The Effect of Race/Ethnicity and MED12 Mutation on the Expression of Long Non-Coding RNAs in Uterine Leiomyoma and Myometrium
Abstract
1. Introduction
2. Results
2.1. Differential Expression of Race/Ethnicity- and MED12 Mutation-Associated Long Non-Coding RNA Transcripts in Leiomyoma and Matched Myometrium
2.2. Validation of Race/Ethnicity- and MED12 Mutation-Associated Long Non-Coding RNA transcripts in Leiomyoma and Matched Myometrium
3. Discussion
4. Materials and Methods
4.1. Myometrium and Leiomyoma Tissues Collection
4.2. MED12 Mutation Analysis
4.3. RNA Sequencing and Bioinformatic Analysis
4.4. Quantitative RT-PCR
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Islam, M.S.; Ciavattini, A.; Petraglia, F.; Castellucci, M.; Ciarmela, P. Extracellular matrix in uterine leiomyoma pathogenesis: A potential target for future therapeutics. Hum. Reprod. Update 2018, 24, 59–85. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.K.; Holthouser, K.; Segars, J.H.; Leppert, P.C. Recent scientific advances in leiomyoma (uterine fibroids) research facilitates better understanding and management. F1000Res 2015, 4, 183. [Google Scholar] [CrossRef] [PubMed]
- Chegini, N. Proinflammatory and profibrotic mediators: Principal effectors of leiomyoma development as a fibrotic disorder. Semin. Reprod. Med. 2010, 28, 180–203. [Google Scholar] [CrossRef] [PubMed]
- Segars, J.H.; Parrott, E.C.; Nagel, J.D.; Guo, X.C.; Gao, X.; Birnbaum, L.S.; Pinn, V.W.; Dixon, D. Proceedings from the Third National Institutes of Health International Congress on Advances in Uterine Leiomyoma Research: Comprehensive review, conference summary and future recommendations. Hum. Reprod. Update 2014, 20, 309–333. [Google Scholar] [CrossRef]
- Stewart, E.A.; Cookson, C.L.; Gandolfo, R.A.; Schulze-Rath, R. Epidemiology of uterine fibroids: A systematic review. BJOG 2017, 124, 1501–1512. [Google Scholar] [CrossRef]
- Wise, L.A.; Laughlin-Tommaso, S.K. Epidemiology of Uterine Fibroids: From Menarche to Menopause. Clin. Obstet. Gynecol. 2016, 59, 2–24. [Google Scholar] [CrossRef]
- Marsh, E.E.; Ekpo, G.E.; Cardozo, E.R.; Brocks, M.; Dune, T.; Cohen, L.S. Racial differences in fibroid prevalence and ultrasound findings in asymptomatic young women (18–30 years old): A pilot study. Fertil. Steril. 2013, 99, 1951–1957. [Google Scholar] [CrossRef]
- Yatsenko, S.A.; Mittal, P.; Wood-Trageser, M.A.; Jones, M.W.; Surti, U.; Edwards, R.P.; Sood, A.K.; Rajkovic, A. Highly heterogeneous genomic landscape of uterine leiomyomas by whole exome sequencing and genome-wide arrays. Fertil. Steril. 2017, 107, 457–466.e9. [Google Scholar] [CrossRef]
- Gallagher, C.S.; Morton, C.C. Genetic Association Studies in Uterine Fibroids: Risk Alleles Presage the Path to Personalized Therapies. Semin. Reprod. Med. 2016, 34, 235–241. [Google Scholar] [CrossRef]
- Liegl-Atzwanger, B.; Heitzer, E.; Flicker, K.; Muller, S.; Ulz, P.; Saglam, O.; Tavassoli, F.; Devouassoux-Shisheboran, M.; Geigl, J.; Moinfar, F. Exploring chromosomal abnormalities and genetic changes in uterine smooth muscle tumors. Mod. Pathol. 2016, 29, 1262–1277. [Google Scholar] [CrossRef]
- Makinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Markowski, D.N.; Bartnitzke, S.; Löning, T.; Drieschner, N.; Helmke, B.M.; Bullerdiek, J. MED12 mutations in uterine fibroids--their relationship to cytogenetic subgroups. Int. J. Cancer 2012, 131, 1528–1536. [Google Scholar] [CrossRef]
- Al-Hendy, A.; Laknaur, A.; Diamond, M.P.; Ismail, N.; Boyer, T.G.; Halder, S.K. Silencing Med12 Gene Reduces Proliferation of Human Leiomyoma Cells Mediated via Wnt/beta-Catenin Signaling Pathway. Endocrinology 2017, 158, 592–603. [Google Scholar] [PubMed]
- El Andaloussi, A.; Al-Hendy, A.; Ismail, N.; Boyer, T.G.; Halder, S.K. Introduction of Somatic Mutation in MED12 Induces Wnt4/β-Catenin and Disrupts Autophagy in Human Uterine Myometrial Cell. Reprod. Sci. 2020, 27, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Verger, A.; Monté, D.; Villeret, V. Twenty years of Mediator complex structural studies. Biochem. Soc. Trans. 2019, 47, 399–410. [Google Scholar] [CrossRef]
- Zhang, S.; O’Regan, R.; Xu, W. The emerging role of mediator complex subunit 12 in tumorigenesis and response to chemotherapeutics. Cancer 2020, 126, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Kim, S.; Ishii, S.; Boyer, T.G. Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol. Cell. Biol. 2006, 26, 8667–8682. [Google Scholar] [CrossRef]
- Huang, S.; Holzel, M.; Knijnenburg, T.; Schlicker, A.; Roepman, P.; McDermott, U.; Garnett, M.; Grernrum, W.; Sun, C.; Prahallad, A.; et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell 2012, 151, 937–950. [Google Scholar] [CrossRef]
- Chuang, T.D.; Ton, N.; Rysling, S.; Quintanilla, D.; Boos, D.; Gao, J.; McSwiggin, H.; Yan, W.; Khorram, O. The Influence of Race/Ethnicity on the Transcriptomic Landscape of Uterine Fibroids. Int. J. Mol. Sci. 2023, 24, 13441. [Google Scholar] [CrossRef]
- Chuang, T.D.; Gao, J.; Quintanilla, D.; McSwiggin, H.; Boos, D.; Yan, W.; Khorram, O. Differential Expression of MED12-Associated Coding RNA Transcripts in Uterine Leiomyomas. Int. J. Mol. Sci. 2023, 24, 3742. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Li, H.; Yu, Q.; Xiao, W.; Wang, D.O. LncRNA-mediated DNA methylation: An emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. 2022, 41, 100. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Khorram, O. Expression Profiling of lncRNAs, miRNAs, and mRNAs and Their Differential Expression in Leiomyoma Using Next-Generation RNA Sequencing. Reprod. Sci. 2018, 25, 246–255. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, X.; Dong, R.; Liu, X.; Li, Y.; Lu, S.; Xu, L.; Wang, Y.; Wang, X.; Hou, D.; et al. Integrated analysis of long noncoding RNAs and mRNAs reveals their potential roles in the pathogenesis of uterine leiomyomas. Oncotarget 2014, 5, 8625–8636. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Xie, Y.; Yan, W.; Khorram, O. Next-generation sequencing reveals differentially expressed small noncoding RNAs in uterine leiomyoma. Fertil. Steril. 2018, 109, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Rehan, A.; Khorram, O. Functional role of the long noncoding RNA X-inactive specific transcript in leiomyoma pathogenesis. Fertil. Steril. 2021, 115, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Long Noncoding RNA MIAT Modulates the Extracellular Matrix Deposition in Leiomyomas by Sponging MiR-29 Family. Endocrinology 2021, 162, bqab186. [Google Scholar] [CrossRef]
