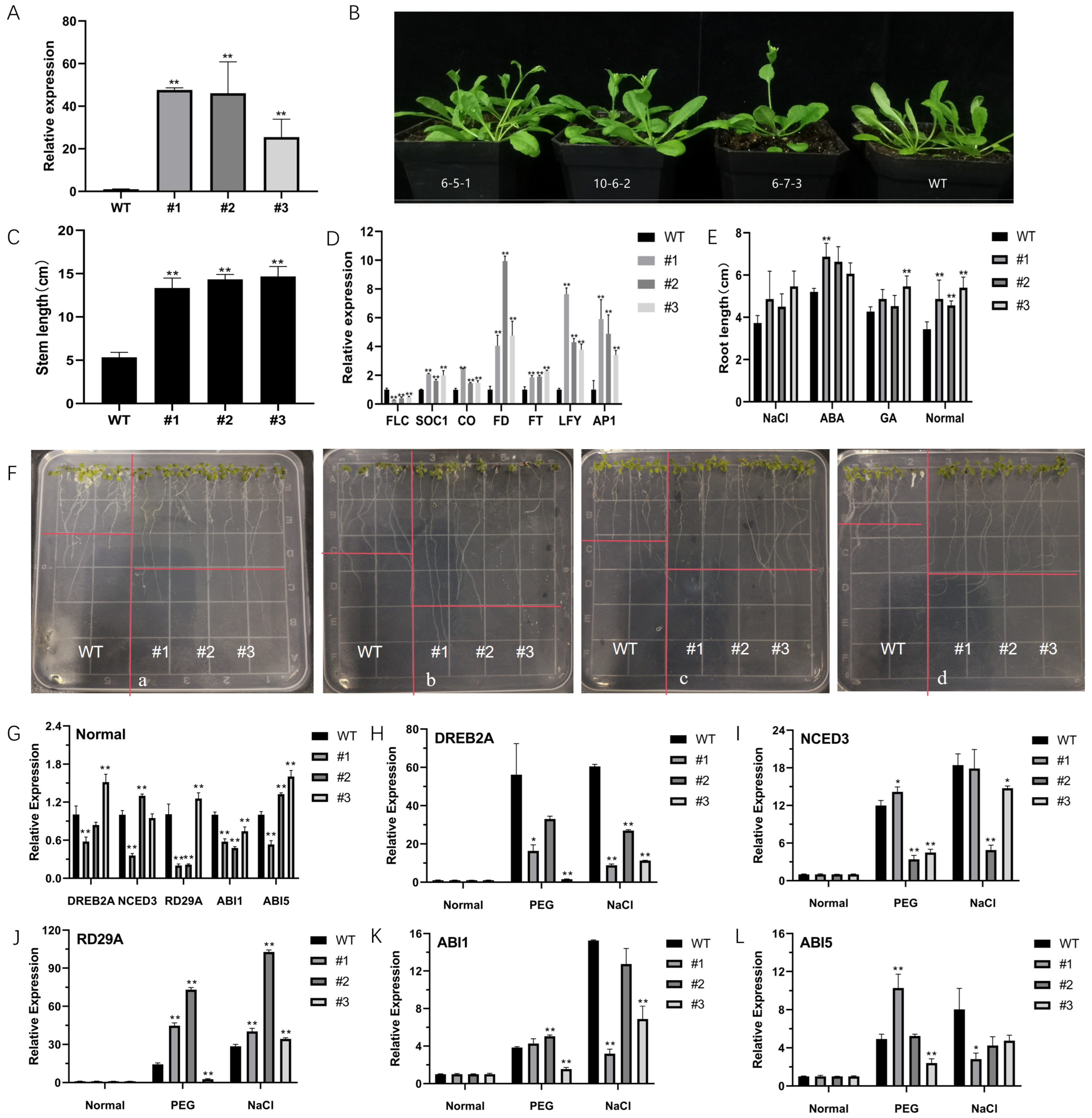
2.3. Morphological Characteristics of CsNF-YC6 Transgenic Arabidopsis Plants
2.4. Overexpression of CsNF-YC6 Enhances Stress Tolerance to Abiotic Factors
3. Discussion
3.1. Evolutionary Analysis and Molecular Characterization of the CsNF-YC Gene
3.2. CsNF-YC Genes Play an Important Role in Tea Plant Growth and Development and Response to Abiotic Stresses
3.3. Early Flowering and Improved Stress Tolerance by Overexpression of CsNF-YC6
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
4.2. Identification, Phylogenetic Tree Construction, Gene Structure, Motif Analysis and Multiple Comparisons
4.3. Quantitative Real-Time PCR
4.4. Cloning and Physicochemical Property Analysis of CsNF-YC Gene
4.5. Expression Vector Construction and Agrobacterium-Mediated Transformation of Arabidopsis thaliana
4.6. Abiotic Stresses Treatment
4.7. Morphological Characteristics of Transgenic Arabidopsis Plants
4.8. Measurement of Physiological Indicators
4.9. Experimental Data Processing and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gusmaroli, G.; Tonelli, C.; Mantovani, R. Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene 2001, 264, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Baudin, M.; Laloum, T.; Lepage, A.; Rípodas, C.; Ariel, F.; Frances, L.; Crespi, M.; Gamas, P.; Blanco, F.A.; Zanetti, M.E.; et al. A Phylogenetically Conserved Group of Nuclear Factor-Y Transcription Factors Interact to Control Nodulation in Legumes. Plant Physiol. 2015, 169, 2761–2773. [Google Scholar] [CrossRef] [PubMed]
- Dolfini, D.; Gatta, R.; Mantovani, R. NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 2011, 47, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.; Tayrose, G.; Holt, B.F. Tissue-Specific Expression Patterns of Arabidopsis NF-Y Transcription Factors Suggest Potential for Extensive Combinatorial Complexity. Plant Physiol. 2008, 149, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, N.; Zhu, X.; Ma, R.; Liu, S.; Wang, X.; Yang, J.; Si, H. Genome-Wide Analysis of NF-Y Genes in Potato and Functional Identification of StNF-YC9 in Drought Tolerance. Front. Plant Sci. 2021, 12, 749688. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, T.J.; McIntyre, C.L.; Collet, C.; Xue, G.-P. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 2007, 65, 77–92. [Google Scholar] [CrossRef]
- Li, M.; Li, G.; Liu, W.; Dong, X.; Zhang, A. Genome-wide analysis of the NF-Y gene family in peach (Prunus persica L.). BMC Genom. 2019, 20, 1–15. [Google Scholar] [CrossRef]
- Sinha, S.; Kim, I.S.; Sohn, K.Y.; de Crombrugghe, B.; Maity, S.N. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate function-al domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 1996, 16, 328–337. [Google Scholar] [CrossRef]
