Lifespan Extension by Retrotransposons under Conditions of Mild Stress Requires Genes Involved in tRNA Modifications and Nucleotide Metabolism
Abstract
1. Introduction
2. Results and Discussion
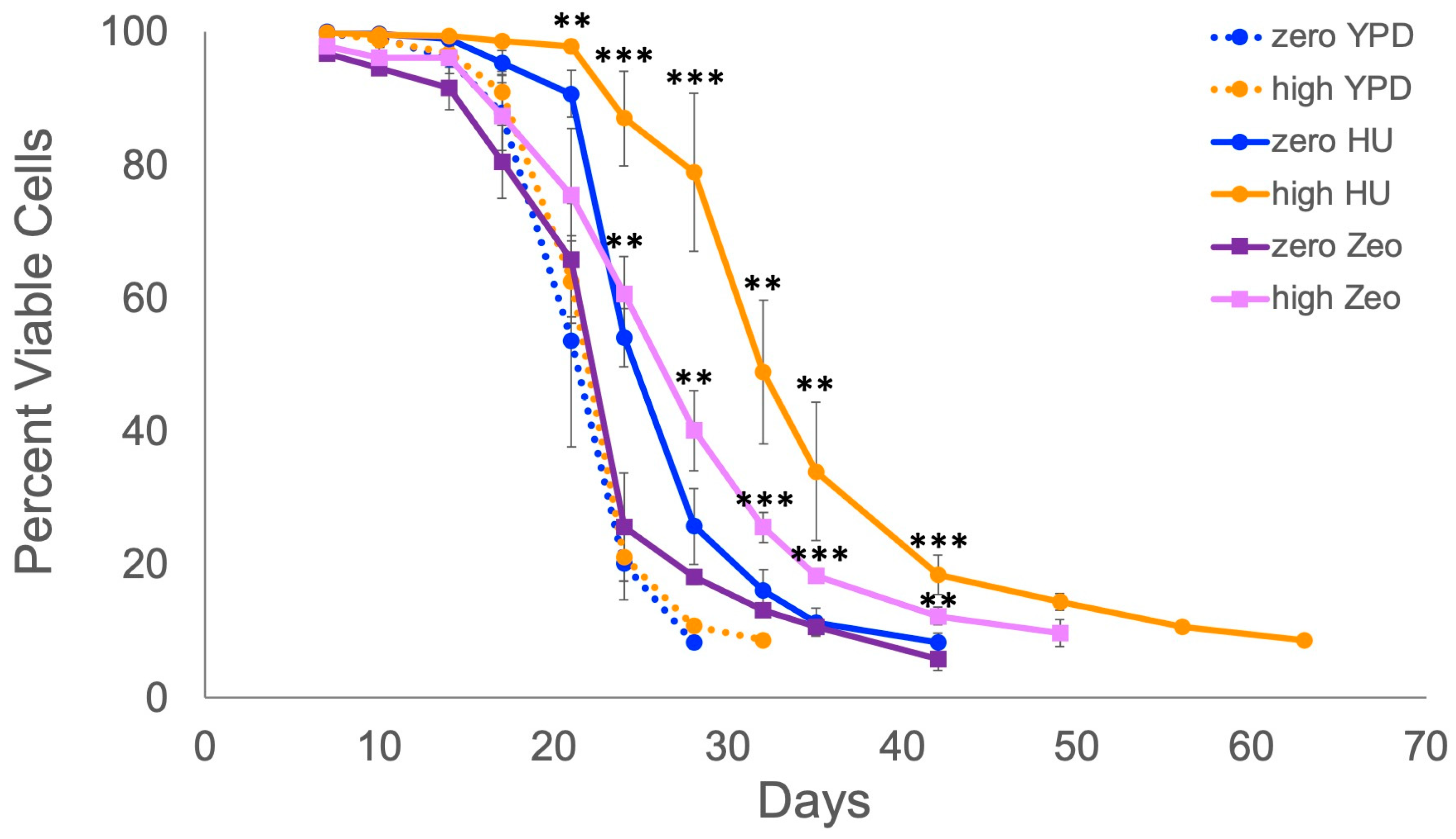
2.1. Retrotransposons Extend Lifespan When Cells Are Exposed to Mild DNA Replication or DNA Damage Stress
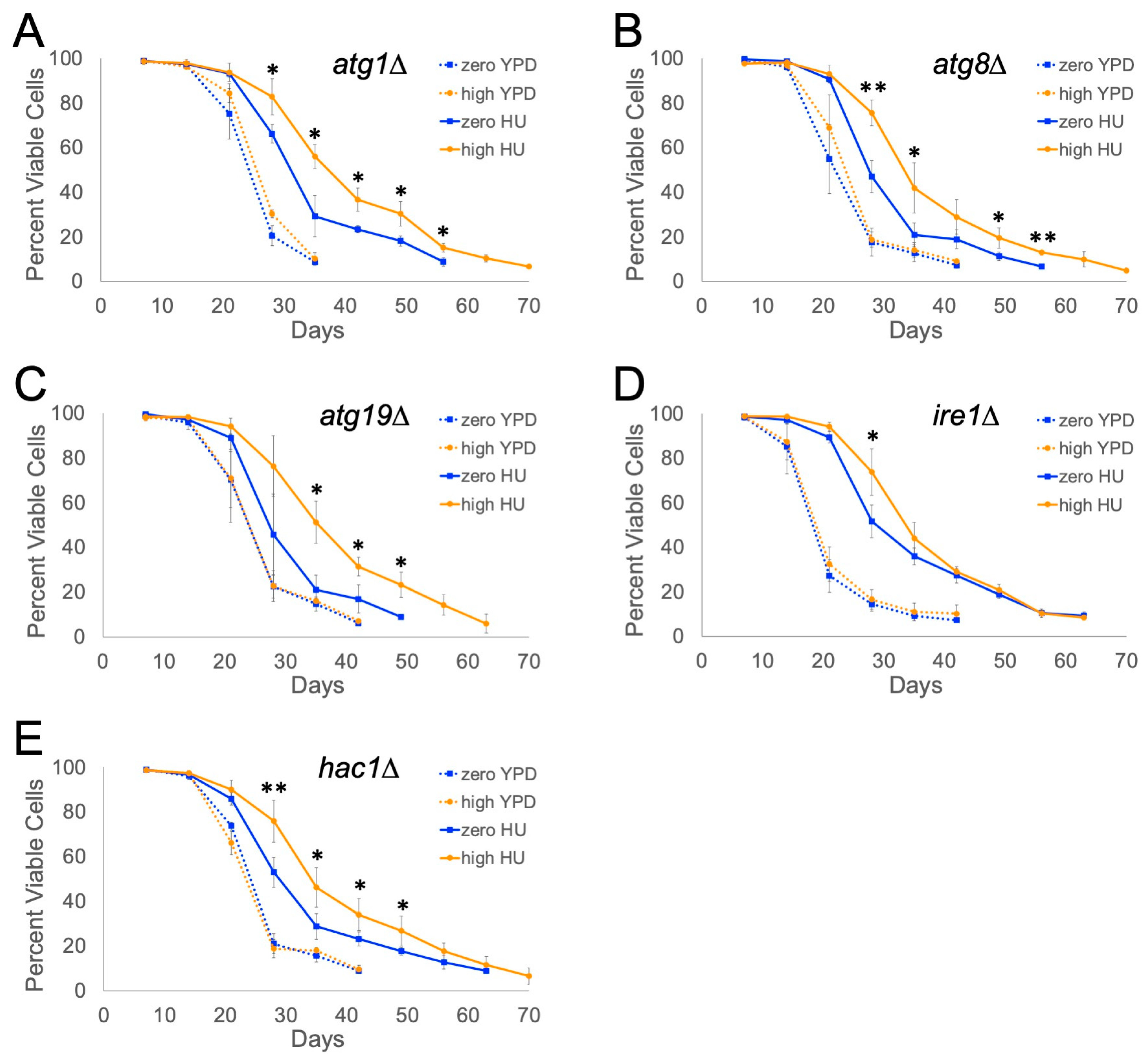
2.2. IRE1 and HAC1 Gene Functions Are Partly Required for Ty1-Dependent Lifespan Extension
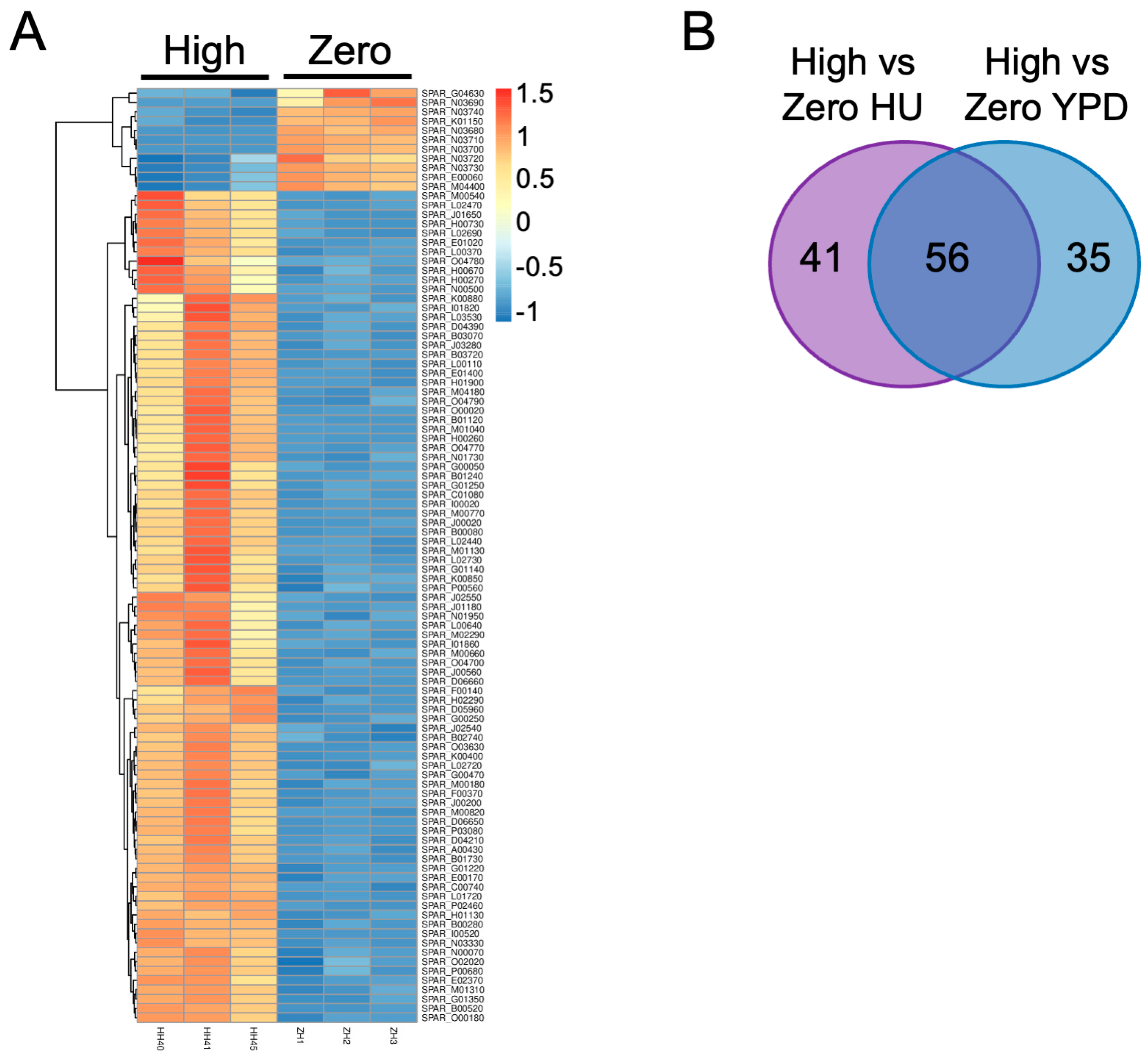
2.3. Differential Gene Expression in the Presence of Ty1 and HU
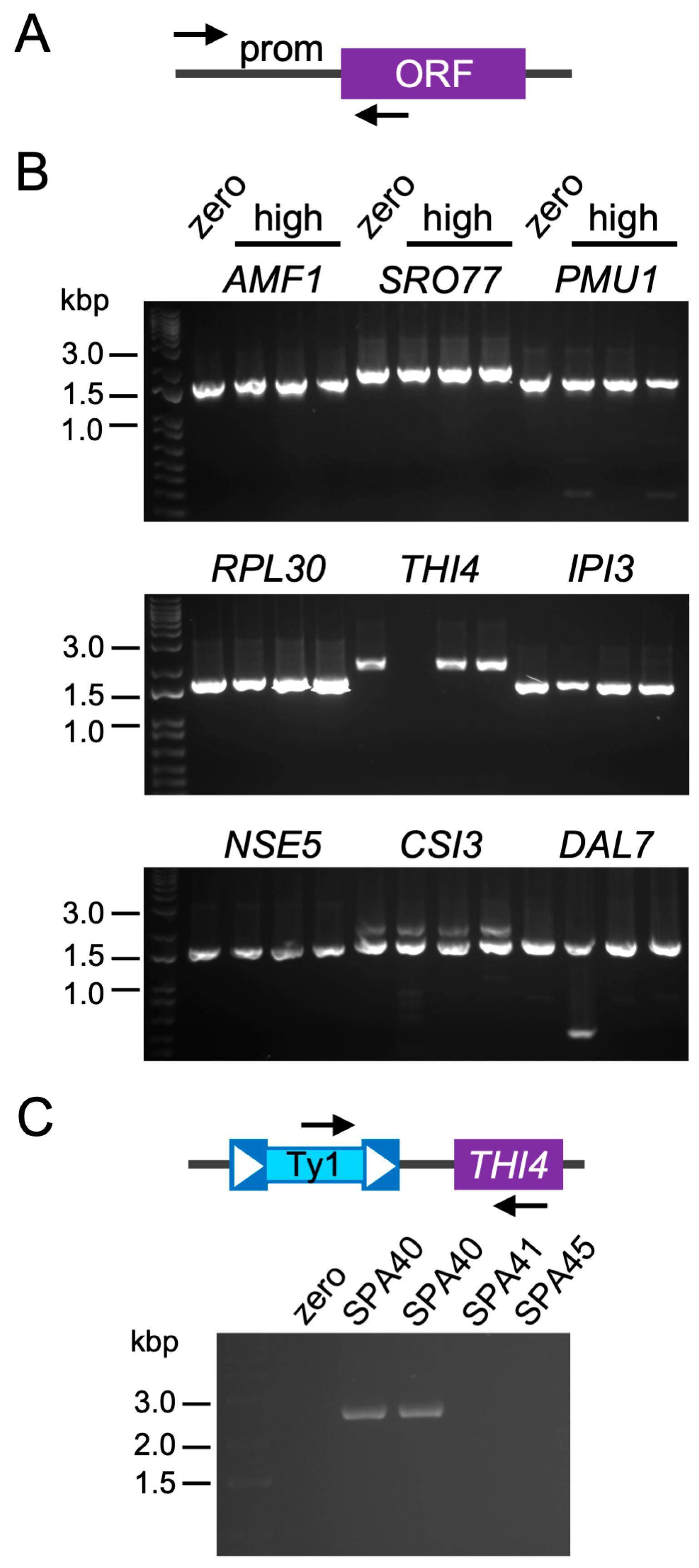
2.4. Genes Overexpressed in High-Copy Ty1 Strains Treated with HU Contribute to Lifespan Extension
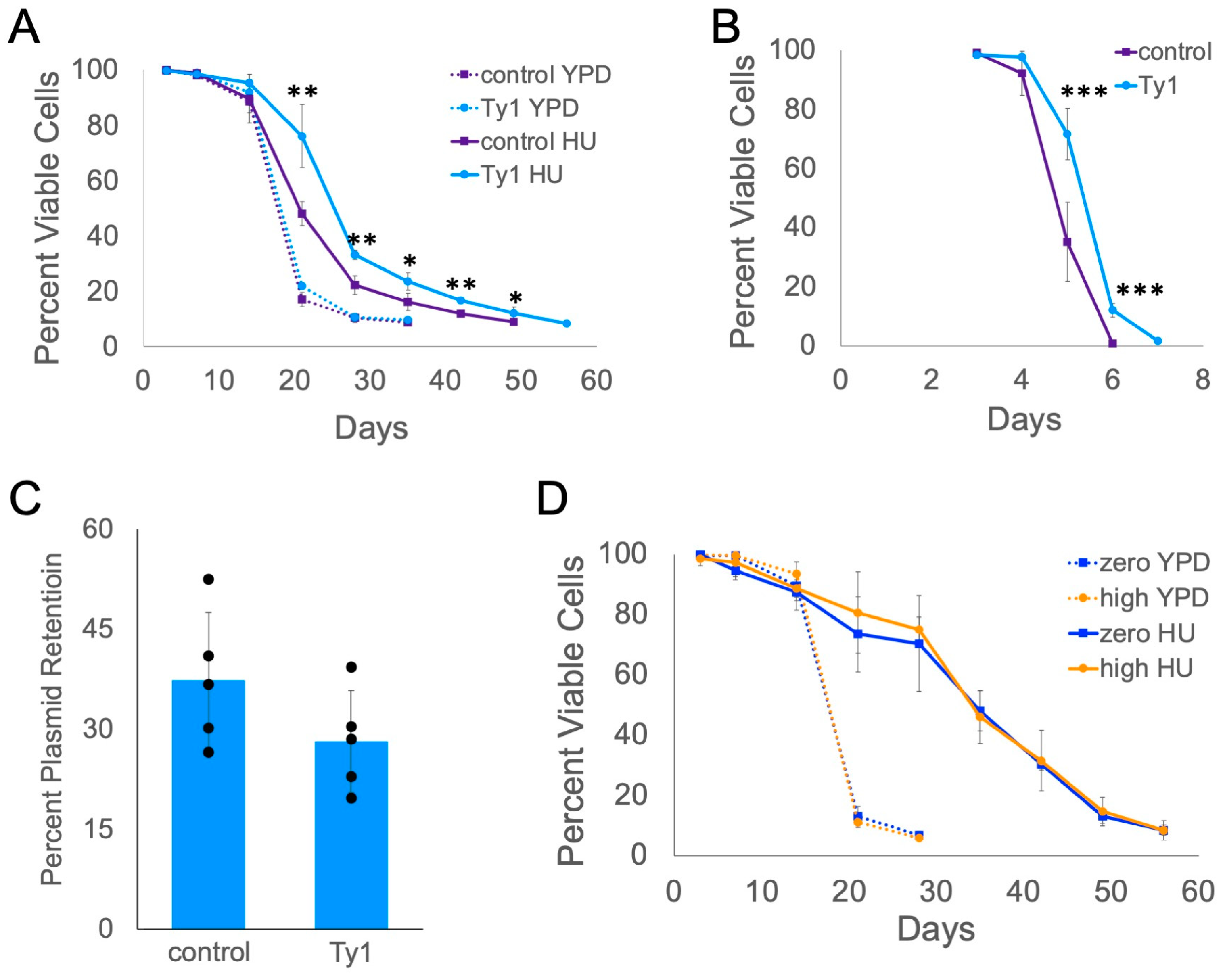
2.5. Ty1 Expression Causes Lifespan Extension
2.6. Ty1 Insertions Are Not Typically Nearby Promoters of Overexpressed Genes
3. Materials and Methods
3.1. Yeast Strains and Media
3.2. Chronological Lifespan Determination
3.3. RNA Extraction for RT-PCR and RNA Sequencing
3.4. RT-PCR for HAC1 Splicing
3.5. RNA Sequencing and Analysis
3.6. Plasmid Construction and Expression of Ty1 in the Zero-Copy Ty1 Strain
3.7. PCR for Differentially Expressed Gene Sequences
3.8. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of Aging: An Expanding Universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Burhans, W.C.; Curcio, M.J. Retrotransposition Is Associated with Genome Instability during Chronological Aging. Proc. Natl. Acad. Sci. USA 2011, 108, 20376–20381. [Google Scholar] [CrossRef] [PubMed]
- Dennis, S.; Sheth, U.; Feldman, J.L.; English, K.A.; Priess, J.R. C. elegans Germ Cells Show Temperature and Age-Dependent Expression of Cer1, a Gypsy/Ty3-Related Retrotransposon. PLoS Pathog. 2012, 8, e1002591. [Google Scholar] [CrossRef]
- De Cecco, M.; Criscione, S.W.; Peckham, E.J.; Hillenmeyer, S.; Hamm, E.A.; Manivannan, J.; Peterson, A.L.; Kreiling, J.A.; Neretti, N.; Sedivy, J.M. Genomes of Replicatively Senescent Cells Undergo Global Epigenetic Changes Leading to Gene Silencing and Activation of Transposable Elements. Aging Cell 2013, 12, 247–256. [Google Scholar] [CrossRef]
- De Cecco, M.; Criscione, S.W.; Peterson, A.L.; Neretti, N.; Sedivy, J.M.; Kreiling, J.A. Transposable Elements Become Active and Mobile in the Genomes of Aging Mammalian Somatic Tissues. Aging 2013, 5, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Prazak, L.; Chatterjee, N.; Grüninger, S.; Krug, L.; Theodorou, D.; Dubnau, J. Activation of Transposable Elements during Aging and Neuronal Decline in Drosophila. Nat. Neurosci. 2013, 16, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chen, K.; Xia, Z.; Chavez, M.; Pal, S.; Seol, J.-H.; Chen, C.-C.; Li, W.; Tyler, J.K. Nucleosome Loss Leads to Global Transcriptional Up-Regulation and Genomic Instability during Yeast Aging. Genes Dev. 2014, 28, 396–408. [Google Scholar] [CrossRef]
