Genome-Wide Association Study Identifies Rice Panicle Blast-Resistant Gene Pb4 Encoding a Wall-Associated Kinase
Abstract
1. Introduction
2. Results
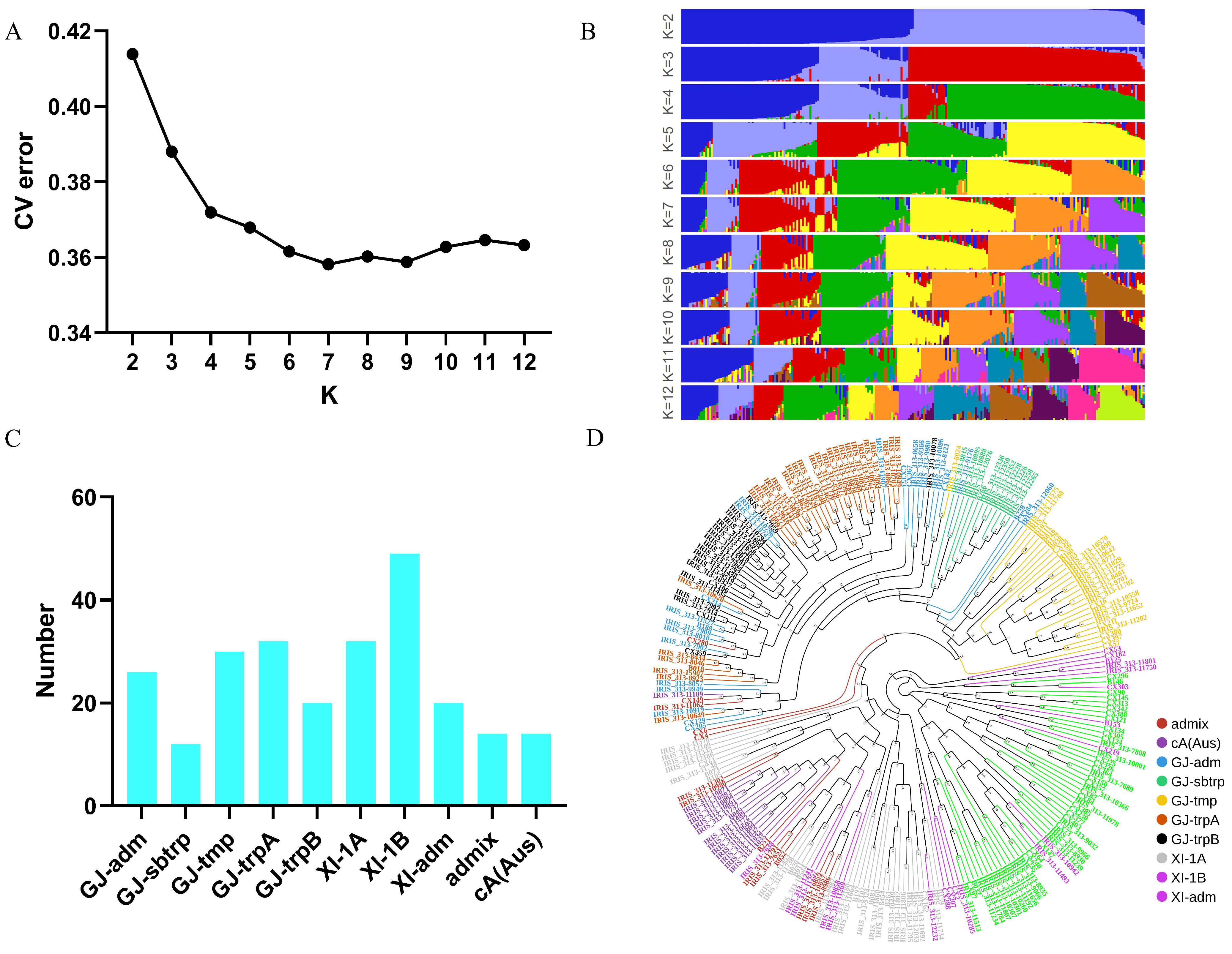
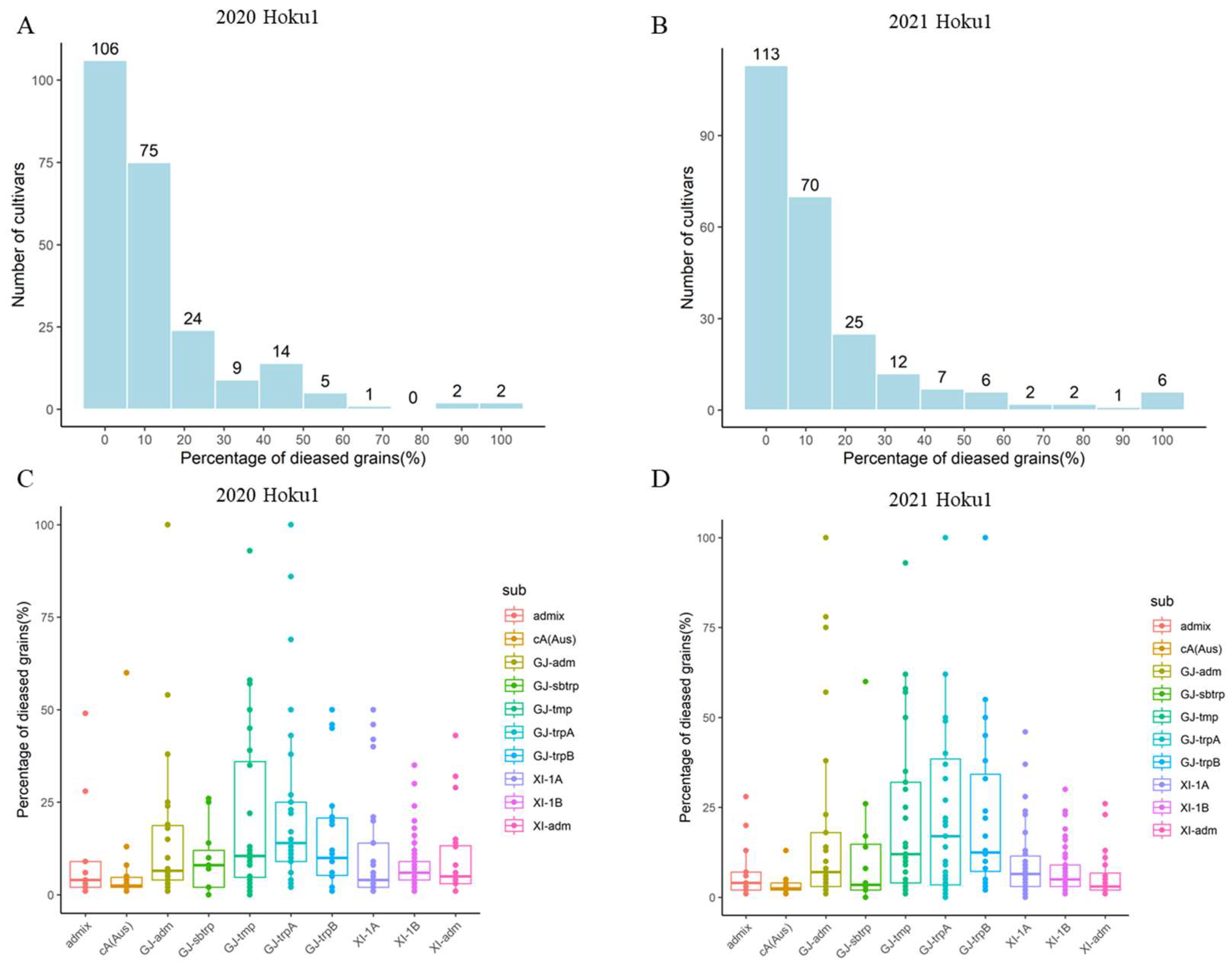
2.1. Structure and Phenotype of the Population
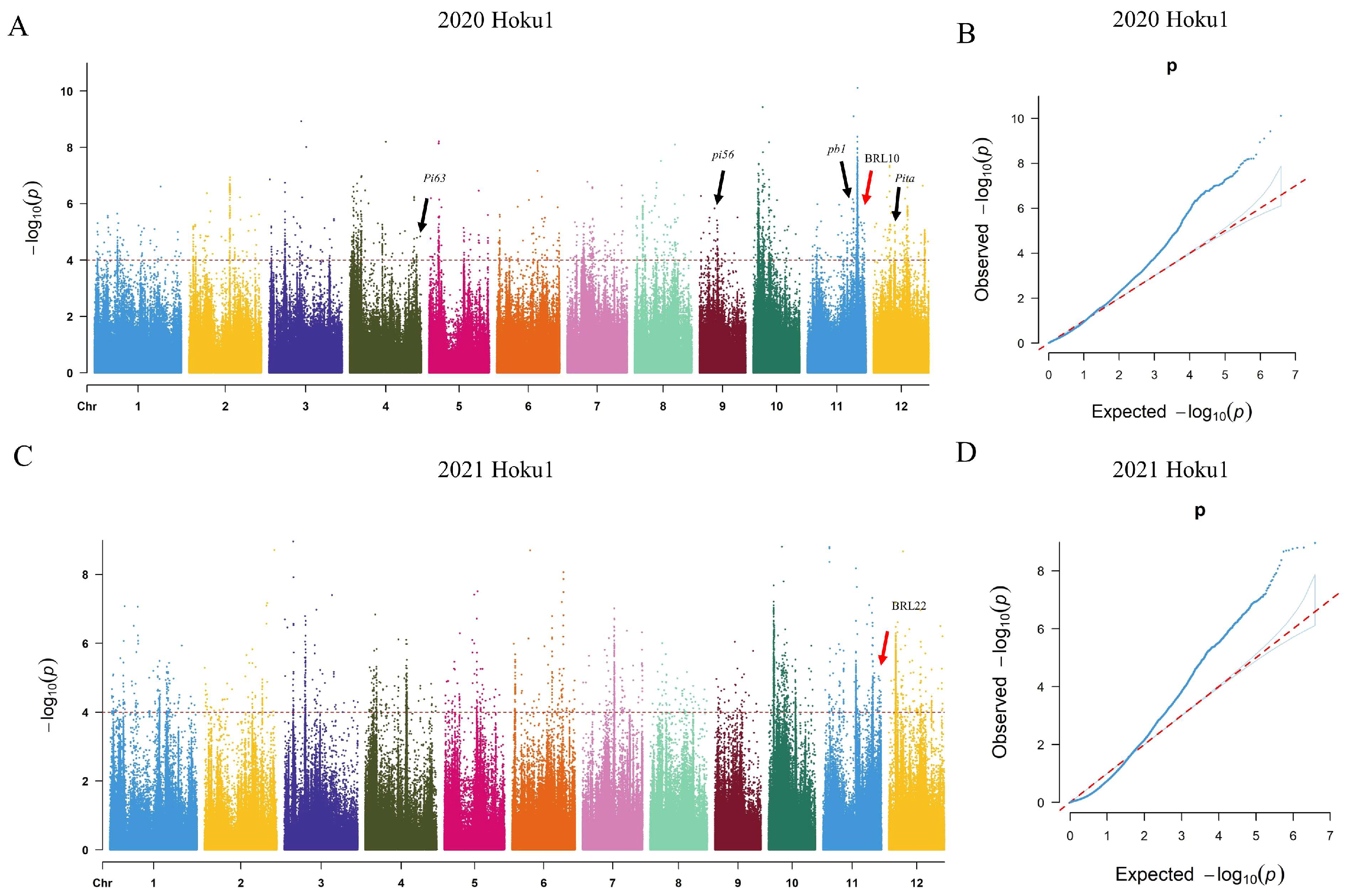
2.2. Identification of Blast-Resistant Loci in Whole Genome
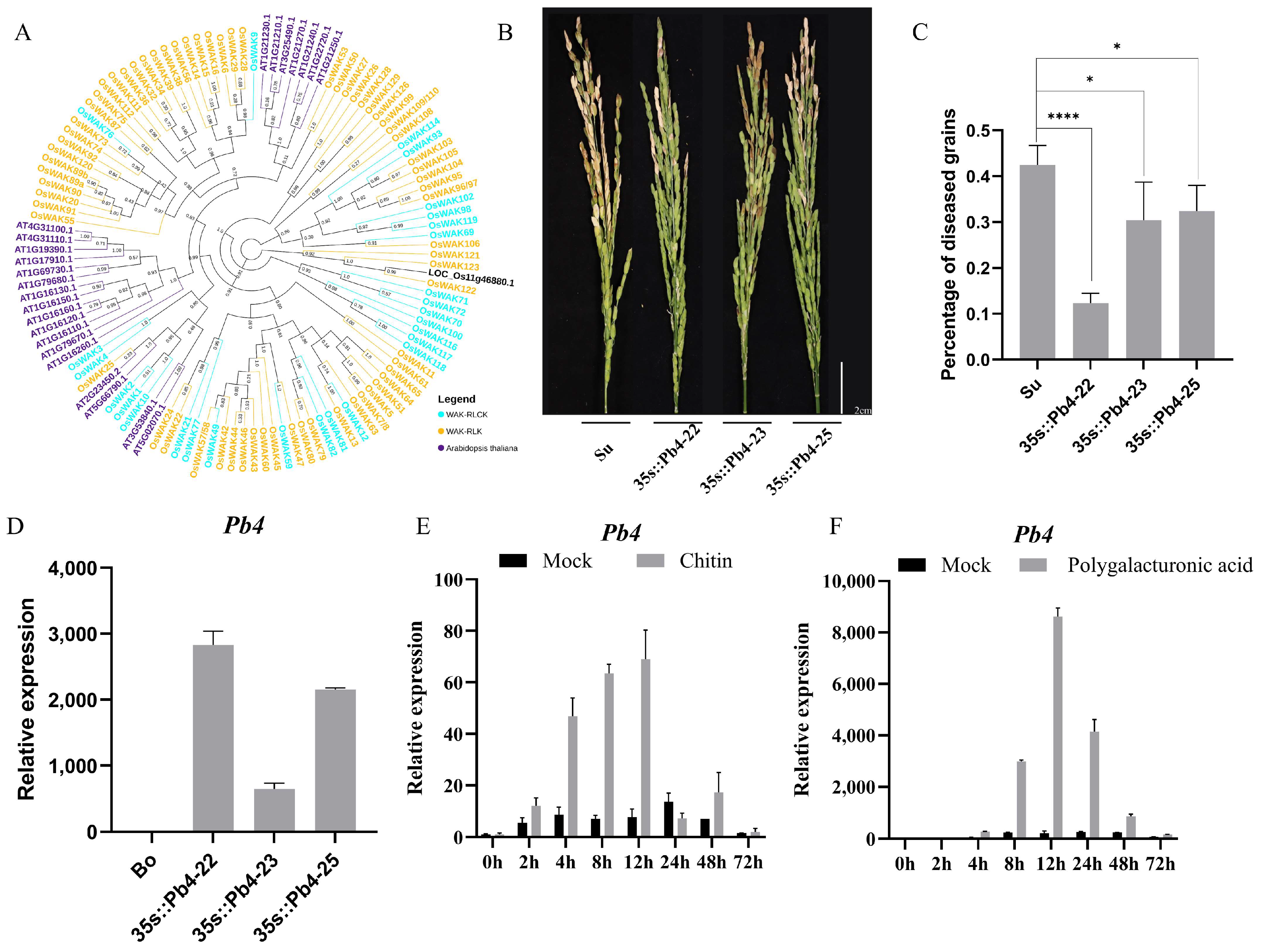
2.3. Analysis of the Candidate Genes in BRL10 and BRL22
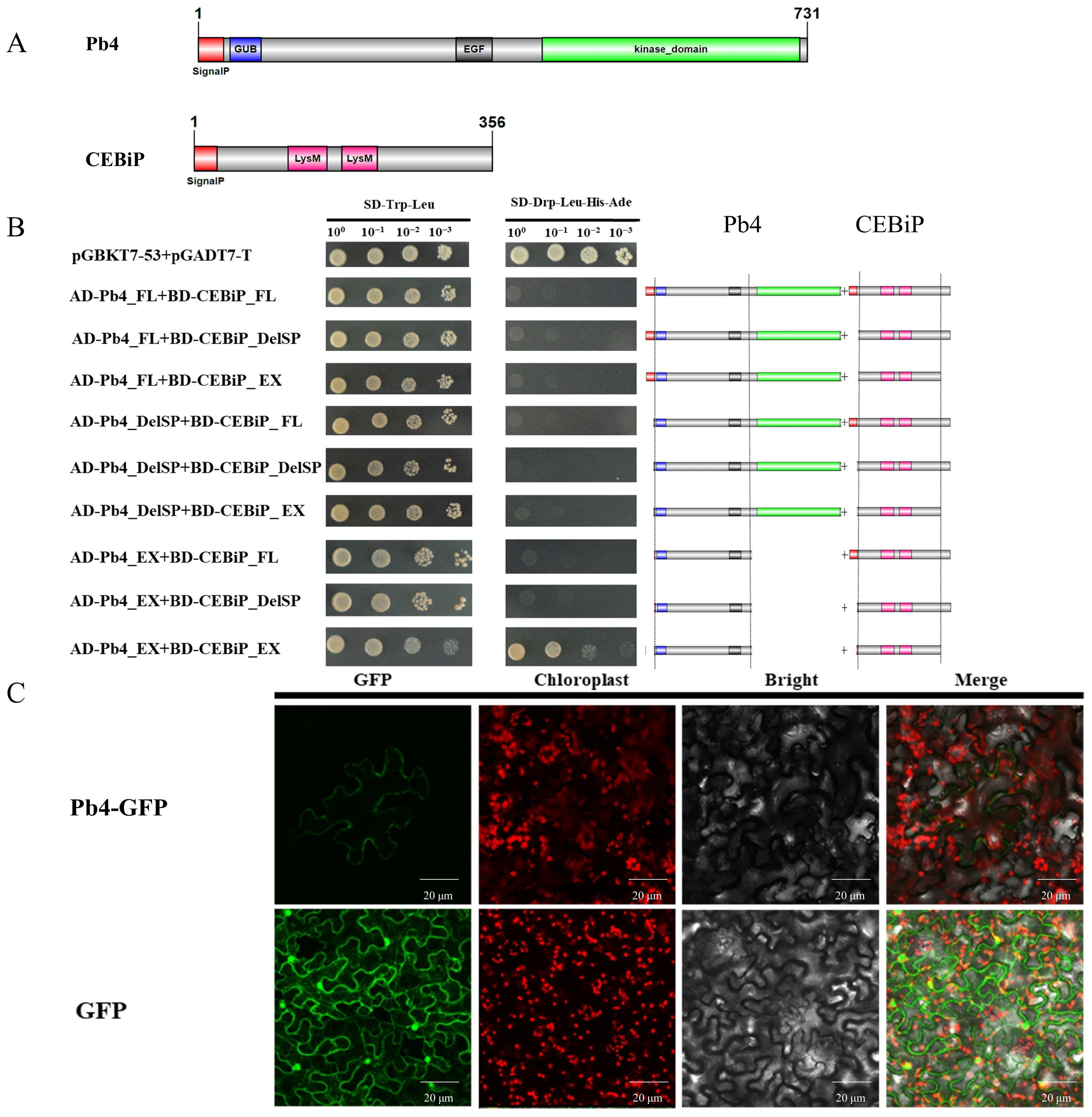
2.4. Pb4 Positively Regulates Resistance to Rice Blast
3. Discussion
4. Materials and Methods
4.1. Plant and Fungal Materials
4.2. Population Structure and GWAS Analysis
4.3. RNA Extraction and Real-Time PCR
4.4. Construction of Transgenic Rice Plants
4.5. Subcellular Localization
4.6. Y2H Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, Y.; Ning, Y.; He, Z.; Wang, G.L. Exploiting Broad-Spectrum Disease Resistance in Crops: From Molecular Dissection to Breeding. Annu. Rev. Plant Biol. 2020, 71, 575–603. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T.; et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010, 64, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ma, L.; Wang, X.; Zhao, Z.; Wang, W.; Fan, Y.; Liu, K.; Jiang, T.; Xiong, Z.; Song, Q.; et al. Genome-Wide Association Study Identifies a Rice Panicle Blast Resistance Gene, Pb2, Encoding NLR Protein. Int. J. Mol. Sci. 2022, 23, 5668. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yu, Y.; Li, C.; Wang, P.; Liu, K.; Ma, W.; Wang, W.; Fan, Y.; Xiong, Z.; Jiang, T.; et al. Genome-Wide Association Study Identifies a Rice Panicle Blast Resistance Gene Pb3 Encoding NLR Protein. Int. J. Mol. Sci. 2022, 23, 14032. [Google Scholar] [CrossRef]
- Chen, J.; Shi, Y.; Liu, W.; Chai, R.; Fu, Y.; Zhuang, J.; Wu, J. A Pid3 allele from rice cultivar Gumei2 confers resistance to Magnaporthe oryzae. J. Genet. Genom. 2011, 38, 209–216. [Google Scholar] [CrossRef]
