Generation and Application of Monoclonal Antibodies against Porcine S100A8, S100A9, and S100A12 Proteins Using Hybridoma Technology
Abstract
1. Introduction
2. Results
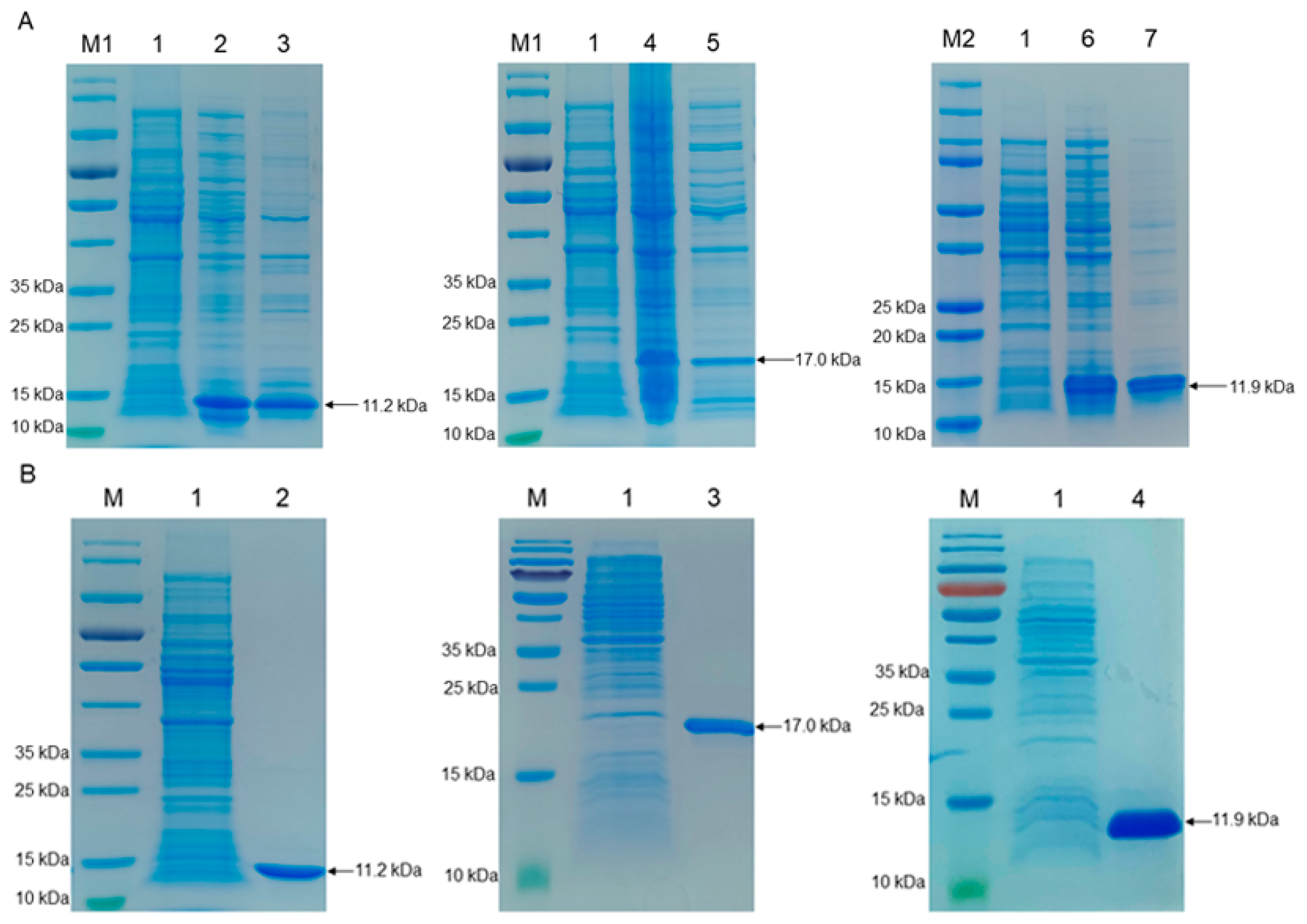
2.1. Expression and Purification of the Recombinant S100A8, S100A9, and S100A12 Proteins
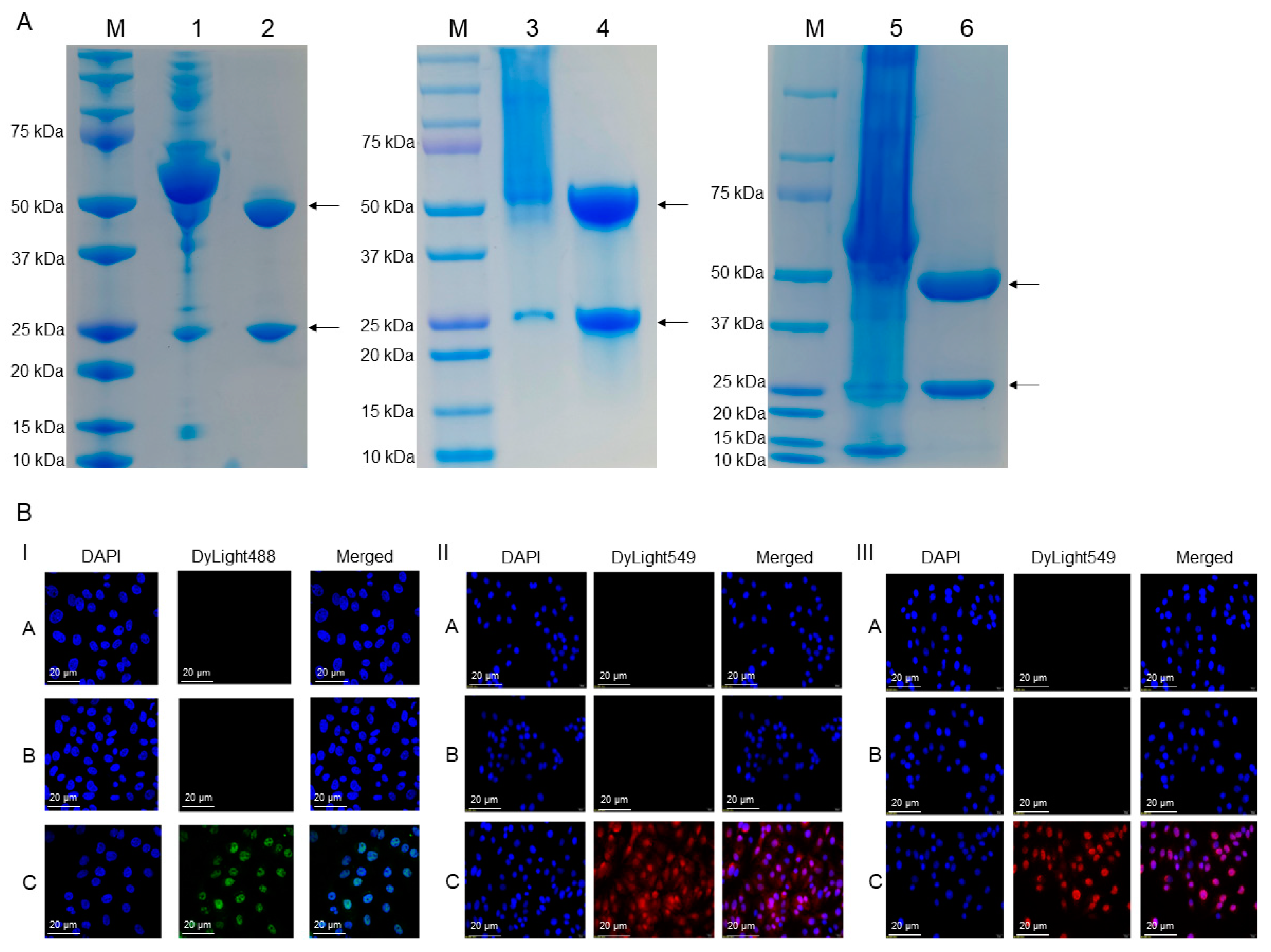
2.2. Preparation and Production of mAbs
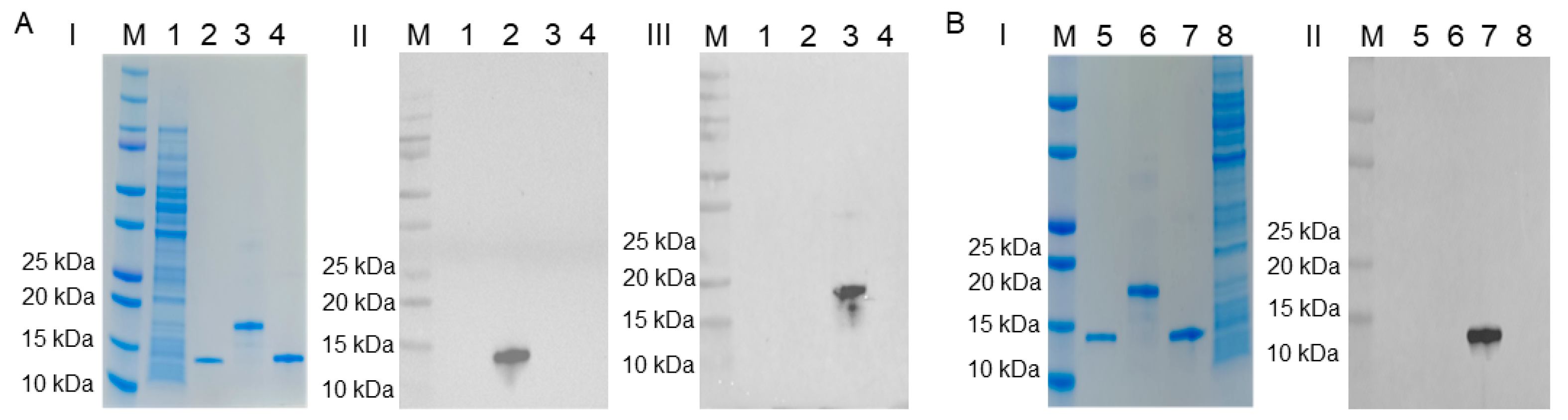
2.3. Characterization of mAbs against Porcine S100A8, S100A9, and S100A12 Proteins
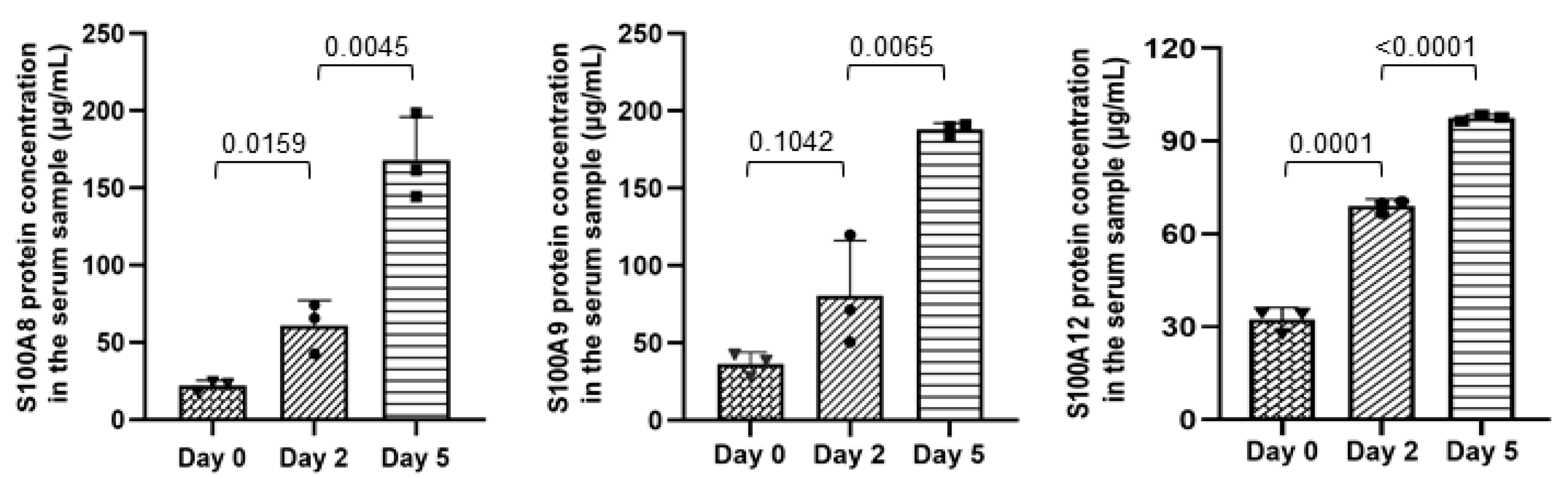
2.4. Application of mAbs to Test S100A8, S100A9, and S100A12 Content in the Serum Samples of Piglets
3. Discussion
4. Materials and Methods
4.1. Animals, Cell Lines, and Culture Conditions
4.2. S100A8, S100A9, and S100A12 Gene Cloning and Expression
4.3. Animals Immunization and Hybridoma Technology to Produce mAbs
4.4. Construction of Stable Cell Lines for S100A8, S100A9, and S100A12 Protein Expression
4.5. Characterization of mAbs against the S100A8, S100A9, and S100A12 Proteins
4.6. Determination of S100A8, S100A9, and S100A12 Protein Contents in the Serum Using mAbs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gonzalez, L.L.; Garrie, K.; Turner, M.D. Role of S100 proteins in health and disease. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118677. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Ji, X.; Yan, L.; Lian, S.; Chen, Z.; Luo, Y. Roles of S100A8, S100A9 and S100A12 in infection, inflammation and immunity. Immunology 2023. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, C.C.; Skaar, E.P. Nutritional immunity: The battle for nutrient metals at the host-pathogen interface. Nat. Rev. Microbiol. 2022, 20, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Zackular, J.P.; Chazin, W.J.; Skaar, E.P. Nutritional immunity: S100 proteins at the host-pathogen interface. J. Biol. Chem. 2015, 290, 18991–18998. [Google Scholar] [CrossRef] [PubMed]