- Patterson, M.N.; Scannapieco, A.E.; Au, P.H.; Dorsey, S.; Royer, C.A.; Maxwell, P.H. Preferential Retrotransposition in Aging Yeast Mother Cells Is Correlated with Increased Genome Instability. DNA Repair 2015, 34, 18–27. [Google Scholar] [CrossRef]
- Chang, Y.-H.; Keegan, R.M.; Prazak, L.; Dubnau, J. Cellular Labeling of Endogenous Retrovirus Replication (CLEVR) Reveals de Novo Insertions of the Gypsy Retrotransposable Element in Cell Culture and in Both Neurons and Glial Cells of Aging Fruit Flies. PLoS Biol. 2019, 17, e3000278. [Google Scholar] [CrossRef]
- Krug, L.; Chatterjee, N.; Borges-Monroy, R.; Hearn, S.; Liao, W.-W.; Morrill, K.; Prazak, L.; Rozhkov, N.; Theodorou, D.; Hammell, M.; et al. Retrotransposon Activation Contributes to Neurodegeneration in a Drosophila TDP-43 Model of ALS. PLoS Genet. 2017, 13, e1006635. [Google Scholar] [CrossRef]
- Kaneko, H.; Dridi, S.; Tarallo, V.; Gelfand, B.D.; Fowler, B.J.; Cho, W.G.; Kleinman, M.E.; Ponicsan, S.L.; Hauswirth, W.W.; Chiodo, V.A.; et al. DICER1 Deficit Induces Alu RNA Toxicity in Age-Related Macular Degeneration. Nature 2011, 471, 325–330. [Google Scholar] [CrossRef]
- Tarallo, V.; Hirano, Y.; Gelfand, B.D.; Dridi, S.; Kerur, N.; Kim, Y.; Cho, W.G.; Kaneko, H.; Fowler, B.J.; Bogdanovich, S.; et al. DICER1 Loss and Alu RNA Induce Age-Related Macular Degeneration via the NLRP3 Inflammasome and MyD88. Cell 2012, 149, 847–859. [Google Scholar] [CrossRef]
- Kim, Y.; Tarallo, V.; Kerur, N.; Yasuma, T.; Gelfand, B.D.; Bastos-Carvalho, A.; Hirano, Y.; Yasuma, R.; Mizutani, T.; Fowler, B.J.; et al. DICER1/Alu RNA Dysmetabolism Induces Caspase-8–Mediated Cell Death in Age-Related Macular Degeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 16082–16087. [Google Scholar] [CrossRef]
- Fukuda, S.; Varshney, A.; Fowler, B.J.; Wang, S.-B.; Narendran, S.; Ambati, K.; Yasuma, T.; Magagnoli, J.; Leung, H.; Hirahara, S.; et al. Cytoplasmic Synthesis of Endogenous Alu Complementary DNA via Reverse Transcription and Implications in Age-Related Macular Degeneration. Proc. Natl. Acad. Sci. USA 2021, 118, e2022751118. [Google Scholar] [CrossRef]