- Ma, J.; Lei, C.; Xu, X.; Hao, K.; Wang, J.; Cheng, Z.; Ma, X.; Ma, J.; Zhou, K.; Zhang, X.; et al. Pi64, Encoding a Novel CC-NBS-LRR Protein, Confers Resistance to Leaf and Neck Blast in Rice. Mol. Plant Microbe Interact. 2015, 28, 558–568. [Google Scholar] [CrossRef]
- Devi, S.; Singh, K.; Umakanth, B.; Vishalakshi, B.; Rao, K.V.S.; Suneel, B.; Sharma, S.K.; Kadambari, G.K.M.; Prasad, M.S.; Senguttvel, P.; et al. Identification and Characterization of a Large Effect QTL from Oryza glumaepatula Revealed Pi68(t) as Putative Candidate Gene for Rice Blast Resistance. Rice 2020, 13, 17. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, H.; Chern, M.; Yin, J.; Chen, Y.; Wang, J.; Zhu, X.; Chen, Z.; Yuan, C.; Zhao, W.; et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar] [CrossRef]
- Gao, M.; He, Y.; Yin, X.; Zhong, X.; Yan, B.; Wu, Y.; Chen, J.; Li, X.; Zhai, K.; Huang, Y.; et al. Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 2021, 184, 5391–5404.e17. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G.; et al. AB-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cao, Y.; Yang, Z.; Xu, C.; Li, X.; Wang, S.; Zhang, Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004, 37, 517–527. [Google Scholar] [CrossRef]
- Hu, K.; Cao, J.; Zhang, J.; Xia, F.; Ke, Y.; Zhang, H.; Xie, W.; Liu, H.; Cui, Y.; Cao, Y.; et al. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, V.; Gupta, A.K. Wall associated kinases from plants—An overview. Physiol. Mol. Biol. Plants 2008, 14, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Verica, J.A.; He, Z.H. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 2002, 129, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Park, A.R.; Cho, S.K.; Yun, U.J.; Jin, M.Y.; Lee, S.H.; Sachetto-Martins, G.; Park, O.K. Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J. Biol. Chem. 2001, 276, 26688–26693. [Google Scholar] [CrossRef]
- Decreux, A.; Thomas, A.; Spies, B.; Brasseur, R.; Van Cutsem, P.; Messiaen, J. In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry 2006, 67, 1068–1079. [Google Scholar] [CrossRef]