- Yu, T.F.; Liu, Y.; Fu, J.D.; Ma, J.; Fang, Z.W.; Chen, J.; Zheng, L.; Lu, Z.W.; Zhou, Y.B.; Chen, M.; et al. The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 2021, 19, 2589–2605. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M.E.; Rípodas, C.; Niebel, A. Plant NF-Y transcription factors: Key players in plant-microbe interactions, root development and adaptation to stress. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yao, Y.; Wen, S.; Bin, J.; Tan, Q.; Lou, J.; Xie, L.; Zeng, R.; Guo, H.; Zhang, Z.; et al. Genome-Wide Characterization of Chrysanthemum indicum Nuclear Factor Y, Subunit C Gene Family Reveals the Roles of CiNF-YCs in Flowering Regulation. Int. J. Mol. Sci. 2022, 23, 12812. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Liu, C.; Zhao, J.; Ye, X.; Wu, Q.; Yao, T.; Peng, L.; Zou, L.; Zhao, G. Genome-wide analysis of the NF-Y gene family and their roles in relation to fruit devel-opment in Tartary buckwheat (Fagopyrum tataricum). Int. J. Biol. Macromol. 2021, 190, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Fornari, M.; Calvenzani, V.; Masiero, S.; Tonelli, C.; Petroni, K. The Arabidopsis NF-YA3 and NF-YA8 genes are functionally re-dundant and are required in early embryogenesis. PLoS ONE 2013, 8, e82043. [Google Scholar] [CrossRef]
- E, Z.; Li, T.; Zhang, H.; Liu, Z.; Deng, H.; Sharma, S.; Wei, X.; Wang, L.; Niu, B.; Chen, C. A group of nuclear factor Y transcription factors are sub-functionalized during endosperm development in monocots. J. Exp. Bot. 2018, 69, 2495–2510. [Google Scholar] [CrossRef]
- Hackenberg, D.; Keetman, U.; Grimm, B. Homologous NF-YC2 Subunit from Arabidopsis and Tobacco Is Activated by Photooxidative Stress and Induces Flowering. Int. J. Mol. Sci. 2012, 13, 3458–3477. [Google Scholar] [CrossRef]
- Lv, X.; Zeng, X.; Hu, H.; Chen, L.; Zhang, F.; Liu, R.; Liu, Y.; Zhou, X.; Wang, C.; Wu, Z.; et al. Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO–NF–Y master transcription factor complex. Plant Cell 2021, 33, 1182–1195. [Google Scholar] [CrossRef]
- Myers, Z.A.; Kumimoto, R.W.; Siriwardana, C.L.; Gayler, K.K.; Risinger, J.R.; Pezzetta, D.; Iii, B.F.H. NUCLEAR FACTOR Y, Subunit C (NF-YC) Transcription Factors Are Positive Regulators of Photomorphogenesis in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006333. [Google Scholar] [CrossRef]
