In Vitro Profiling of the Antiviral Peptide TAT-I24
Abstract
1. Introduction
2. Results and Discussion
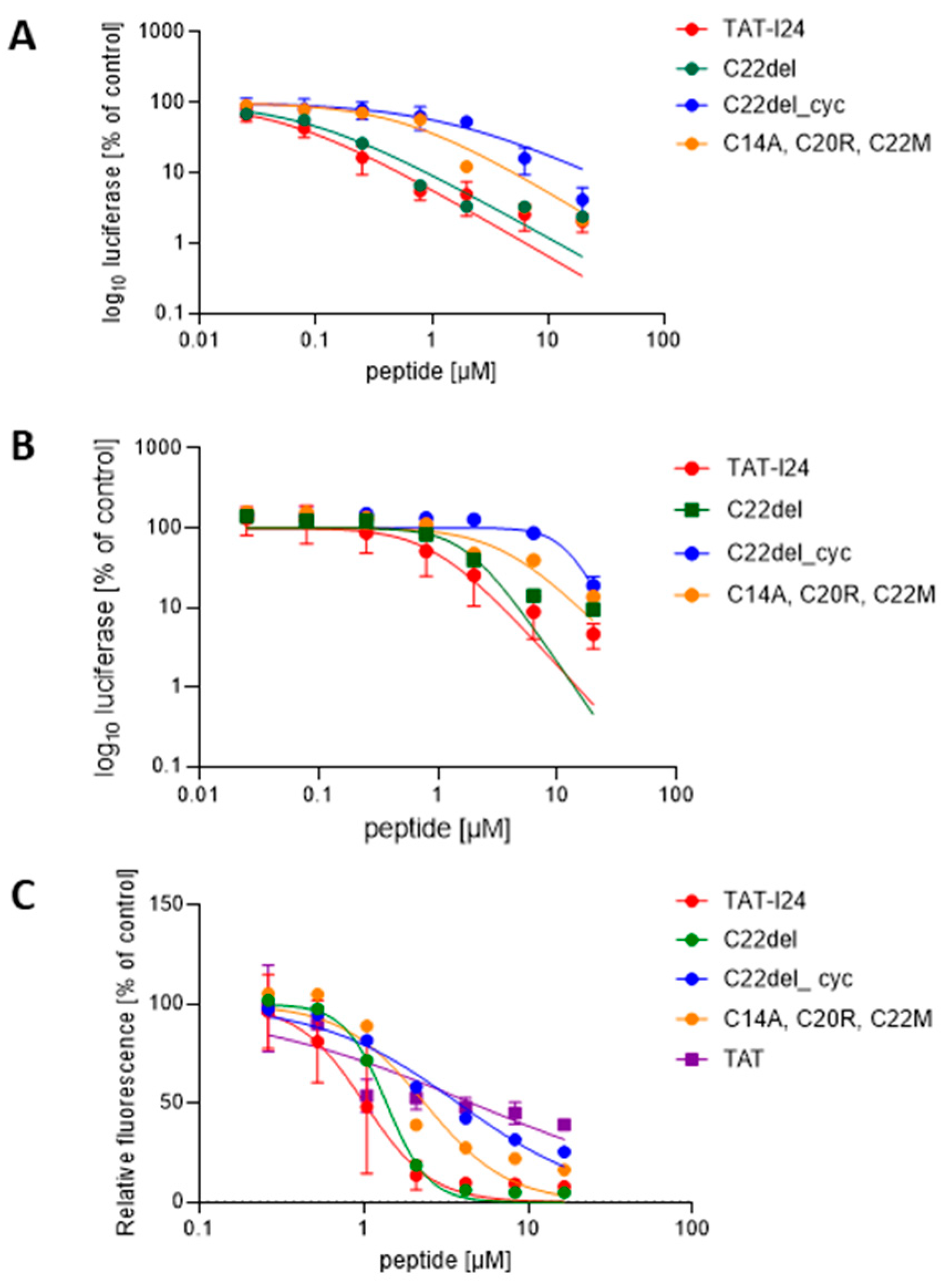