- Fukuda, S.; Narendran, S.; Varshney, A.; Nagasaka, Y.; Wang, S.; Ambati, K.; Apicella, I.; Pereira, F.; Fowler, B.J.; Yasuma, T.; et al. Alu Complementary DNA Is Enriched in Atrophic Macular Degeneration and Triggers Retinal Pigmented Epithelium Toxicity via Cytosolic Innate Immunity. Sci. Adv. 2021, 7, eabj3658. [Google Scholar] [CrossRef]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. LINE-1 Derepression in Senescent Cells Triggers Interferon and Inflammaging. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Wu, Z.; Ren, J.; Fan, Y.; Sun, L.; Cao, G.; Niu, Y.; Zhang, B.; Ji, Q.; et al. Resurrection of Endogenous Retroviruses during Aging Reinforces Senescence. Cell 2023, 186, 287–304.e26. [Google Scholar] [CrossRef]
- Schneider, K.L.; Hao, X.; Keuenhof, K.S.; Berglund, L.L.; Fischbach, A.; Ahmadpour, D.; Chawla, S.; Gómez, P.; Höög, J.L.; Widlund, P.O.; et al. Elimination of Virus-like Particles Reduces Protein Aggregation and Extends Replicative Lifespan in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2024, 121, e2313538121. [Google Scholar] [CrossRef]
- VanHoute, D.; Maxwell, P.H. Extension of Saccharomyces paradoxus Chronological Lifespan by Retrotransposons in Certain Media Conditions Is Associated with Changes in Reactive Oxygen Species. Genetics 2014, 198, 531–545. [Google Scholar] [CrossRef]
- Curcio, M.J.; Lutz, S.; Lesage, P. The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae. Microbiol. Spectr. 2015, 3, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Sampaio-Marques, B.; Burhans, W.C.; Ludovico, P. Yeast at the Forefront of Research on Ageing and Age-Related Diseases. Prog. Mol. Subcell. Biol. 2019, 58, 217–242. [Google Scholar] [CrossRef]
- Burtner, C.R.; Murakami, C.J.; Kennedy, B.K.; Kaeberlein, M. A Molecular Mechanism of Chronological Aging in Yeast. Cell Cycle 2009, 8, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Morimoto, M.; Kondo, C.; Ohsumi, Y. Selective Autophagy Regulates Insertional Mutagenesis by the Ty1 Retrotransposon in Saccharomyces cerevisiae. Dev. Cell 2011, 21, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Kumsta, C.; Hansen, M. Hormetic Heat Shock and HSF-1 Overexpression Improve C. elegans Survival and Proteostasis by Inducing Autophagy. Autophagy 2017, 13, 1076–1077. [Google Scholar] [CrossRef]
- Agathokleous, E.; Kitao, M.; Calabrese, E.J. Environmental Hormesis and Its Fundamental Biological Basis: Rewriting the History of Toxicology. Environ. Res. 2018, 165, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Tyler, J.K.; Johnson, J.E. The Role of Autophagy in the Regulation of Yeast Life Span. Ann. N. Y. Acad. Sci. 2018, 1418, 31–43. [Google Scholar] [CrossRef]
- Li, B.; Dong, L.; Meng, W.; Xiong, S.-Y.; Wu, G.-S.; Ma, W.-Z.; Luo, H.-R. Phloretic Acid Requires the Insulin/IGF-1 Pathway and Autophagy to Enhance Stress Resistance and Extend the Lifespan of Caenorhabditis elegans. Front. Pharmacol. 2024, 15, 1384227. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef]