- Damo, S.M.; Kehl-Fie, T.E.; Sugitani, N.; Holt, M.E.; Rathi, S.; Murphy, W.J.; Zhang, Y.; Betz, C.; Hench, L.; Fritz, G.; et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. USA 2013, 110, 3841–3846. [Google Scholar] [CrossRef]
- Sreejit, G.; Flynn, M.C.; Patil, M.; Krishnamurthy, P.; Murphy, A.J.; Nagareddy, P.R. S100 family proteins in inflammation and beyond. Adv. Clin. Chem. 2020, 98, 173–231. [Google Scholar]
- Lee, D.G.; Woo, J.W.; Kwok, S.K.; Cho, M.L.; Park, S.H. MRP8 promotes Th17 differentiation via upregulation of IL-6 production by fibroblast-like synoviocytes in rheumatoid arthritis. Exp. Mol. Med. 2013, 45, e20. [Google Scholar] [CrossRef]
- Lewis, J.D. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology 2011, 140, 1817–1826.e2. [Google Scholar] [CrossRef]
- Kang, K.Y.; Woo, J.W.; Park, S.H. S100A8/A9 as a biomarker for synovial inflammation and joint damage in patients with rheumatoid arthritis. Korean J. Intern. Med. 2014, 29, 12–19. [Google Scholar] [CrossRef]
- Rosen, T.; Nolan, E.M. Metal Sequestration and antimicrobial activity of human calprotectin are pH-dependent. Biochemistry 2020, 59, 2468–2478. [Google Scholar] [CrossRef]
- Wang, Q.; Aleshintsev, A.; Jose, A.N.; Aramini, J.M.; Gupta, R. Calcium regulates S100A12 zinc sequestration by limiting structural variations. ChemBioChem 2020, 21, 1372–1382. [Google Scholar] [CrossRef]
- Shastri, Y.M.; Bergis, D.; Povse, N.; Schäfer, V.; Shastri, S.; Weindel, M.; Ackermann, H.; Stein, J. Prospective multicenter study evaluating fecal calprotectin in adult acute bacterial diarrhea. Am. J. Med. 2008, 121, 1099–1106. [Google Scholar] [CrossRef]