- Falahati, Z.; Mohseni-Dargah, M.; Mirfakhraie, R. Emerging Roles of Long Non-coding RNAs in Uterine Leiomyoma Pathogenesis: A Review. Reprod. Sci. 2022, 29, 1086–1101. [Google Scholar] [CrossRef]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Differential Expression of Super-Enhancer-Associated Long Non-coding RNAs in Uterine Leiomyomas. Reprod. Sci. 2022, 29, 2960–2976. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Dey, B.K.; Mueller, A.C.; Dutta, A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription 2014, 5, e944014. [Google Scholar] [CrossRef] [PubMed]
- Ergun, S.; Oztuzcu, S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumor Biol. 2015, 36, 3129–3136. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Bradner, J.E.; Hnisz, D.; Young, R.A. Transcriptional Addiction in Cancer. Cell 2017, 168, 629–643. [Google Scholar] [CrossRef] [PubMed]
- Soibam, B. Super-lncRNAs: Identification of lncRNAs that target super-enhancers via RNA:DNA:DNA triplex formation. RNA 2017, 23, 1729–1742. [Google Scholar] [CrossRef]
- Kiss, T.; Fayet-Lebaron, E.; Jady, B.E. Box H/ACA small ribonucleoproteins. Mol. Cell 2010, 37, 597–606. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Yang, J.H.; Li, J.H.; Shao, P.; Zhou, H.; Chen, Y.Q.; Qu, L.H. starBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011, 39, D202–D209. [Google Scholar] [CrossRef]
- Aissani, B.; Zhang, K.; Wiener, H. Follow-up to genome-wide linkage and admixture mapping studies implicates components of the extracellular matrix in susceptibility to and size of uterine fibroids. Fertil. Steril. 2015, 103, 528–534.e13. [Google Scholar] [CrossRef]
- Chiang, S. Recent advances in smooth muscle tumors with PGR and PLAG1 gene fusions and myofibroblastic uterine neoplasms. Genes Chromosomes Cancer 2021, 60, 138–146. [Google Scholar] [CrossRef]
- Conconi, D.; Redaelli, S.; Lissoni, A.A.; Cilibrasi, C.; Perego, P.; Gautiero, E.; Sala, E.; Paderno, M.; Dalprà, L.; Landoni, F.; et al. Genomic and Epigenomic Profile of Uterine Smooth Muscle Tumors of Uncertain Malignant Potential (STUMPs) Revealed Similarities and Differences with Leiomyomas and Leiomyosarcomas. Int. J. Mol. Sci. 2021, 22, 1580. [Google Scholar] [CrossRef] [PubMed]
- Kerins, M.J.; Milligan, J.; Wohlschlegel, J.A.; Ooi, A. Fumarate hydratase inactivation in hereditary leiomyomatosis and renal cell cancer is synthetic lethal with ferroptosis induction. Cancer Sci. 2018, 109, 2757–2766. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Khorram, O. miR-200c Regulates IL8 Expression by Targeting IKBKB: A Potential Mediator of Inflammation in Leiomyoma Pathogenesis. PLoS ONE 2014, 9, e95370. [Google Scholar] [CrossRef] [PubMed]