- Cabrera, J.C.; Boland, A.; Messiaen, J.; Cambier, P.; Van Cutsem, P. Egg box conformation of oligogalacturonides: The time-dependent stabilization of the elicitor-active conformation increases its biological activity. Glycobiology 2008, 18, 473–482. [Google Scholar] [CrossRef]
- Kohorn, B.D.; Johansen, S.; Shishido, A.; Todorova, T.; Martinez, R.; Defeo, E.; Obregon, P. Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 2009, 60, 974–982. [Google Scholar] [CrossRef]
- Zhai, K.; Deng, Y.; Liang, D.; Tang, J.; Liu, J.; Yan, B.; Yin, X.; Lin, H.; Chen, F.; Yang, D.; et al. RRM Transcription Factors Interact with NLRs and Regulate Broad-Spectrum Blast Resistance in Rice. Mol. Cell 2019, 74, 996–1009.e7. [Google Scholar] [CrossRef] [PubMed]
- Harkenrider, M.; Sharma, R.; De Vleesschauwer, D.; Tsao, L.; Zhang, X.; Chern, M.; Canlas, P.; Zuo, S.; Ronald, P.C. Overexpression of Rice Wall-Associated Kinase 25 (OsWAK25) Alters Resistance to Bacterial and Fungal Pathogens. PLoS ONE 2016, 11, e0147310. [Google Scholar] [CrossRef] [PubMed]
- Demirjian, C.; Vailleau, F.; Berthome, R.; Roux, F. Genome-wide association studies in plant pathosystems: Success or failure? Trends Plant Sci. 2023, 28, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.H.; Kang, H.; Xu, Y.; Peng, Y.; Wang, D.; Gao, L.; Wang, X.; Ning, Y.; Wu, J.; Liu, W.; et al. Genome-wide association study identifies an NLR gene that confers partial resistance to Magnaporthe oryzae in rice. Plant Biotechnol. J. 2020, 18, 1376–1383. [Google Scholar] [CrossRef]
- Xu, Y.; Bai, L.; Liu, M.; Liu, Y.; Peng, S.; Hu, P.; Wang, D.; Liu, Q.; Yan, S.; Gao, L.; et al. Identification of two novel rice S genes through combination of association and transcription analyses with gene-editing technology. Plant Biotechnol. J. 2023, 21, 1628–1641. [Google Scholar] [CrossRef]