- Kaiser, T.; Langhorst, J.; Wittkowski, H.; Becker, K.; Friedrich, A.W.; Rueffer, A.; Dobos, G.J.; Roth, J.; Foell, D. Faecal S100A12 as a non-invasive marker distinguishing inflammatory bowel disease from irritable bowel syndrome. Gut 2007, 56, 1706–1713. [Google Scholar] [CrossRef]
- Chen, H.; Cheng, L.; Yang, S.; Liu, X.; Liu, Y.; Tang, J.; Li, X.; He, Q.; Zhao, S. Molecular characterization, induced expression, and transcriptional regulation of porcine S100A12 gene. Mol. Immunol. 2010, 47, 1601–1607. [Google Scholar] [CrossRef]
- Chen, H.; Lunney, J.K.; Cheng, L.; Li, X.; Cao, J.; Zhu, M.; Zhao, S. Porcine S100A8 and S100A9: Molecular characterizations and crucial functions in response to Haemophilus parasuis infection. Dev. Comp. Immunol. 2011, 35, 490–500. [Google Scholar] [CrossRef]
- Li, X.; Tang, J.; Xu, J.; Zhu, M.; Cao, J.; Liu, Y.; Yu, M.; Zhao, S. The inflammation-related gene S100A12 is positively regulated by C/EBPβ and AP-1 in pigs. Int. J. Mol. Sci. 2014, 15, 13802–13816. [Google Scholar] [CrossRef]
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef]
- Weiner, G.J. Building better monoclonal antibody-based therapeutics. Nat. Rev. Cancer 2015, 15, 361–370. [Google Scholar] [CrossRef]
- El Miedany, Y. MABS: Targeted therapy tailored to the patient’s need. Br. J. Nurs. 2015, 24, S4–S13. [Google Scholar] [CrossRef]
- Castelli, M.S.; McGonigle, P.; Hornby, P.J. The pharmacology and therapeutic applications of monoclonal antibodies. Pharmacol. Res. Perspect. 2019, 7, e00535. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Jia, X.; Johansson, P.; Wang, C.; Moskalenko, R.; Steinau, A.; Forsgren, L.; Wågberg, T.; Svensson, J.; Zetterberg, H.; et al. Pro-inflammatory S100A9 protein as a robust biomarker differentiating early stages of cognitive impairment in Alzheimer’s disease. ACS Chem. Neurosci. 2016, 7, 34–39. [Google Scholar] [CrossRef]