- Cheong, H.; Nair, U.; Geng, J.; Klionsky, D.J. The Atg1 Kinase Complex Is Involved in the Regulation of Protein Recruitment to Initiate Sequestering Vesicle Formation for Nonspecific Autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 2008, 19, 668–681. [Google Scholar] [CrossRef]
- Xie, Z.; Nair, U.; Klionsky, D.J. Atg8 Controls Phagophore Expansion during Autophagosome Formation. Mol. Biol. Cell 2008, 19, 3290–3298. [Google Scholar] [CrossRef] [PubMed]
- Doh, J.H.; Lutz, S.; Curcio, M.J. Co-Translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-like Particle Assembly Sites. PLoS Genet. 2014, 10, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Welihinda, A.A.; Kaufman, R.J. The Unfolded Protein Response Pathway in Saccharomyces cerevisiae. Oligomerization and Trans-Phosphorylation of Ire1p (Ern1p) Are Required for Kinase Activation. J. Biol. Chem. 1996, 271, 18181–18187. [Google Scholar] [CrossRef]
- Sidrauski, C.; Walter, P. The Transmembrane Kinase Ire1p Is a Site-Specific Endonuclease That Initiates mRNA Splicing in the Unfolded Protein Response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Kawahara, T.; Yanagi, H.; Yura, T.; Mori, K. Endoplasmic Reticulum Stress-Induced mRNA Splicing Permits Synthesis of Transcription Factor Hac1p/Ern4p That Activates the Unfolded Protein Response. Mol. Biol. Cell 1997, 8, 1845–1862. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Nikawa, J.-I.; Yamashita, S. IRE1 Encodes a Putative Protein Kinase Containing a Membrane-Spanning Domain and Is Required for Inositol Phototrophy in Saccharomyces cerevisiae. Mol. Microbiol. 1992, 6, 1441–1446. [Google Scholar] [CrossRef]
- Dakshinamurthy, A.; Nyswaner, K.M.; Farabaugh, P.J.; Garfinkel, D.J. BUD22 Affects Ty1 Retrotransposition and Ribosome Biogenesis in Saccharomyces cerevisiae. Genetics 2010, 185, 1193–1205. [Google Scholar] [CrossRef]
- Suresh, S.; Ahn, H.W.; Joshi, K.; Dakshinamurthy, A.; Kananganat, A.; Garfinkel, D.J.; Farabaugh, P.J. Ribosomal Protein and Biogenesis Factors Affect Multiple Steps during Movement of the Saccharomyces cerevisiae Ty1 Retrotransposon. Mob. DNA 2015, 6, 22. [Google Scholar] [CrossRef]
- Ahn, H.W.; Tucker, J.M.; Arribere, J.A.; Garfinkel, D.J. Ribosome Biogenesis Modulates Ty1 Copy Number Control in Saccharomyces cerevisiae. Genetics 2017, 207, 1441–1456. [Google Scholar] [CrossRef]