- Kim, S.K.; Park, H.Y.; Jang, Y.H.; Lee, K.C.; Chung, Y.S.; Lee, J.H.; Kim, J.K. OsNF-YC2 and OsNF-YC4 proteins inhibit flowering under long-day conditions in rice. Planta 2016, 243, 563–576. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Zhang, Y.; Siefers, N.; Holt, B.F. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010, 63, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yuan, Y.; Zhang, W.; Song, T.; Hou, X.; Kong, L.; Cui, G. Overexpression of an NF-YC2 gene confers alkali tolerance to transgenic alfalfa (Medicago sativa L.). Front. Plant Sci. 2022, 13, 960160. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhao, Y.; Zhuo, C.; Lu, S.; Guo, Z. Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 2015, 13, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Wang, L.; Zhou, B.; Liu, Q.; Chen, S.; Sun, D.; Zou, Y.; Chen, W.; Li, P.; Tang, Q. Comparative transcriptional analysis reveled genes related to short winter-dormancy regulation in Camellia sinensis. Plant Growth Regul. 2020, 92, 401–415. [Google Scholar] [CrossRef]
- Ren, C.; Zhang, Z.; Wang, Y.; Li, S.; Liang, Z. Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.). BMC Genom. 2016, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Y.; Zhu, J.; Ma, W.; Li, Z.; Bi, Z.; Sun, C.; Bai, J.; Zhang, J.; Liu, Y. Genome-Wide Identification and Analysis of the NF-Y Gene Family in Potato (Solanum tu-berosum L.). Front. Genet. 2021, 12, 739989. [Google Scholar] [CrossRef]
- Liu, R.; Wu, M.; Liu, H.L.; Gao, Y.M.; Chen, J.; Yan, H.W.; Xiang, Y. Genome-wide identification and expression analysis of the NF-Y transcription factor family in Populus. Plant Physiol. 2021, 171, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zheng, Y.; Guo, Y.; Chen, X.; Sun, Y.; Yang, J.; Ye, N. Identification, expression, and putative target gene analysis of nuclear factor-Y (NF-Y) transcription factors in tea plant (Camellia sinensis). Planta 2019, 250, 1671–1686. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Zhang, C.; Zou, H.; Wu, Z. Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem. Biophys. Res. Commun. 2016, 478, 752–758. [Google Scholar] [CrossRef]
- Yue, Y.; Zhang, D.; Dou, H.; Liu, X.; Gao, L.; Chen, Y.; Fan, G.; Zhang, Y.; Wu, Y. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in tea plant. Res. Sq. 2019, 12, 1237. [Google Scholar]
- Hwang, Y.H.; Kim, S.K.; Lee, K.C.; Chung, Y.S.; Lee, J.H.; Kim, J.K. Functional conservation of rice OsNF-YB/YC and Arabidopsis AtNF-YB/YC proteins in the regulation of flowering time. Plant Cell Rep. 2016, 35, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, W.; Chen, Z.; Han, B.; Haque, M.E.; Liu, A. Gene structure, expression pattern and interaction of Nuclear Factor-Y family in castor bean (Ricinus communis). Planta 2017, 247, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, C.; Yao, M.; Chen, L. The tea plant CsWRKY26 promotes drought tolerance in transgenic Arabidopsis plants. Beverage Plant Res. 2021, 1, 3. [Google Scholar] [CrossRef]
- Andrés, F.; Porri, A.; Torti, S.; Mateos, J.; Romera-Branchat, M.; García-Martínez, J.L.; Fornara, F.; Gregis, V.; Kater, M.M.; Coupland, G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 2014, 111, E2760–E2769. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC–RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef]