- Däbritz, J.; Musci, J.; Foell, D. Diagnostic utility of faecal biomarkers in patients with irritable bowel syndrome. World J. Gastroenterol. 2014, 20, 363–375. [Google Scholar] [CrossRef]
- Shabani, F.; Farasat, A.; Mahdavi, M.; Gheibi, N. Calprotectin (S100A8/S100A9): A key protein between inflammation and cancer. Inflamm. Res. 2018, 67, 801–812. [Google Scholar] [CrossRef]
- Roszkowski, L.; Jaszczyk, B.; Plebańczyk, M.; Ciechomska, M. S100A8 and S100A12 proteins as biomarkers of high disease activity in patients with Rheumatoid Arthritis that can be regulated by epigenetic drugs. Int. J. Mol. Sci. 2022, 24, 710. [Google Scholar] [CrossRef]
- Wang, Q.; Aleshintsev, A.; Bolton, D.; Zhuang, J.; Brenowitz, M.; Gupta, R. Ca(II) and Zn(II) cooperate to modulate the structure and self-assembly of S100A12. Biochemistry 2019, 58, 2269–2281. [Google Scholar] [CrossRef]
- Nolan, E.M.; Peet, J.J.Y. Post-translational modifications on the metal-sequestering protein calprotectin. Biometals 2023, 36, 817–828. [Google Scholar] [CrossRef]
- Ghavami, S.; Eshragi, M.; Ande, S.R.; Chazin, W.J.; Klonisch, T.; Halayko, A.J.; McNeill, K.D.; Hashemi, M.; Kerkhoff, C.; Los, M. S100A8/A9 induces autophagy and apoptosis via ROS-mediated cross-talk between mitochondria and lysosomes that involves BNIP3. Cell Res. 2010, 20, 314–331. [Google Scholar] [CrossRef]
- Yang, X.; Xia, P.; Zhang, Y.; Lian, S.; Li, H.; Zhu, G.; Wang, P. Photothermal Nano-antibiotic for effective treatment of multidrug-resistant bacterial infection. ACS Appl. Bio Mater. 2020, 3, 5395–5406. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Zemskova, M.Y.; Rystsov, G.K.; Vologzhannikova, A.A.; Deryusheva, E.I.; Rastrygina, V.A.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N.; et al. Specific S100 proteins bind tumor necrosis factor and inhibit its activity. Int. J. Mol. Sci. 2022, 23, 15956. [Google Scholar] [CrossRef] [PubMed]