- Bryan, G.T.; Wu, K.S.; Farrall, L.; Jia, Y.; Hershey, H.P.; McAdams, S.A.; Faulk, K.N.; Donaldson, G.K.; Tarchini, R.; Valent, B. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2033–2046. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, B.; Zhu, X.; Yang, J.; Bordeos, A.; Wang, G.; Leach, J.E.; Leung, H. Fine-mapping and molecular marker development for Pi56(t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor. Appl. Genet. 2013, 126, 985–998. [Google Scholar] [CrossRef]
- Xu, X.; Hayashi, N.; Wang, C.T.; Fukuoka, S.; Kawasaki, S.; Takatsuji, H.; Jiang, C.J. Rice blast resistance gene Pikahei-1(t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol. Breed. 2014, 34, 691–700. [Google Scholar] [CrossRef]
- Li, C.; Wang, D.; Peng, S.; Chen, Y.; Su, P.; Chen, J.; Zheng, L.; Tan, X.; Liu, J.; Xiao, Y.; et al. Genome-wide association mapping of resistance against rice blast strains in South China and identification of a new Pik allele. Rice 2019, 12, 47. [Google Scholar] [CrossRef]
- Delteil, A.; Gobbato, E.; Cayrol, B.; Estevan, J.; Michel-Romiti, C.; Dievart, A.; Kroj, T.; Morel, J.B. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016, 16, 17. [Google Scholar] [CrossRef]
- Kouzai, Y.; Mochizuki, S.; Nakajima, K.; Desaki, Y.; Hayafune, M.; Miyazaki, H.; Yokotani, N.; Ozawa, K.; Minami, E.; Kaku, H.; et al. Targeted Gene Disruption of Reveals Its Indispensable Role in Chitin Perception and Involvement in the Peptidoglycan Response and Immunity in Rice. Mol. Plant Microbe Interact. 2014, 27, 975–982. [Google Scholar] [CrossRef]
- Kalia, S.; Rathour, R. Current status on mapping of genes for resistance to leaf- and neck-blast disease in rice. 3 Biotech. 2019, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Wu, Y.Y.; Li, A.L. Strategy for Use of Rice Blast Resistance Genes in Rice Molecular Breeding. Rice Sci. 2020, 27, 263–277. [Google Scholar]