- Dichtl, B.; Tollervey, D. Pop3p Is Essential for the Activity of the RNase MRP and RNase P Ribonucleoproteins in Vivo. EMBO J. 1997, 16, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J.E.; Exinger, F.; Erbs, P.; Jund, R. New Insights into the Pyrimidine Salvage Pathway of Saccharomyces cerevisiae: Requirement of Six Genes for Cytidine Metabolism. Curr. Genet. 1999, 36, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Bayly, A.M.; Berglez, J.M.; Patel, O.; Castelli, L.A.; Hankins, E.G.; Coloe, P.; Hopkins Sibley, C.; Macreadie, I.G. Folic Acid Utilisation Related to Sulfa Drug Resistance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2001, 204, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Woods, R.A.; Jackson, I.E. The Accumulation of Glycinamide Ribotide by ade3 and ade8 Mutants of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1973, 53, 787–793. [Google Scholar] [CrossRef]
- Carter, A.T.; Beiche, F.; Hove-Jensen, B.; Narbad, A.; Barker, P.J.; Schweizer, L.M.; Schweizer, M. PRS1 Is a Key Member of the Gene Family Encoding Phosphoribosylpyrophosphate Synthetase in Saccharomyces cerevisiae. Mol. Gen. Genet. 1997, 254, 148–156. [Google Scholar] [CrossRef]
- Deng, W.; Babu, I.R.; Su, D.; Yin, S.; Begley, T.J.; Dedon, P.C. Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet. 2015, 11, e1005706. [Google Scholar] [CrossRef] [PubMed]
- Begley, U.; Dyavaiah, M.; Patil, A.; Rooney, J.P.; DiRenzo, D.; Young, C.M.; Conklin, D.S.; Zitomer, R.S.; Begley, T.J. Trm9 Catalyzed tRNA Modifications Link Translation to the DNA Damage Response. Mol. Cell 2007, 28, 860–870. [Google Scholar] [CrossRef]
- Tavares, J.F.; Davis, N.K.; Poim, A.; Reis, A.; Kellner, S.; Sousa, I.; Soares, A.R.; Moura, G.M.R.; Dedon, P.C.; Santos, M. tRNA-Modifying Enzyme Mutations Induce Codon-Specific Mistranslation and Protein Aggregation in Yeast. RNA Biol. 2021, 18, 563–575. [Google Scholar] [CrossRef]
- Rattray, A.J.; Shafer, B.K.; Garfinkel, D.J. The Saccharomyces cerevisiae DNA Recombination and Repair Functions of the RAD52 Epistasis Group Inhibit Ty1 Transposition. Genetics 2000, 154, 543–556. [Google Scholar] [CrossRef]
- Scholes, D.T.; Banerjee, M.; Bowen, B.; Curcio, M.J. Multiple Regulators of Ty1 Transposition in Saccharomyces cerevisiae Have Conserved Roles in Genome Maintenance. Genetics 2001, 159, 1449–1465. [Google Scholar] [CrossRef]
- Curcio, M.J.; Kenny, A.E.; Moore, S.; Garfinkel, D.J.; Weintraub, M.; Gamache, E.R.; Scholes, D.T. S-Phase Checkpoint Pathways Stimulate the Mobility of the Retrovirus-like Transposon Ty1. Mol. Cell. Biol. 2007, 27, 8874–8885. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Noma, A.; Sakaguchi, Y.; Suzuki, T. Mechanistic Characterization of the Sulfur-Relay System for Eukaryotic 2-Thiouridine Biogenesis at tRNA Wobble Positions. Nucleic Acids Res. 2009, 37, 1335–1352. [Google Scholar] [CrossRef]
- Todeschini, A.-L.; Morillon, A.; Springer, M.; Lesage, P. Severe Adenine Starvation Activates Ty1 Transcription and Retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005, 25, 7459–7472. [Google Scholar] [CrossRef][Green Version]