- Farokhzadian, J.; Mangolian Shahrbabaki, P.; Bagheri, V. S100A12-CD36 axis: A novel player in the pathogenesis of atherosclerosis? Cytokine 2019, 122, 154104. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Franco, F.; Tsui, Y.C.; Xie, X.; Trefny, M.P.; Zappasodi, R.; Mohmood, S.R.; Fernández-García, J.; Tsai, C.H.; Schulze, I.; et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020, 21, 298–308. [Google Scholar] [CrossRef]
- Shank, J.M.; Kelley, B.R.; Jackson, J.W.; Tweedie, J.L.; Franklin, D.; Damo, S.M.; Gaddy, J.A.; Murphy, C.N.; Johnson, J.G. The host antimicrobial protein calgranulin C participates in the control of Campylobacter jejuni growth via zinc sequestration. Infect. Immun. 2018, 86, e00234-18. [Google Scholar] [CrossRef]
- Gaddy, J.A.; Radin, J.N.; Loh, J.T.; Piazuelo, M.B.; Kehl-Fie, T.E.; Delgado, A.G.; Ilca, F.T.; Peek, R.M.; Cover, T.L.; Chazin, W.J.; et al. The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration. PLoS Pathog. 2014, 10, e1004450. [Google Scholar] [CrossRef] [PubMed]
- Corbin, B.D.; Seeley, E.H.; Raab, A.; Feldmann, J.; Miller, M.R.; Torres, V.J.; Anderson, K.L.; Dattilo, B.M.; Dunman, P.M.; Gerads, R.; et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 2008, 319, 962–965. [Google Scholar] [CrossRef]
- Haley, K.P.; Delgado, A.G.; Piazuelo, M.B.; Mortensen, B.L.; Correa, P.; Damo, S.M.; Chazin, W.J.; Skaar, E.P.; Gaddy, J.A. The human antimicrobial protein calgranulin C participates in control of Helicobacter pylori growth and regulation of virulence. Infect. Immun. 2015, 83, 2944–2956. [Google Scholar] [CrossRef]