- Paul, E.N.; Grey, J.A.; Carpenter, T.J.; Madaj, Z.B.; Lau, K.H.; Givan, S.A.; Burns, G.W.; Chandler, R.L.; Wegienka, G.R.; Shen, H.; et al. Transcriptome and DNA methylome analyses reveal underlying mechanisms for the racial disparity in uterine fibroids. JCI Insight 2022, 7, e160274. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell. Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Jiang, Y.; Wang, Z.; Zhang, N.; Al-Hendy, A.; Mamillapalli, R.; Kallen, A.N.; Kodaman, P.; Taylor, H.S.; Li, D.; et al. H19 lncRNA identified as a master regulator of genes that drive uterine leiomyomas. Oncogene 2019, 38, 5356–5366. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, X.; Wang, F.; Zhang, N.; Zhang, Y.; Ao, Z.; He, F. Long noncoding RNA TPTEP1 suppresses diabetic retinopathy by reducing oxidative stress and targeting the miR-489-3p/NRF2 axis. Acta Biochim. Pol. 2023, 70, 45–50. [Google Scholar] [CrossRef]
- Tang, T.; Wang, L.X.; Yang, M.L.; Zhang, R.M. lncRNA TPTEP1 inhibits stemness and radioresistance of glioma through miR-106a-5p-mediated P38 MAPK signaling. Mol. Med. Rep. 2020, 22, 4857–4867. [Google Scholar] [CrossRef]
- Sun, X.; Qiu, X. LncRNA TPTEP1 inhibited the proliferation and metastasis of non-small cell lung cancer cells by targeting miR-761/LATS2 axis. Am. J. Transl. Res. 2021, 13, 8653–8669. [Google Scholar]
- Dong, Y.; Wang, Q.; Sun, J.; Liu, H.; Wang, H. Long non-coding RNA TPTEP1 exerts inhibitory effects on hepatocellular carcinoma by impairing microRNA-454-3p-mediated DLG5 downregulation. Dig. Liver Dis. 2022, 54, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Zhuang, H.W.; Wang, J.; Shen, Y.; Bu, Y.Z.; Guan, B.G.; Xu, F.; Dou, J. Long noncoding RNA CA3-AS1 suppresses gastric cancer migration and invasion by sponging miR-93-5p and targeting BTG3. Gene Ther. 2022, 29, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yang, Z.; Lin, B. Overexpression of long non coding RNA CA3-AS1 suppresses proliferation, invasion and promotes apoptosis via miRNA-93/PTEN axis in colorectal cancer. Gene 2019, 687, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Shuai, Y.; Wang, H.; Li, H.; Li, Y. Exosome-mediated long noncoding RNA (lncRNA) PART1 suppresses malignant progression of oral squamous cell carcinoma via miR-17-5p/SOCS6 axis. Turk. J. Med. Sci. 2023, 53, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Song, X.; Yu, H.; Wang, J.; Huang, L.; Zhou, Y.; He, X. Mechanism of Exosomal LncRNA PART1 in Esophageal Cancer Angiogenesis by Targeting miR-302a-3p/CDC25A Axis. Technol. Cancer Res. Treat. 2023, 22, 15330338231184327. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Qiao, L.; Fan, H.; Liao, C.; Zheng, J.; Wang, W.; Ma, X.; Yang, M.; Sun, X.; Zhao, W. Long non-coding RNA MSC-AS1 facilitates the proliferation and glycolysis of gastric cancer cells by regulating PFKFB3 expression. Int. J. Med. Sci. 2021, 18, 546–554. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wang, X.; Du, M.; Dong, Y. LncRNA MSC-AS1 Promotes Colorectal Cancer Progression by Regulating miR-325/TRIM14 Axis. J. Oncol. 2021, 2021, 9954214. [Google Scholar] [CrossRef]
- Cao, C.; Zhong, Q.; Lu, L.; Huang, B.; Li, J.; Meng, L.; Wei, H. Long noncoding RNA MSC-AS1 promotes hepatocellular carcinoma oncogenesis via inducing the expression of phosphoglycerate kinase 1. Cancer Med. 2020, 9, 5174–5184. [Google Scholar] [CrossRef]