- Chen, H.L.; Chen, B.T.; Zhang, D.P.; Xie, Y.F.; Zhang, Q. Pathotypes of Pyricularia grisea in Rice Fields of Central and Southern China. Plant Dis. 2001, 85, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Berruyer, R.; Adreit, H.; Milazzo, J.; Gaillard, S.; Berger, A.; Dioh, W.; Lebrun, M.H.; Tharreau, D. Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 2003, 107, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Chen, D.; Yi, G.H.; Wang, G.L.; Ronald, P.C. Genetic and physical mapping of Pi5(t), a locus associated with broad-spectrum resistance to rice blast. Mol. Genet. Genom. 2003, 269, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Liu, G.; Zhou, B.; Bellizzi, M.; Zeng, L.; Dai, L.; Han, B.; Wang, G.L. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 2006, 172, 1901–1914. [Google Scholar] [CrossRef]
- Zhou, B.; Qu, S.; Liu, G.; Dolan, M.; Sakai, H.; Lu, G.; Bellizzi, M.; Wang, G.L. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 2006, 19, 1216–1228. [Google Scholar] [CrossRef]
- Jeung, J.U.; Kim, B.R.; Cho, Y.C.; Han, S.S.; Moon, H.P.; Lee, Y.T.; Jena, K.K. A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor. Appl. Genet. 2007, 115, 1163–1177. [Google Scholar] [CrossRef]
- Thakur, S.; Singh, P.K.; Das, A.; Rathour, R.; Variar, M.; Prashanthi, S.K.; Singh, A.K.; Singh, U.D.; Chand, D.; Singh, N.K.; et al. Extensive sequence variation in rice blast resistance gene Pi54 makes it broad spectrum in nature. Front. Plant Sci. 2015, 6, 345. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Rebaque, D.; Del Hierro, I.; Lopez, G.; Bacete, L.; Vilaplana, F.; Dallabernardina, P.; Pfrengle, F.; Jorda, L.; Sanchez-Vallet, A.; Perez, R.; et al. Cell wall-derived mixed-linked beta-1,3/1,4-glucans trigger immune responses and disease resistance in plants. Plant J. 2021, 106, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.; Gauthier, A.; Bezier, A.; Poinssot, B.; Joubert, J.M.; Pugin, A.; Heyraud, A.; Baillieul, F. Elicitor and resistance-inducing activities of beta-1,4 cellodextrins in grapevine, comparison with beta-1,3 glucans and alpha-1,4 oligogalacturonides. J. Exp. Bot. 2007, 58, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Casasoli, M.; Spadoni, S.; Lilley, K.S.; Cervone, F.; De Lorenzo, G.; Mattei, B. Identification