- Hou, X.L.; Zhou, J.N.; Liu, C.; Liu, L.; Shen, L.S.; Yu, H. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 2014, 5, 4601. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Wang, Y.; Zhu, J.; Zhang, Y.; Hou, H. Genomic Organization, Phylogenetic Comparison, and Differential Expression of the Nuclear Factor-Y Gene Family in Apple (Malus domestica). Plants 2020, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Y.; Li, T.; Ni, C.; Bai, X.; Lin, R.; Xiao, K. TaNF-YA7-5B, a gene encoding nuclear factor Y (NF-Y) subunit A in Triticum aestivum, confers plant tolerance to PEG-inducing dehydration simulating drought through modulating osmotic stress-associated phys-iological processes. Plant Physiol. Biochem.. 2022, 188, 81–96. [Google Scholar] [CrossRef]
- Ma, X.-J.; Yu, T.-F.; Li, X.H.; Cao, X.-Y.; Ma, J.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Zhang, J.H.; et al. Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants. BMC Plant Biol. 2020, 20, 123. [Google Scholar] [CrossRef]
- Li, X.; Zhang, G.; Liang, Y.; Hu, L.; Zhu, B.; Qi, D.; Cui, S.; Zhao, H. TCP7 interacts with Nuclear Factor-Ys to promote flowering by directly regulating SOC1 in Arabidopsis. Plant J. 2021, 108, 1493–1506. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Wei, X.; Hu, X.; Zhang, P.; Chang, K.; Zhang, D.; Chen, W.; Tang, D.; Tang, Q.; Li, P.; et al. Genome-Wide Analysis of Nuclear factor-YC Genes in the Tea Plant (Camellia sinensis) and Functional Identification of CsNF-YC6. Int. J. Mol. Sci. 2024, 25, 836. https://doi.org/10.3390/ijms25020836
Chen S, Wei X, Hu X, Zhang P, Chang K, Zhang D, Chen W, Tang D, Tang Q, Li P, et al. Genome-Wide Analysis of Nuclear factor-YC Genes in the Tea Plant (Camellia sinensis) and Functional Identification of CsNF-YC6. International Journal of Molecular Sciences. 2024; 25(2):836. https://doi.org/10.3390/ijms25020836
Chicago/Turabian StyleChen, Shengxiang, Xujiao Wei, Xiaoli Hu, Peng Zhang, Kailin Chang, Dongyang Zhang, Wei Chen, Dandan Tang, Qian Tang, Pinwu Li, and et al. 2024. "Genome-Wide Analysis of Nuclear factor-YC Genes in the Tea Plant (Camellia sinensis) and Functional Identification of CsNF-YC6" International Journal of Molecular Sciences 25, no. 2: 836. https://doi.org/10.3390/ijms25020836
APA StyleChen, S., Wei, X., Hu, X., Zhang, P., Chang, K., Zhang, D., Chen, W., Tang, D., Tang, Q., Li, P., & Tan, L. (2024). Genome-Wide Analysis of Nuclear factor-YC Genes in the Tea Plant (Camellia sinensis) and Functional Identification of CsNF-YC6. International Journal of Molecular Sciences, 25(2), 836. https://doi.org/10.3390/ijms25020836