- Kou, X.; Zhu, J.; Xie, X.; Hao, M.; Zhao, Y. Expression of lncRNA MSC-AS1 in hepatocellular carcinoma cell lines and its effect on proliferation, apoptosis, and migration. Turk. J. Gastroenterol. 2020, 31, 860–867. [Google Scholar] [CrossRef]
- Tian, T.; Luo, B.; Shen, G.; Ji, G. LncRNA MSC-AS1, as an oncogene in melanoma, promotes the proliferation and glutaminolysis by regulating the miR-330-3p/YAP1 axis. Anti-Cancer. Drugs 2022, 33, 1012–1023. [Google Scholar] [CrossRef]
- Zhao, Y.; Yuan, D.; Zhu, D.; Xu, T.; Huang, A.; Jiang, L.; Liu, C.; Qian, H.; Bu, X. LncRNA-MSC-AS1 inhibits the ovarian cancer progression by targeting miR-425-5p. J. Ovarian Res. 2021, 14, 109. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, P.; Yang, W.; Shan, Y.; Zhang, Q.; Wu, H. The role of lncRNA MSC-AS1/miR-29b-3p axis-mediated CDK14 modulation in pancreatic cancer proliferation and Gemcitabine-induced apoptosis. Cancer Biol. Ther. 2019, 20, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Khorram, O. Mechanisms underlying aberrant expression of miR-29c in uterine leiomyoma. Fertil. Steril. 2016, 105, 236–245.e1. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.-D.; Khorram, O. Cross-talk between miR-29c and transforming growth factor-β3 is mediated by an epigenetic mechanism in leiomyoma. Fertil. Steril. 2019, 112, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Khorram, O. Regulation of Cell Cycle Regulatory Proteins by MicroRNAs in Uterine Leiomyoma. Reprod. Sci. 2019, 26, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Shoorei, H.; Hussen, B.M.; Abdullah, S.R.; Poornajaf, Y.; Taheri, M.; Samsami, M. LncRNA SNHG12: A budding star in human diseases. Pathol. Res. Pract. 2023, 251, 154897. [Google Scholar] [CrossRef] [PubMed]
- Toprani, S.M.; Kelkar Mane, V. Role of DNA damage and repair mechanisms in uterine fibroid/leiomyomas: A review. Biol. Reprod. 2021, 104, 58–70. [Google Scholar] [CrossRef]
- Wang, H.; Yu, S.; Peng, H.; Shu, Y.; Zhang, W.; Zhu, Q.; Wu, Y.; Xu, Y.; Yan, J.; Xiang, H. Long noncoding RNA Linc00337 functions as an E2F1 co-activator and promotes cell proliferation in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 216. [Google Scholar] [CrossRef]
- Hu, B.; Wang, X.; Li, L. Long noncoding RNA LINC00337 promote gastric cancer proliferation through repressing p21 mediated by EZH2. Am. J. Transl. Res. 2019, 11, 3238–3245. [Google Scholar]
- Zhang, R.N.; Wu, D.M.; Wu, L.P.; Gao, G.W. LncRNA LINC00337 sponges mir-1285-3p to promote proliferation and metastasis of lung adenocarcinoma cells by upregulating YTHDF1. Cancer Cell Int. 2021, 21, 550. [Google Scholar] [CrossRef]
- Xu, X.; Nie, J.; Lu, L.; Du, C.; Meng, F.; Song, D. LINC00337 promotes tumor angiogenesis in colorectal cancer by recruiting DNMT1, which suppresses the expression of CNN1. Cancer Gene Ther. 2021, 28, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gong, J.; Lu, J.; Chen, J.; Zhou, Y.; Li, T.; Ding, L. Long noncoding RNA LINC00337 accelerates the non-small-cell lung cancer progression through inhibiting TIMP2 by recruiting DNMT1. Am. J. Transl. Res. 2019, 11, 6075–6083. [Google Scholar] [PubMed]