by 2-D DIGE of apoplastic proteins regulated by oligogalacturonides in Arabidopsis thaliana. Proteomics 2008, 8, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.A.; Li, S.; Lin, A.Z.; Boutrot, F.; Grossmann, G.; Zipfel, C.; Somerville, S.C. Cellulose-Derived Oligomers Act as Damage-Associated Molecular Patterns and Trigger Defense-Like Responses. Plant Physiol. 2017, 173, 2383–2398. [Google Scholar] [CrossRef] [PubMed]
- Denoux, C.; Galletti, R.; Mammarella, N.; Gopalan, S.; Werck, D.; De Lorenzo, G.; Ferrari, S.; Ausubel, F.M.; Dewdney, J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 2008, 1, 423–445. [Google Scholar] [CrossRef]
- Davis, K.R.; Darvill, A.G.; Albersheim, P.; Dell, A. Host-Pathogen Interactions: XXIX. Oligogalacturonides Released from Sodium Polypectate by Endopolygalacturonic Acid Lyase Are Elicitors of Phytoalexins in Soybean. Plant Physiol. 1986, 80, 568–577. [Google Scholar] [CrossRef]
- Simpson, S.D.; Ashford, D.A.; Harvey, D.J.; Bowles, D.J. Short chain oligogalacturonides induce ethylene production and expression of the gene encoding aminocyclopropane 1-carboxylic acid oxidase in tomato plants. Glycobiology 1998, 8, 579–583. [Google Scholar] [CrossRef]
- Galletti, R.; Denoux, C.; Gambetta, S.; Dewdney, J.; Ausubel, F.M.; De Lorenzo, G.; Ferrari, S. The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 2008, 148, 1695–1706. [Google Scholar] [CrossRef]
- Galletti, R.; Ferrari, S.; De Lorenzo, G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 2011, 157, 804–814. [Google Scholar] [CrossRef]
- Fang, N.; Wei, X.; Shen, L.; Yu, Y.; Li, M.; Yin, C.; He, W.; Guan, C.; Chen, H.; Zhang, H.; et al. Fine mapping of a panicle blast resistance gene Pb-bd1 in Japonica landrace Bodao and its application in rice breeding. Rice 2019, 12, 18. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Dong, S.S.; He, W.M.; Ji, J.J.; Zhang, C.; Guo, Y.; Yang, T.L. LDBlockShow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2021, 22, bbaa227. [Google Scholar] [CrossRef]