- Rolfes, R.J.; Hinnebusch, A.G. Translation of the Yeast Transcriptional Activator GCN4 Is Stimulated by Purine Limitation: Implications for Activation of the Protein Kinase GCN2. Mol. Cell. Biol. 1993, 13, 5099–5111. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Meyer, M.R.; Jackson, B.M.; Slade, D.; Roberts, C.; Hinnebusch, A.G.; Marton, M.J. Transcriptional Profiling Shows That Gcn4p Is a Master Regulator of Gene Expression during Amino Acid Starvation in Yeast. Mol. Cell. Biol. 2001, 21, 4347–4368. [Google Scholar] [CrossRef] [PubMed]
- Kokina, A.; Tanilas, K.; Ozolina, Z.; Pleiko, K.; Shvirksts, K.; Vamza, I.; Liepins, J. Purine Auxotrophic Starvation Evokes Phenotype Similar to Stationary Phase Cells in Budding Yeast. J. Fungi 2021, 8, 29. [Google Scholar] [CrossRef]
- Winston, F.; Durbin, K.J.; Fink, G.R. The SPT3 Gene Is Required for Normal Transcription of Ty Elements in S. cerevisiae. Cell 1984, 39, 675–682. [Google Scholar] [CrossRef]
- Boeke, J.D.; Styles, C.A.; Fink, G.R. Saccharomyces cerevisiae SPT3 Gene Is Required for Transposition and Transpositional Recombination of Chromosomal Ty Elements. Mol. Cell. Biol. 1986, 6, 3575–3581. [Google Scholar] [CrossRef]
- Huisinga, K.L.; Pugh, B.F. A Genome-Wide Housekeeping Role for TFIID and a Highly Regulated Stress-Related Role for SAGA in Saccharomyces cerevisiae. Mol. Cell 2004, 13, 573–585. [Google Scholar] [CrossRef]
- Zanton, S.J.; Pugh, B.F. Changes in Genomewide Occupancy of Core Transcriptional Regulators during Heat Stress. Proc. Natl. Acad. Sci. USA 2004, 101, 16843–16848. [Google Scholar] [CrossRef] [PubMed]
- Amberg, D.C.; Burke, D.; Strathern, J.N. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2005; ISBN 978-0-87969-728-0. [Google Scholar]
- Garfinkel, D.J.; Nyswaner, K.; Wang, J.; Cho, J.-Y. Post-Transcriptional Cosuppression of Ty1 Retrotransposition. Genetics 2003, 165, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-C.; Dawson, A.; Henderson, A.C.; Lockyer, E.J.; Read, E.; Sritharan, G.; Ryan, M.; Sgroi, M.; Ngou, P.M.; Woodruff, R.; et al. Nutrient Supplements Boost Yeast Transformation Efficiency. Sci. Rep. 2016, 6, 35738. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, R.S.; Hieter, P. A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of Genomic DNA from Yeasts for PCR-Based Applications. Biotechniques 2011, 50, 325–328. [Google Scholar] [CrossRef]




| High- vs. Zero-Copy Ty1 in YPD with HU | High- vs. Zero-Copy Ty1 in YPD | ||||
|---|---|---|---|---|---|
| GO Term | NES 1 | FDR 2 | GO Term | NES | FDR |
| Iron ion homeostasis | 1.58 | 0.19 | S-adenosylmethionine dependent methyltransferase activity | 1.73 | 0.06 |