- Chuang, T.D.; Khorram, O. Tranilast Inhibits Genes Functionally Involved in Cell Proliferation, Fibrosis, and Epigenetic Regulation and Epigenetically Induces miR-29c Expression in Leiomyoma Cells. Reprod. Sci. 2017, 24, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhang, X.; Fang, K.; Guo, Z.; Li, L. LINC00536 Promotes Breast Cancer Progression by Regulating ROCK1 via Sponging of miR-214-5p. Biochem. Genet. 2023, 61, 1163–1184. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.F.; Wang, W.; Wang, L.; Tan, J.D. LINC00536 knockdown inhibits breast cancer cells proliferation, invasion, and migration through regulation of the miR-4282/centromere protein F axis. Kaohsiung J. Med. Sci. 2022, 38, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.G.; Song, H.Y.; Zhang, K.P.; Liu, L.M.; Jiang, Y.T.; Liu, S. LINC00536 promotes hepatocellular carcinoma progression via the miR-203b-5p/VEGFA axis. Neoplasma 2022, 69, 136–144. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Qin, Z.; Wei, Y.; Deng, Z.; Zhu, C.; Tang, J.; Ma, L. High LINC00536 expression promotes tumor progression and poor prognosis in bladder cancer. Exp. Cell Res. 2019, 378, 32–40. [Google Scholar] [CrossRef]
- Xu, Y.; Dong, M.; Wang, J.; Zhao, W.; Jiao, M. LINC01436 Inhibited miR-585-3p Expression and Upregulated MAPK1 Expression to Promote Gastric Cancer Progression. Dig. Dis. Sci. 2021, 66, 1885–1894. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, G.; He, X.; Chen, S.; Zhang, F.; Fang, X. LINC01436, regulating miR-585 and FBXO11, is an oncogenic lncRNA in the progression of gastric cancer. Cell Biol. Int. 2020, 44, 882–893. [Google Scholar] [CrossRef]
- Mahboobeh, Z.; Pegah, M.; Fatemeh, S.; Elham, K.; Hanieh, A.; Milad, R.; Mohammad, S. lncRNA ZEB2-AS1: A promising biomarker in human cancers. IUBMB Life 2020, 72, 1891–1899. [Google Scholar] [CrossRef]
- Cheng, Z.; Liu, L.; Li, Q. lncRNA ZEB2-AS1 stimulates cardiac hypertrophy by downregulating PTEN. Exp. Ther. Med. 2020, 20, 92. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Yang, X.; Zhang, D.; Luo, J.; Chen, R. Long noncoding RNA LINC01186, regulated by TGF-β/SMAD3, inhibits migration and invasion through Epithelial-Mesenchymal-Transition in lung cancer. Gene 2017, 608, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Duan, H.; Zhang, C.; Zhou, Y.; Gao, R. The LINC01186 suppresses cell proliferation and invasion ability in papillary thyroid carcinoma. Oncol. Lett. 2018, 16, 5639–5644. [Google Scholar] [CrossRef] [PubMed]
- Mittal, P.; Shin, Y.H.; Yatsenko, S.A.; Castro, C.A.; Surti, U.; Rajkovic, A. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J. Clin. Investig. 2015, 125, 3280–3284. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, H.R.; Pasanen, A.; Heikinheimo, O.; Tanskanen, T.; Palin, K.; Tolvanen, J.; Vahteristo, P.; Sjoberg, J.; Pitkanen, E.; Butzow, R.; et al. Multiple clinical characteristics separate MED12-mutation-positive and -negative uterine leiomyomas. Sci. Rep. 2017, 7, 1015. [Google Scholar] [CrossRef] [PubMed]