| Locus | Chr | Position | Top SNP | p-Value | Years | Locus Reference |
|---|---|---|---|---|---|---|
| BRL1 | 3 | 15255983-15434525 | 118389 | 4.86 × 10−5 | 2020 | [30] |
| BRL2 | 4 | 4088528-4505882 | 4088552 | 3.4708 × 10−6 | 2020 | |
| BRL3 | 4 | 32972226-33188952 | 33188952 | 1.626 × 10−5 | 2020 | Pi63 |
| BRL4 | 6 | 1454195-1604484 | 1490561 | 2.461 × 10−6 | 2020 | |
| BRL5 | 9 | 8674158-9304868 | 9069693 | 1.812 × 10−5 | 2020 | pi56 |
| BRL6 | 10 | 4221890-5647528 | 4798719 | 1.5076 × 10−8 | 2020 | |
| BRL7 | 10 | 7667073-8691546 | 7837737 | 6.7119 × 10−9 | 2020 | |
| BRL8 | 11 | 22734774-23104761 | 22905810 | 9.0682 × 10−7 | 2020 | pb1 |
| BRL9 | 11 | 24235297-25570330 | 24926857 | 7.8047 × 10−11 | 2020 | |
| BRL10 | 11 | 28018096-28187112 | 28103725 | 4.9804 × 10−6 | 2020 | |
| BRL11 | 12 | 10653088-10797158 | 10734066 | 9.7844 × 10−6 | 2020 | Pita |
| BRL12 | 12 | 15203722-15771634 | 15673466 | 6.6541 × 10−7 | 2020 | |
| BRL13 | 4 | 4112569-4517761 | 4444275 | 1.648 × 10−6 | 2021 | |
| BRL14 | 4 | 5459995-5745230 | 5590542 | 1.4732 × 10−6 | 2021 | [30] |
| BRL15 | 5 | 7403759-7735177 | 7735177 | 5.0795 × 10−7 | 2021 | [30] |
| BRL16 | 5 | 16077078-16438269 | 16438239 | 3.0808 × 10−8 | 2021 | [25] |
| BRL17 | 6 | 1216276-1514569 | 1490561 | 2.0864 × 10−6 | 2021 | |
| BRL18 | 10 | 4203994-5367946 | 4749066 | 5.9358 × 10−7 | 2021 | |
| BRL19 | 10 | 6200492-9329504 | 6525986 | 1.571 × 10−9 | 2021 | [30] |
| BRL20 | 11 | 8017796-8518194 | 8409165 | 1.5501 × 10−6 | 2021 | [30] |
| BRL21 | 11 | 24202727-24319531 | 24319531 | 1.3067 × 10−7 | 2021 | |
| BRL22 | 11 | 28136278-28187112 | 28186726 | 1.203 × 10−5 | 2021 | |
| BRL23 | 12 | 15535945-16059900 | 15673466 | 1.0786 × 10−7 | 2021 |
| Gene | Annotation | Position | Length |
|---|---|---|---|
| LOC_Os11g46880 | protein kinase domain containing | Chr11:28159898-28165301 | 711 |
| LOC_Os11g46870 | protein kinase | Chr11:28142191-28148337 | 680 |
| LOC_Os11g46900 | wall-associated receptor kinase | Chr11:28174980-28179995 | 707 |