| Glutamine metabolic process | 1.61 | 0.20 | Ribosomal large subunit assembly | 1.61 | 0.08 |
| Establishment of mitotic sister chromatid cohesion3 | 1.59 | 0.20 | Establishment of mitotic sister chromatid cohesion | 1.61 | 0.09 |
| S-adenosylmethionine dependent methyltransferase activity | 1.51 | 0.22 | Translational elongation | 1.68 | 0.09 |
| tRNA modification | 1.51 | 0.22 | Methyltransferase activity | 1.62 | 0.10 |
| Response to stress | 1.52 | 0.23 | Maturation of large subunit rRNA | 1.63 | 0.10 |
| Carbohydrate phosphorylation | 1.53 | 0.24 | Cytosolic large ribosomal subunit | 1.65 | 0.11 |
| Ribosomal large subunit assembly | 1.51 | 0.24 | Methylation | 1.63 | 0.12 |
| Microtubule nucleation | 1.50 | 0.24 | Purine nucleotide biosynthetic process | 1.55 | 0.15 |
| Carbohydrate phosphorylation | 1.56 | 0.15 | |||
| Cytoplasmic translation | 1.55 | 0.16 | |||
| Large ribosomal subunit | 1.57 | 0.16 | |||
| GPI anchor biosynthetic process | 1.56 | 0.16 | |||
| Ran GTPase binding | 1.55 | 0.17 | |||
| Preribosome, large subunit precursor | 1.53 | 0.18 | |||
| Microtubule plus end binding | 1.52 | 0.19 | |||
| Translation elongation factor activity | 1.51 | 0.19 | |||
| One carbon metabolic process | 1.51 | 0.19 | |||
| Condensed chromosome kinetochore | 1.50 | 0.22 | |||
| Nucleoside metabolic process | 1.49 | 0.23 | |||
| High- vs. Zero-Copy Ty1 in YPD with HU | High- vs. Zero-Copy Ty1 in YPD | ||||
|---|---|---|---|---|---|
| Pathway | NES 1 | FDR 2 | Pathway | NES | FDR |
| Fructose and mannose metabolism3 | −1.70 | 0.04 | Fructose and mannose metabolism | −1.66 | 0.12 |
| Pantothenate and CoA biosynthesis | 1.39 | 0.22 | Fatty acid degradation | 1.39 | 0.23 |
| Arginine biosynthesis | 1.52 | 0.23 | Pyrimidine metabolism | 1.42 | 0.23 |
| Thiamine metabolism | 1.48 | 0.23 | GPI anchor biosynthesis | 1.45 | 0.23 |
| Pyrimidine metabolism | 1.40 | 0.24 | Base excision repair | 1.40 | 0.24 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 1.37 | 0.24 | |||
| Mismatch repair | 1.35 | 0.24 | |||
| Tyrosine metabolism | 1.41 | 0.24 | |||
| Gene | Description | Fold Overexpression | q-Value |
|---|---|---|---|
| POP3 | Subunit of RNase MRP and nuclear RNase P | 8.16 | 7.31 × 10-20 |
| NCS2 | Protein required for uridine thiolation of Lys(UUU) and Glu(UUC) tRNAs | 5.18 | 4.52 × 10-13 |
| TRM9 | tRNA methyltransferase | 5.10 | 5.55 × 10-16 |
| CDD1 | Cytidine deaminase | 4.65 | 1.46 × 10-16 |
| FOL3 | Dihydrofolate synthetase, involved in folic acid biosynthesis | 4.43 | 1.66 × 10-17 |
| ARG3 | Ornithine carbamoyltransferase | 4.36 | 1.95 × 10-17 |
| AMF1 | Low affinity NH4+ transporter | 4.21 | 2.84 × 10-14 |