- Turunen, M.; Spaeth, J.M.; Keskitalo, S.; Park, M.J.; Kivioja, T.; Clark, A.D.; Makinen, N.; Gao, F.; Palin, K.; Nurkkala, H.; et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 2014, 7, 654–660. [Google Scholar] [CrossRef]
- Park, M.J.; Shen, H.; Spaeth, J.M.; Tolvanen, J.H.; Failor, C.; Knudtson, J.F.; McLaughlin, J.; Halder, S.K.; Yang, Q.; Bulun, S.E.; et al. Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J. Biol. Chem. 2018, 293, 4870–4882. [Google Scholar] [CrossRef]
- Bray, M.J.; Wellons, M.F.; Jones, S.H.; Torstenson, E.S.; Edwards, T.L.; Velez Edwards, D.R. Transethnic and race-stratified genome-wide association study of fibroid characteristics in African American and European American women. Fertil. Steril. 2018, 110, 737–745.e34. [Google Scholar] [CrossRef]
- Commandeur, A.E.; Styer, A.K.; Teixeira, J.M. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum. Reprod. Update 2015, 21, 593–615. [Google Scholar] [CrossRef]
- Catherino, W.H.; Eltoukhi, H.M.; Al-Hendy, A. Racial and ethnic differences in the pathogenesis and clinical manifestations of uterine leiomyoma. Semin. Reprod. Med. 2013, 31, 370–379. [Google Scholar] [CrossRef]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Tryptophan catabolism is dysregulated in leiomyomas. Fertil. Steril. 2021, 116, 1160–1171. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Further characterization of tryptophan metabolism and its dysregulation in fibroids. F S Sci. 2022, 3, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Panda, H.; Luo, X.; Chegini, N. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr. Relat. Cancer 2012, 19, 541–556. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, X.; Obijuru, L.; Laser, J.; Aris, V.; Lee, P.; Mittal, K.; Soteropoulos, P.; Wei, J.J. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer 2007, 46, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ton, N.; Rysling, S.; Quintanilla, D.; Boos, D.; Khorram, O. Therapeutic Effects of In Vivo Administration of An Inhibitor of Tryptophan 2,3-dioxygenase (680C91) for the Treatment of Fibroids: A Preclinical Study. Fertil. Steril. 2023; online ahead of print. [Google Scholar]
- Chuang, T.D.; Munoz, L.; Quintanilla, D.; Boos, D.; Khorram, O. Therapeutic Effects of Long-Term Administration of Tranilast in an Animal Model for the Treatment of Fibroids. Int. J. Mol. Sci. 2023, 24, 10465. [Google Scholar] [CrossRef]
- Iqbal, A.; Duitama, C.; Metge, F.; Rosskopp, D.; Boucas, J. Flaski, Flaski (3.12.2); Zenodo: Geneva, Switzerland, 2021. [Google Scholar]
- Li, Z.; Zhang, Y.; Fang, J.; Xu, Z.; Zhang, H.; Mao, M.; Chen, Y.; Zhang, L.; Pian, C. NcPath: A novel platform for visualization and enrichment analysis of human non-coding RNA and KEGG signaling pathways. Bioinformatics 2023, 39, btac812. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Almeida, T.A.; Quispe-Ricalde, A.; Montes de Oca, F.; Foronda, P.; Hernandez, M.M. A high-throughput open-array qPCR gene panel to identify housekeeping genes suitable for myometrium and leiomyoma expression analysis. Gynecol. Oncol. 2014, 134, 138–143. [Google Scholar] [CrossRef]
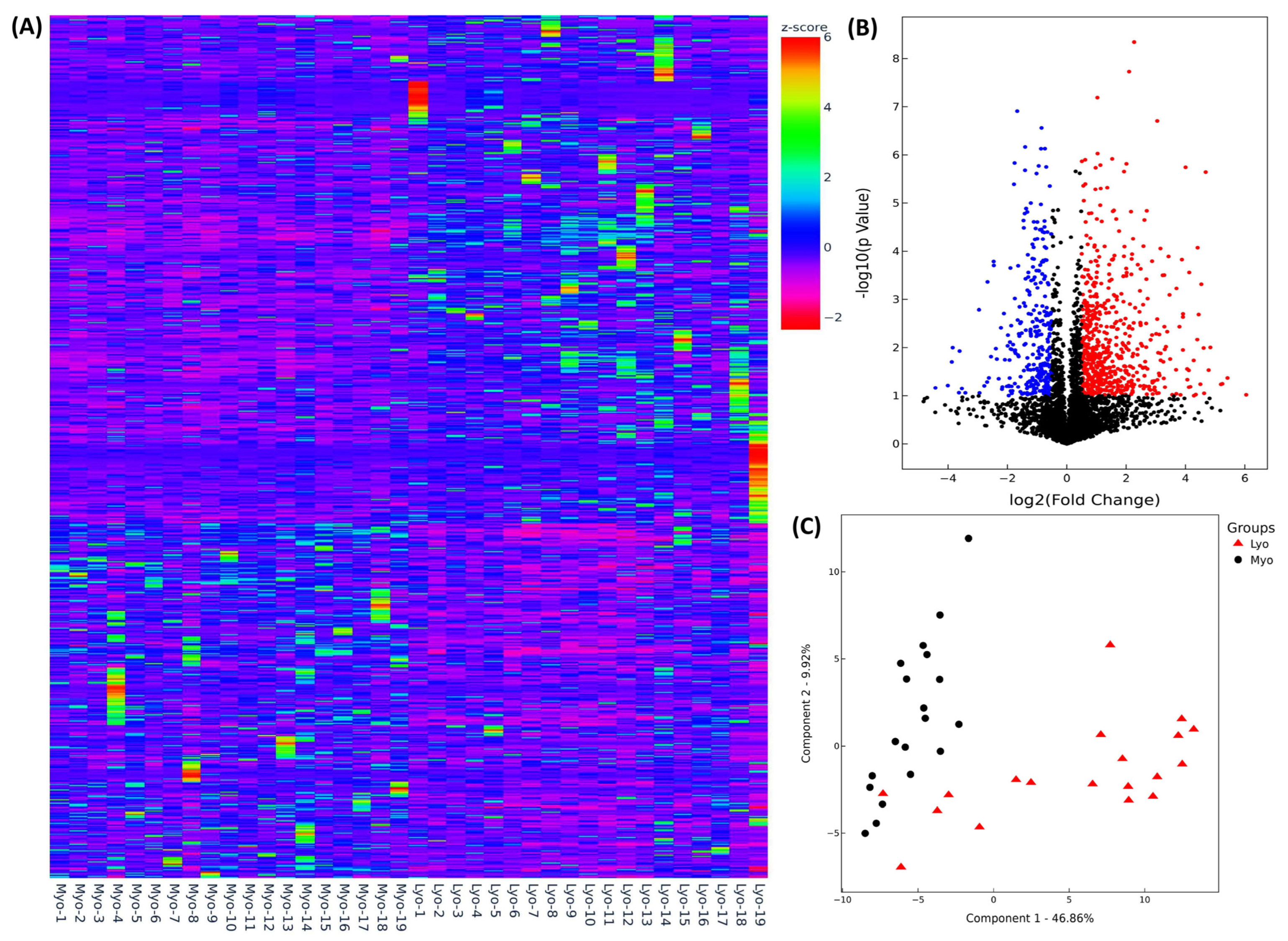
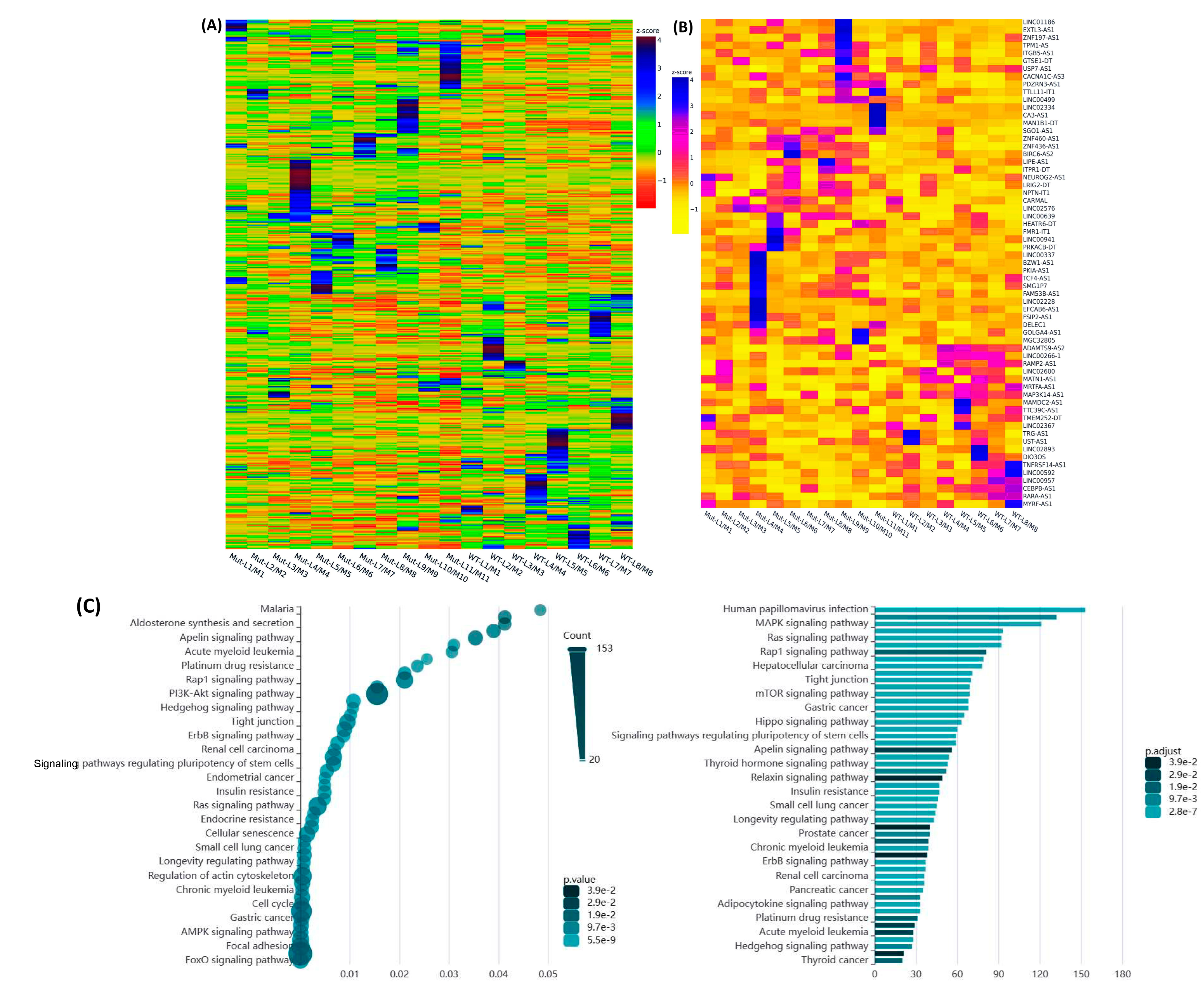