- Xia, P.; Song, Y.; Zou, Y.; Yang, Y.; Zhu, G. F4+ enterotoxigenic Escherichia coli (ETEC) adhesion mediated by the major fimbrial subunit FaeG. J. Basic Microbiol. 2015, 55, 1118–1124. [Google Scholar] [CrossRef]
- Kim, J.P.; Yun, H.; Kim, E.J.; Kim, Y.G.; Lee, C.S.; Ko, B.J.; Kim, B.G.; Jeong, H.J. Generation of a novel monoclonal antibody against inflammatory biomarker S100A8 using hybridoma technology. Biotechnol. Lett. 2023, 45, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, L.; Tissot, N.; Hartmann, D.J.; Cohen, R. Comparison of the results obtained by ELISA and surface plasmon resonance for the determination of antibody affinity. J. Immunol. Methods 2010, 352, 13–22. [Google Scholar] [CrossRef]
- Chen, W.; Liu, W.E.; Li, Y.M.; Luo, S.; Zhong, Y.M. Preparation and preliminary application of monoclonal antibodies to the receptor binding region of Clostridium difficile toxin B. Mol. Med. Rep. 2015, 12, 7712–7720. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xia, P.; Quan, G.; Yang, Y.; Zhao, J.; Wang, Y.; Zhou, M.; Hardwidge, P.R.; Zhu, J.; Liu, S.; Zhu, G. Binding determinants in the interplay between porcine aminopeptidase N and enterotoxigenic Escherichia coli F4 fimbriae. Vet. Res. 2018, 49, 23. [Google Scholar] [CrossRef]
- Tabatabaei, M.S.; Ahmed, M. Enzyme-Linked Immunosorbent Assay (ELISA). Methods Mol. Biol. 2022, 2508, 115–134. [Google Scholar]
| Cells | Titers of Cell Culture Supernatants | Titers of Ascites |
|---|---|---|
| S100A8-C6D2 | 0.4 × 103 | 0.4 × 105 |
| S100A8-C6F6 | 0.4 × 103 | 6.4 × 104 |
| S100A8-C6F7 | 1.6 × 103 | 0.8× 105 |
| S100A8-F3C4 | 0.8 × 103 | 0.4 × 105 |
| S100A8-F8E8 | 1.6 × 103 | 0.8 × 105 |
| S100A8-F8F9 | 1.6 × 103 | 0.8 × 105 |
| S100A8-E11H7 | 0.8 × 103 | 1.6 × 105 |
| S100A9-A97A | 0.8 × 103 | 1.6 × 105 |