| LOC_Os11g46890 | expressed protein | Chr11:28172388-28172985 | 199 |
| SNP | Ref | Alt | Mutant | Gene | Change |
|---|---|---|---|---|---|
| Chr11-28136278 | G | A | Intergenic | LOC_Os11g46860-LOC_Os11g46870 | - |
| Chr11-28141062 | C | T | Intergenic | LOC_Os11g46860-LOC_Os11g46870 | - |
| Chr11-28141136 | G | A | Intergenic | LOC_Os11g46860-LOC_Os11g46870 | - |
| Chr11-28168626 | G | A | Intergenic | LOC_Os11g46880-LOC_Os11g46890 | - |
| Chr11-28172439 | C | T | Nonsynonymous | LOC_Os11g46890 | Gly183Ser |
| Chr11-28186916 | G | T | Intergenic | LOC_Os11g46900-LOC_Os11g46910 | - |
| Chr11-28187112 | C | A | Intergenic | LOC_Os11g46900-LOC_Os11g46910 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Ma, L.; Pan, X.; Tian, P.; Wang, W.; Liu, K.; Xiong, Z.; Li, C.; Wang, Z.; Wang, J.; et al. Genome-Wide Association Study Identifies Rice Panicle Blast-Resistant Gene Pb4 Encoding a Wall-Associated Kinase. Int. J. Mol. Sci. 2024, 25, 830. https://doi.org/10.3390/ijms25020830
Fan Y, Ma L, Pan X, Tian P, Wang W, Liu K, Xiong Z, Li C, Wang Z, Wang J, et al. Genome-Wide Association Study Identifies Rice Panicle Blast-Resistant Gene Pb4 Encoding a Wall-Associated Kinase. International Journal of Molecular Sciences. 2024; 25(2):830. https://doi.org/10.3390/ijms25020830
Chicago/Turabian StyleFan, Yunxin, Lu Ma, Xiaoqian Pan, Pujiang Tian, Wei Wang, Kunquan Liu, Ziwei Xiong, Changqing Li, Zhixue Wang, Jianfei Wang, and et al. 2024. "Genome-Wide Association Study Identifies Rice Panicle Blast-Resistant Gene Pb4 Encoding a Wall-Associated Kinase" International Journal of Molecular Sciences 25, no. 2: 830. https://doi.org/10.3390/ijms25020830
APA StyleFan, Y., Ma, L., Pan, X., Tian, P., Wang, W., Liu, K., Xiong, Z., Li, C., Wang, Z., Wang, J., Zhang, H., & Bao, Y. (2024). Genome-Wide Association Study Identifies Rice Panicle Blast-Resistant Gene Pb4 Encoding a Wall-Associated Kinase. International Journal of Molecular Sciences, 25(2), 830. https://doi.org/10.3390/ijms25020830