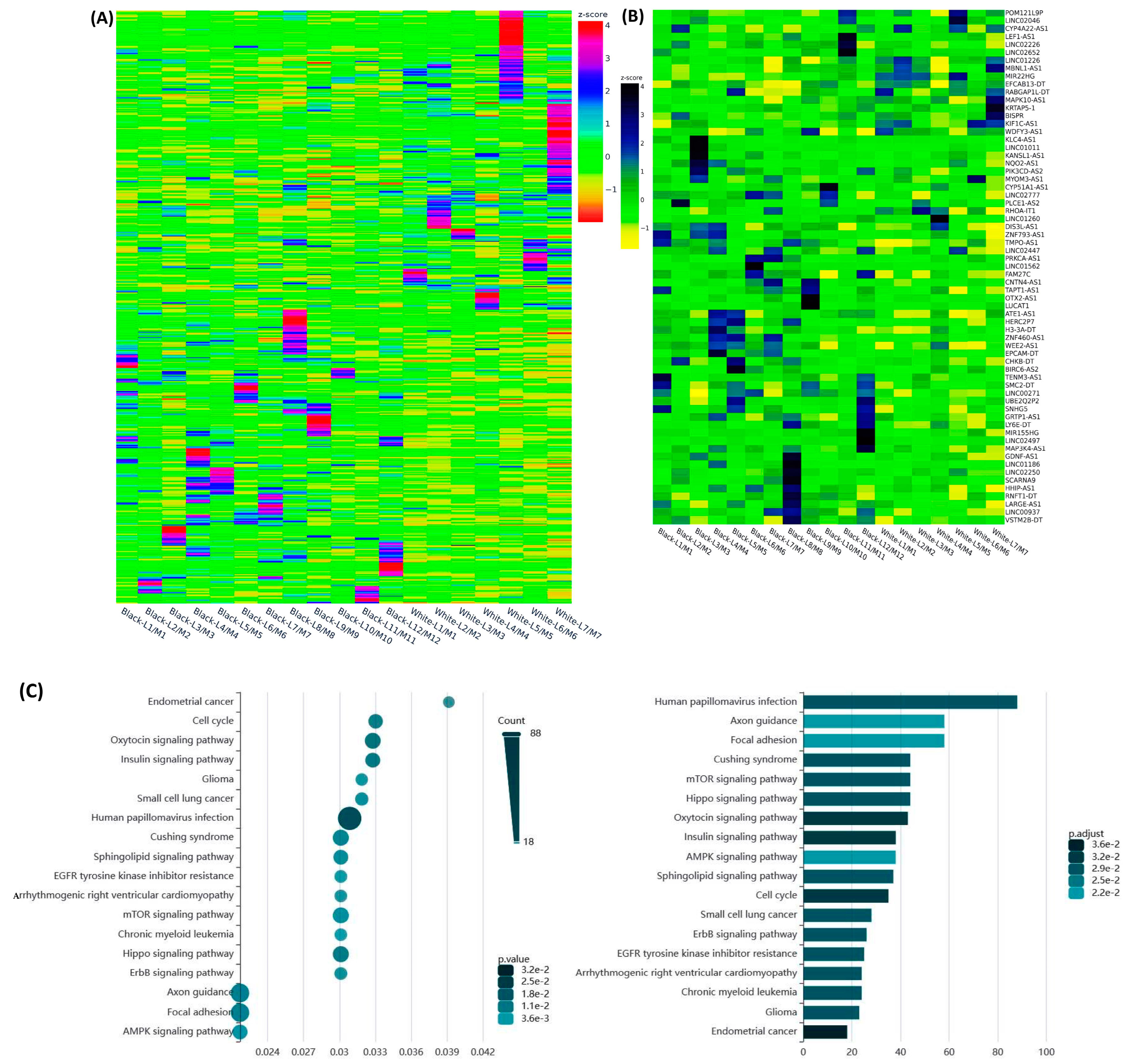
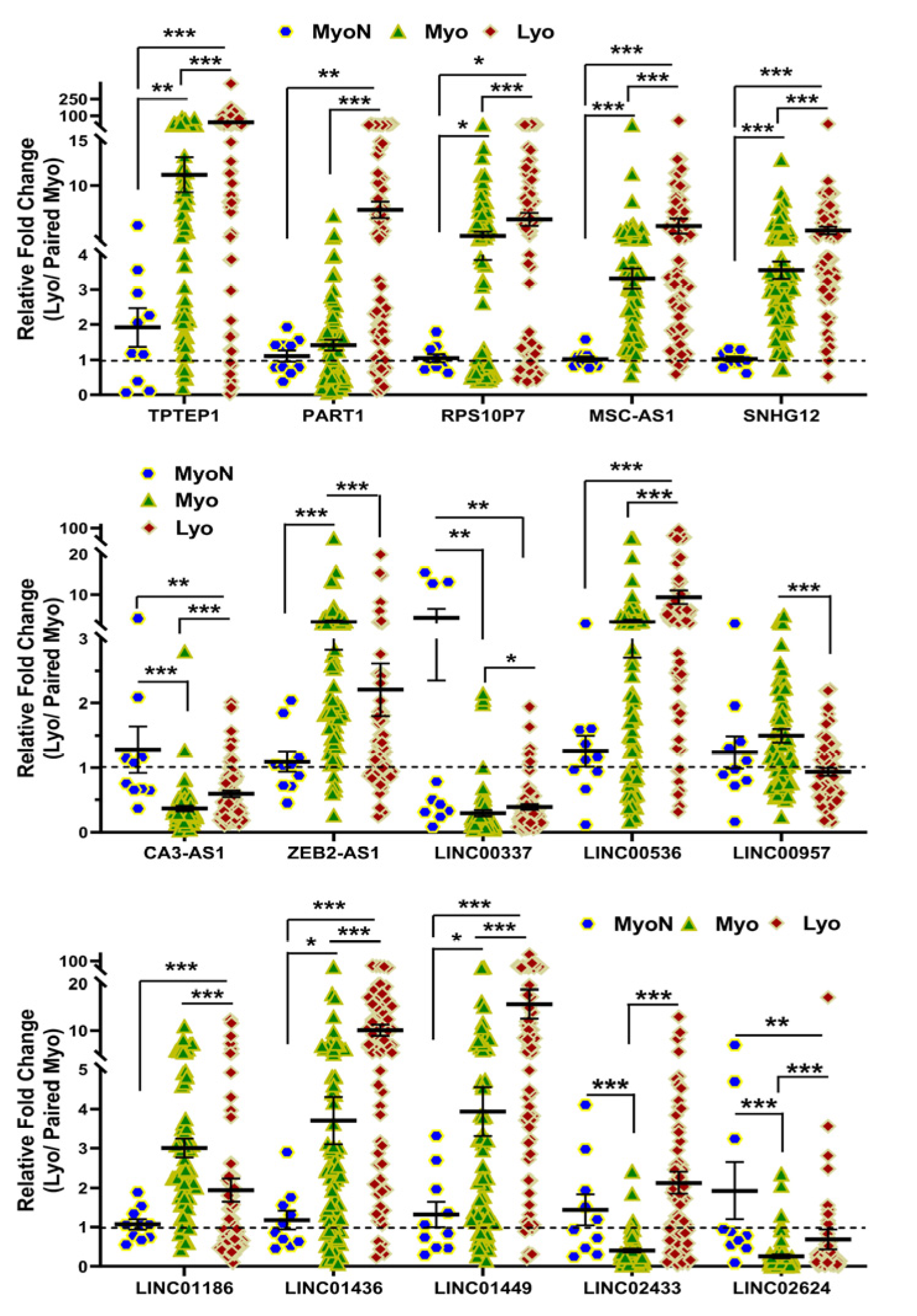
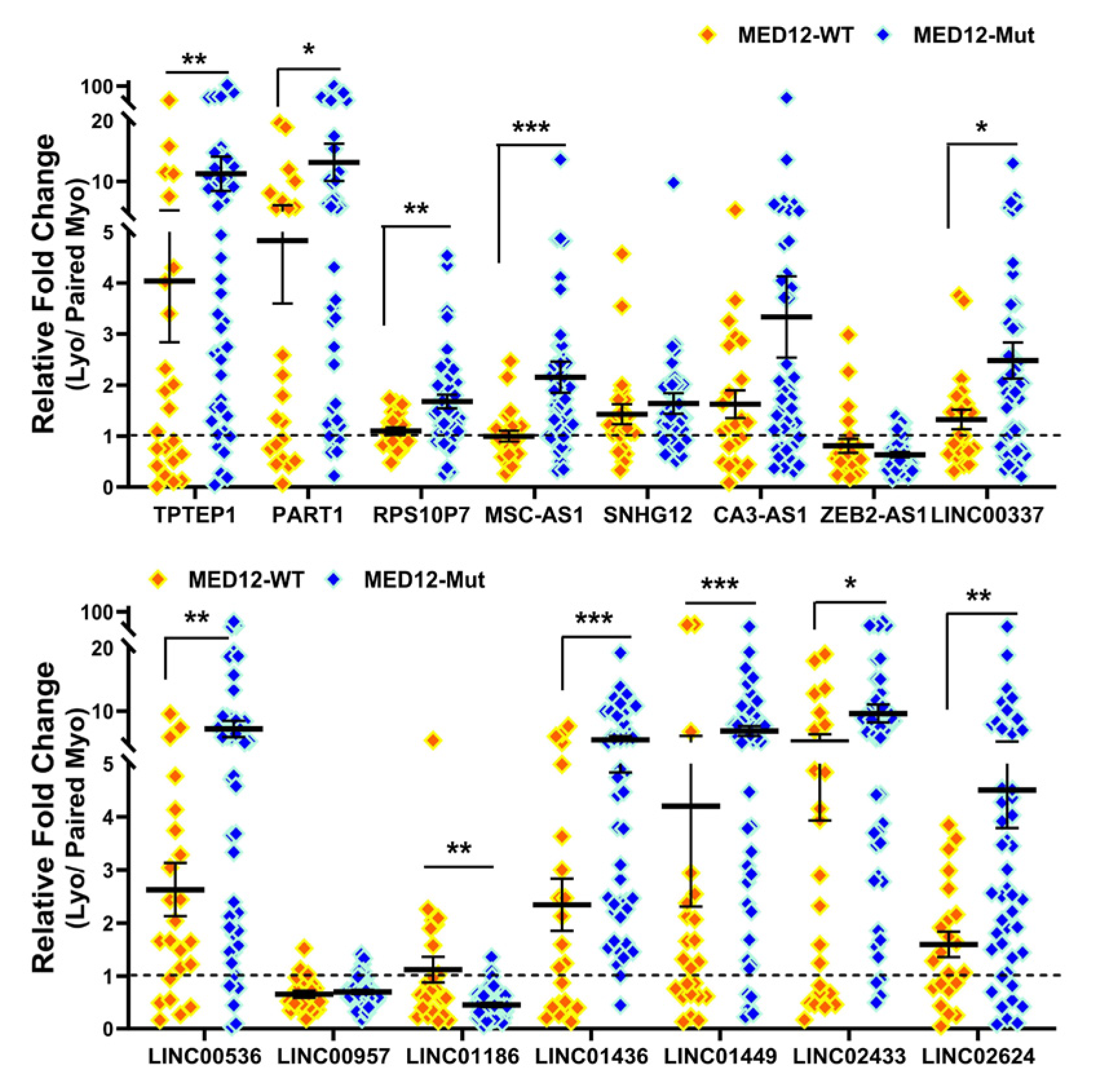
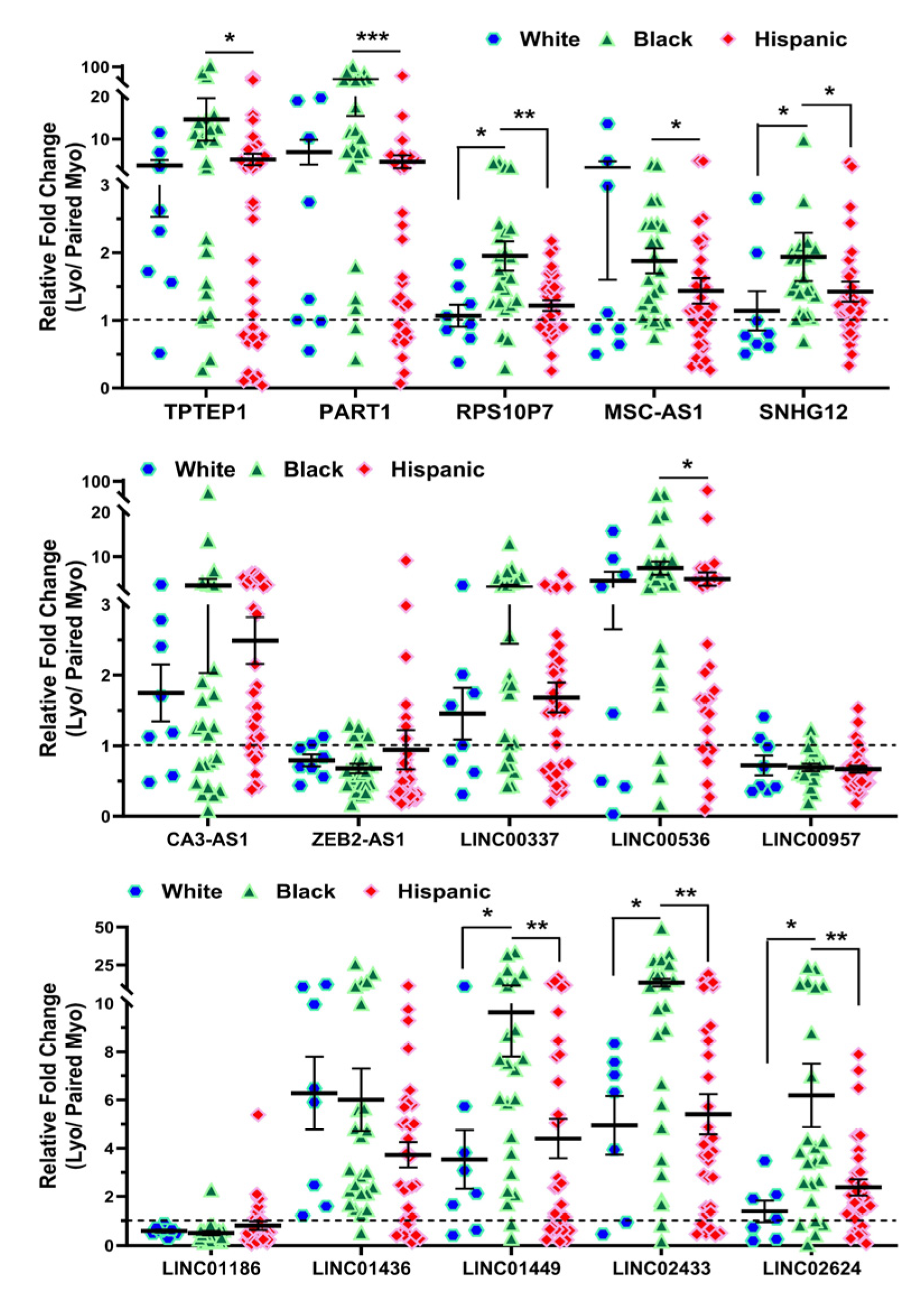
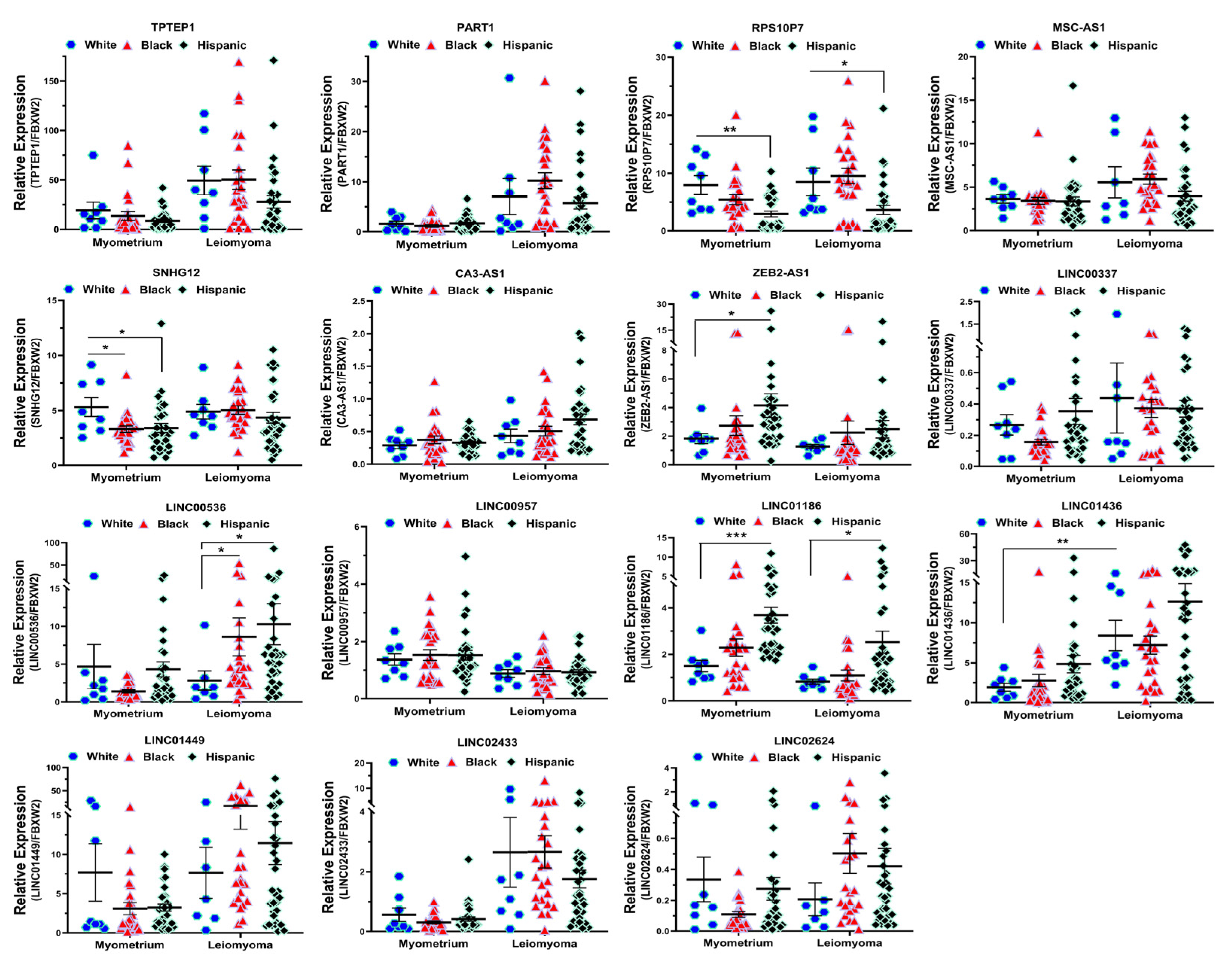
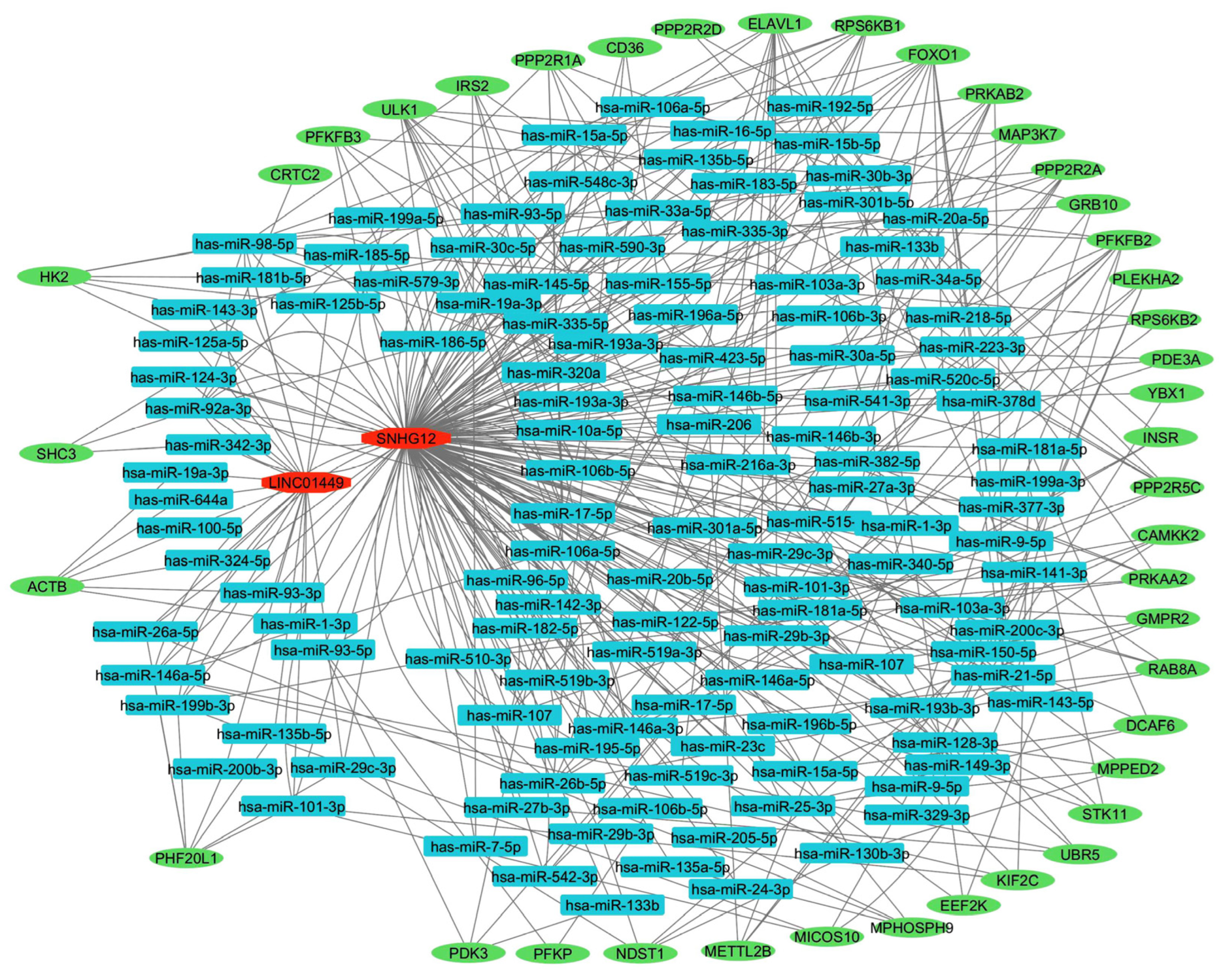
| Symbol | Lyo vs. Myo | MED12-Mut(Lyo/Myo) vs. MED12-WT(Lyo/Myo) | Black(Lyo/Myo) vs. White(Lyo/Myo) | Black(Myo) vs. White(Myo) | Black(Lyo) vs. White(Lyo) |
|---|---|---|---|---|---|
| TPTEP1 | Up (p < 0.001) | Up (p < 0.01) | No Significance | No Significance | No Significance |
| PART1 | Up (p < 0.001) | Up (p < 0.05) | No Significance | No Significance | No Significance |
| RPS10P7 | Up (p < 0.001) | Up (p < 0.01) | Up (p < 0.05) | No Significance | No Significance |
| MSC-AS1 | Up (p < 0.001) | Up (p < 0.001) | No Significance | No Significance | No Significance |
| SNHG12 | Up (p < 0.001) | No Significance | Up (p < 0.05) | Down (p < 0.05) | No Significance |
| CA3-AS1 | Up (p < 0.001) | No Significance | No Significance | No Significance | No Significance |
| ZEB2-AS1 | Down (p < 0.001) | No Significance | No Significance | No Significance | No Significance |
| LINC00337 | Up (p < 0.05) | Up (p < 0.05) | No Significance | No Significance | No Significance |
| LINC00536 | Up (p < 0.001) | Up (p < 0.01) | No Significance | No Significance | Up (p < 0.05) |
| LINC00957 | Down (p < 0.001) | No Significance | No Significance | No Significance | No Significance |
| LINC01186 | Down (p < 0.001) | Down (p < 0.01) | No Significance | No Significance | No Significance |
| LINC01436 | Up (p < 0.001) | Up (p < 0.001) | No Significance | No Significance | No Significance |
| LINC01449 | Up (p < 0.001) | Up (p < 0.001) | Up (p < 0.05) | No Significance | No Significance |
| LINC02433 | Up (p < 0.001) | Up (p < 0.05) | Up (p < 0.05) | No Significance | No Significance |
| LINC02624 | Up (p < 0.01) | Up (p < 0.01) | Up (p < 0.05) | No Significance | No Significance |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, T.-D.; Ton, N.; Rysling, S.; Boos, D.; Khorram, O. The Effect of Race/Ethnicity and MED12 Mutation on the Expression of Long Non-Coding RNAs in Uterine Leiomyoma and Myometrium. Int. J. Mol. Sci. 2024, 25, 1307. https://doi.org/10.3390/ijms25021307
Chuang T-D, Ton N, Rysling S, Boos D, Khorram O. The Effect of Race/Ethnicity and MED12 Mutation on the Expression of Long Non-Coding RNAs in Uterine Leiomyoma and Myometrium. International Journal of Molecular Sciences. 2024; 25(2):1307. https://doi.org/10.3390/ijms25021307
Chicago/Turabian StyleChuang, Tsai-Der, Nhu Ton, Shawn Rysling, Drake Boos, and Omid Khorram. 2024. "The Effect of Race/Ethnicity and MED12 Mutation on the Expression of Long Non-Coding RNAs in Uterine Leiomyoma and Myometrium" International Journal of Molecular Sciences 25, no. 2: 1307. https://doi.org/10.3390/ijms25021307
APA StyleChuang, T.-D., Ton, N., Rysling, S., Boos, D., & Khorram, O. (2024). The Effect of Race/Ethnicity and MED12 Mutation on the Expression of Long Non-Coding RNAs in Uterine Leiomyoma and Myometrium. International Journal of Molecular Sciences, 25(2), 1307. https://doi.org/10.3390/ijms25021307