| S100A9-A912E | 0.8 × 103 | 1.6 × 105 |
| S100A9-A912D | 0.8 × 103 | 0.8 × 105 |
| S100A9-G67B | 0.4 × 103 | 0.8 × 105 |
| S100A9-C92H | 0.8 × 103 | 3.2 × 105 |
| S100A9-18H | 0.8 × 103 | 1.28 × 105 |
| S100A12-C5D10 | 0.2 × 103 | 0.5 × 104 |
| S100A12-D10E2 | 0.4 × 103 | 0.8 × 104 |
| S100A12-D10F10 | 0.2 × 103 | 0.4 × 104 |
| S100A12-F5C6 | 0.2 × 103 | 2.56 × 104 |
| Ig | IgA | IgM | IgG1 | IgG2a | IgG2b | IgG3 | Kappa | Lambda | |
|---|---|---|---|---|---|---|---|---|---|
| S100A8-C6D2 | 0.889 | 0.131 | 0.089 | 3.095 | 0.104 | 0.086 | 0.087 | 0.384 | 0.079 |
| S100A8-C6F6 | 0.917 | 0.096 | 0.105 | 3.1 | 0.163 | 0.109 | 0.084 | 0.369 | 0.086 |
| S100A8-C6F7 | 0.929 | 0.126 | 0.166 | 3.157 | 0.158 | 0.09 | 0.087 | 0.381 | 0.082 |
| S100A8-F3C4 | 1.096 | 0.095 | 0.117 | 3.071 | 0.15 | 0.138 | 0.084 | 0.551 | 0.115 |
| S100A8-F8E8 | 0.928 | 0.093 | 0.09 | 3.003 | 0.116 | 0.35 | 0.104 | 0.368 | 0.096 |
| S100A8-F8F9 | 1.19 | 0.085 | 0.106 | 3.136 | 0.116 | 0.177 | 0.101 | 0.642 | 0.076 |
| S100A8-E11H7 | 1.385 | 0.081 | 0.096 | 3.157 | 0.083 | 0.152 | 0.12 | 0.746 | 0.071 |
| S100A9-A97A | 1.495 | 0.081 | 0.1 | 0.091 | 0.09 | 2.764 | 0.089 | 0.961 | 0.128 |
| S100A9-A912E | 1.614 | 0.089 | 0.102 | 0.091 | 0.09 | 2.845 | 0.09 | 0.999 | 0.091 |
| S100A9-A912D | 1.439 | 0.113 | 0.107 | 0.107 | 0.109 | 2.728 | 0.101 | 0.913 | 0.092 |
| S100A9-G67B | 1.671 | 0.089 | 0.088 | 0.116 | 0.1 | 2.832 | 0.101 | 0.93 | 0.085 |
| S100A9-C92H | 1.687 | 0.132 | 0.097 | 0.11 | 0.101 | 2.893 | 0.122 | 1.168 | 0.082 |
| S100A9-18H | 1.531 | 0.082 | 0.097 | 0.094 | 0.088 | 2.826 | 0.093 | 0.943 | 0.09 |
| S100A12-C5D10 | 0.481 | 0.08 | 0.097 | 2.859 | 0.088 | 0.127 | 0.092 | 0.293 | 0.095 |
| S100A12-D10E2 | 0.505 | 0.125 | 0.096 | 2.927 | 0.095 | 0.09 | 0.086 | 0.31 | 0.093 |