| SRO77 | Protein with roles in exocytosis and cation homeostasis | 4.21 | 3.19 × 10-34 |
| DAL1 | Allantoinase | 4.20 | 3.67 × 10-17 |
| PMU1 | Phosphomutase | 4.10 | 3.85 × 10-18 |
| MOG1 | Conserved nuclear protein that interacts with GTP-Gsp1p | 4.09 | 1.31 × 10-17 |
| RHO3 | Non-essential small GTPase of the Rho/Rac family of Ras-like proteins | 3.78 | 1.91 × 10-14 |
| THR1 | Homoserine kinase | 3.62 | 2.51 × 10-13 |
| TAH18 | NADPH-dependent diflavin reductase | 3.43 | 2.62 × 10-17 |
| TUB4 | Gamma-tubulin | 3.37 | 3.20 × 10-24 |
| SPAR_D04210 | Hypothetical protein, homology to S. cerevisiae SVF1-like proteins encoded by YDR222W and YLR225C | 3.29 | 4.94 × 10-25 |
| SWD3 | Essential subunit of the COMPASS (Set1C) complex | 3.27 | 3.16 × 10-15 |
| YPT31 | Rab family GTPase | 3.27 | 8.91 × 10-18 |
| ADE8 | Phosphoribosyl-glycinamide transformylase | 3.24 | 2.24 × 10-25 |
| ACO2 | Mitochondrial aconitase isozyme | 3.24 | 2.02 × 10-21 |
| HIF1 | Non-essential component of the HAT-B histone acetyltransferase complex | 3.23 | 3.42 × 10-13 |
| RIB3 | 3,4-dihydroxy-2-butanone-4-phosphate synthase (DHBP synthase) | 3.21 | 5.63 × 10-30 |
| NCS6 | Protein required for uridine thiolation of Gln, Lys, and Glu tRNAs | 3.10 | 5.00 × 10-16 |
| PRS1 | 5-phospho-ribosyl-1(alpha)-pyrophosphate synthetase | 3.02 | 1.06 × 10-32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maxwell, P.H.; Mahmood, M.; Villanueva, M.; Devine, K.; Avery, N. Lifespan Extension by Retrotransposons under Conditions of Mild Stress Requires Genes Involved in tRNA Modifications and Nucleotide Metabolism. Int. J. Mol. Sci. 2024, 25, 10593. https://doi.org/10.3390/ijms251910593
Maxwell PH, Mahmood M, Villanueva M, Devine K, Avery N. Lifespan Extension by Retrotransposons under Conditions of Mild Stress Requires Genes Involved in tRNA Modifications and Nucleotide Metabolism. International Journal of Molecular Sciences. 2024; 25(19):10593. https://doi.org/10.3390/ijms251910593
Chicago/Turabian StyleMaxwell, Patrick H., Mustafa Mahmood, Maya Villanueva, Kaitlyn Devine, and Nina Avery. 2024. "Lifespan Extension by Retrotransposons under Conditions of Mild Stress Requires Genes Involved in tRNA Modifications and Nucleotide Metabolism" International Journal of Molecular Sciences 25, no. 19: 10593. https://doi.org/10.3390/ijms251910593
APA StyleMaxwell, P. H., Mahmood, M., Villanueva, M., Devine, K., & Avery, N. (2024). Lifespan Extension by Retrotransposons under Conditions of Mild Stress Requires Genes Involved in tRNA Modifications and Nucleotide Metabolism. International Journal of Molecular Sciences, 25(19), 10593. https://doi.org/10.3390/ijms251910593