| S100A12-D10F10 | 0.514 | 0.089 | 0.124 | 2.898 | 0.117 | 0.095 | 0.092 | 0.291 | 0.081 |
| S100A12-F5C6 | 0.588 | 0.091 | 0.098 | 2.903 | 0.087 | 0.096 | 0.101 | 0.346 | 0.082 |
| Primers | Primer Sequence 1 | Length (bp) |
|---|---|---|
| S100A8-F | 5′-CTAGCTAGCGCCACCATGCTGACGG-3′ | 270 bp |
| S100A8-R | 5′-CCGAAGCTTCTCTTTGTGGATG-3′ | |
| S100A9-F | 5′-CCGCTAGCGCCACCATGGCGGACC-3′ | 435 bp |
| S100A9-R | 5′-CGCAAGCTTGTGGCTGTGGCCA-3′ | |
| S100A12-F | 5′-CTAGCTAGCGCCACCATGACTAAGC-3′ | 279 bp |
| S100A12-R | 5′-CCAAGCTTCTCCTTGTGGATGTTG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, P.; Ma, X.; Yan, L.; Lian, S.; Li, X.; Luo, Y.; Chen, Z.; Ji, X. Generation and Application of Monoclonal Antibodies against Porcine S100A8, S100A9, and S100A12 Proteins Using Hybridoma Technology. Int. J. Mol. Sci. 2024, 25, 1029. https://doi.org/10.3390/ijms25021029
Xia P, Ma X, Yan L, Lian S, Li X, Luo Y, Chen Z, Ji X. Generation and Application of Monoclonal Antibodies against Porcine S100A8, S100A9, and S100A12 Proteins Using Hybridoma Technology. International Journal of Molecular Sciences. 2024; 25(2):1029. https://doi.org/10.3390/ijms25021029
Chicago/Turabian StyleXia, Pengpeng, Xin Ma, Li Yan, Siqi Lian, Xiangyu Li, Yi Luo, Ziyue Chen, and Xingduo Ji. 2024. "Generation and Application of Monoclonal Antibodies against Porcine S100A8, S100A9, and S100A12 Proteins Using Hybridoma Technology" International Journal of Molecular Sciences 25, no. 2: 1029. https://doi.org/10.3390/ijms25021029
APA StyleXia, P., Ma, X., Yan, L., Lian, S., Li, X., Luo, Y., Chen, Z., & Ji, X. (2024). Generation and Application of Monoclonal Antibodies against Porcine S100A8, S100A9, and S100A12 Proteins Using Hybridoma Technology. International Journal of Molecular Sciences, 25(2), 1029. https://doi.org/10.3390/ijms25021029