2.1. Effect of Cysteine Replacements on Antiviral Activity of TAT-I24
2.2. Computational Biology Support
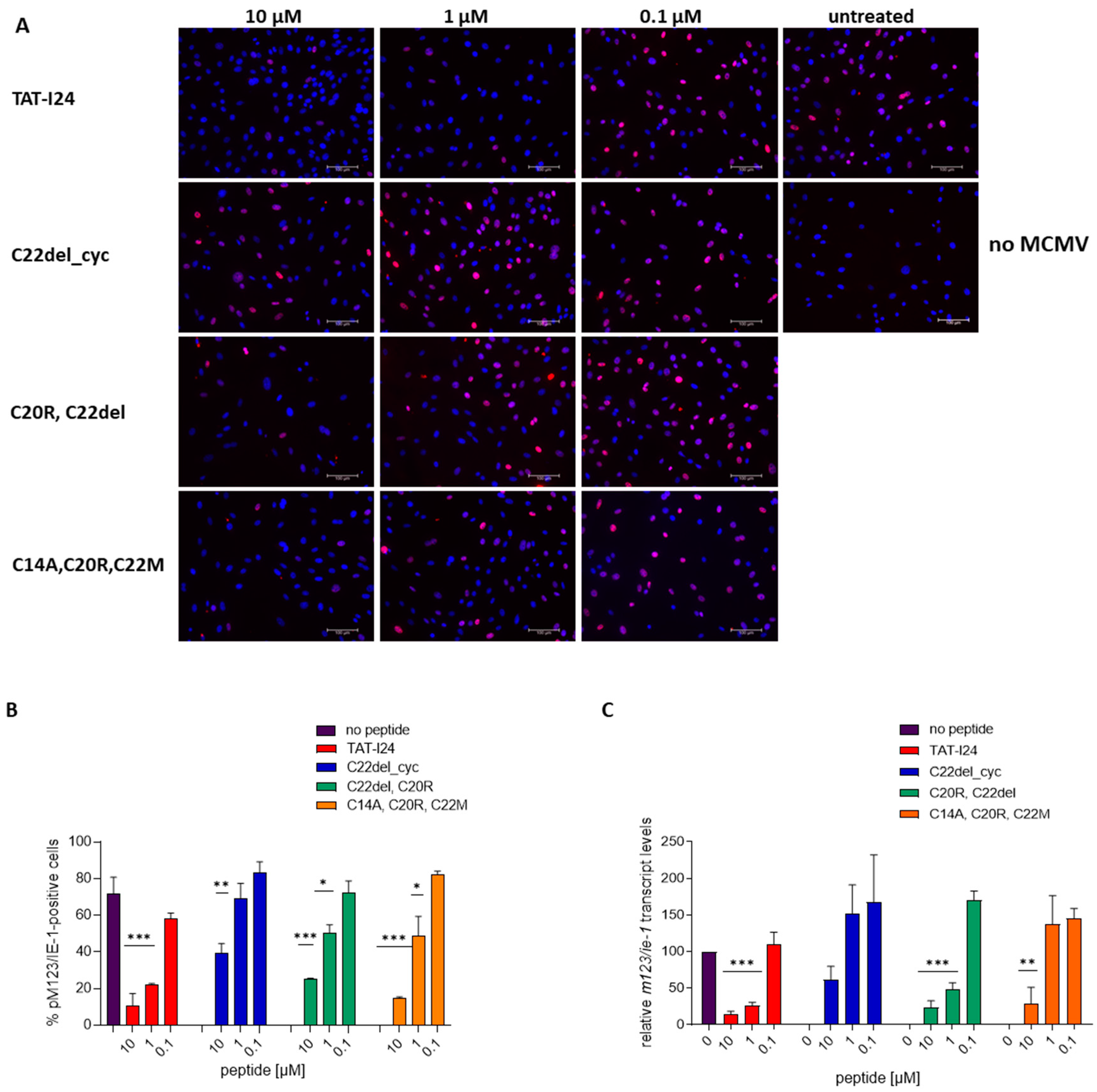
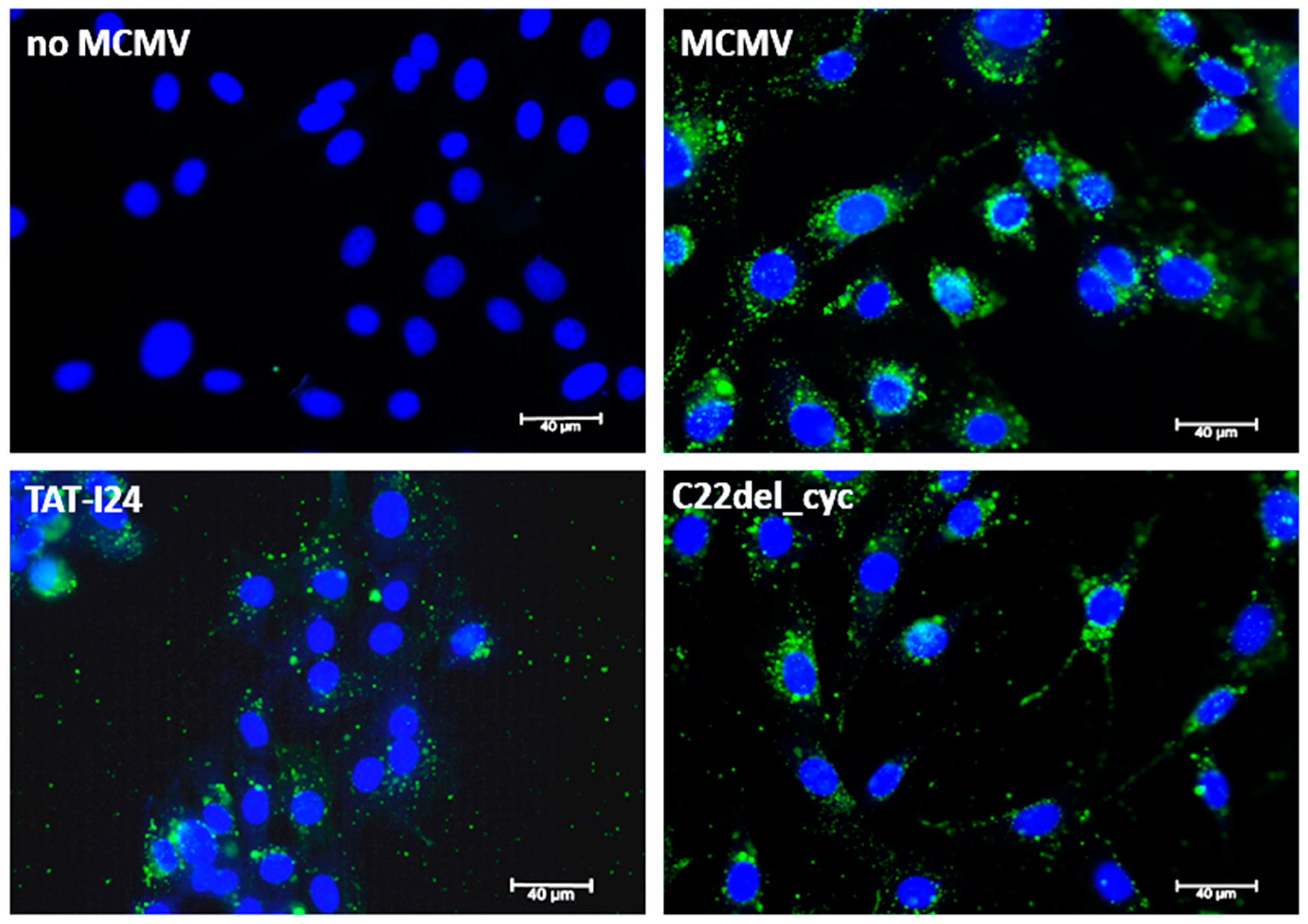
2.3. Effects of Peptide Variants on Early Stages of Virus Infection
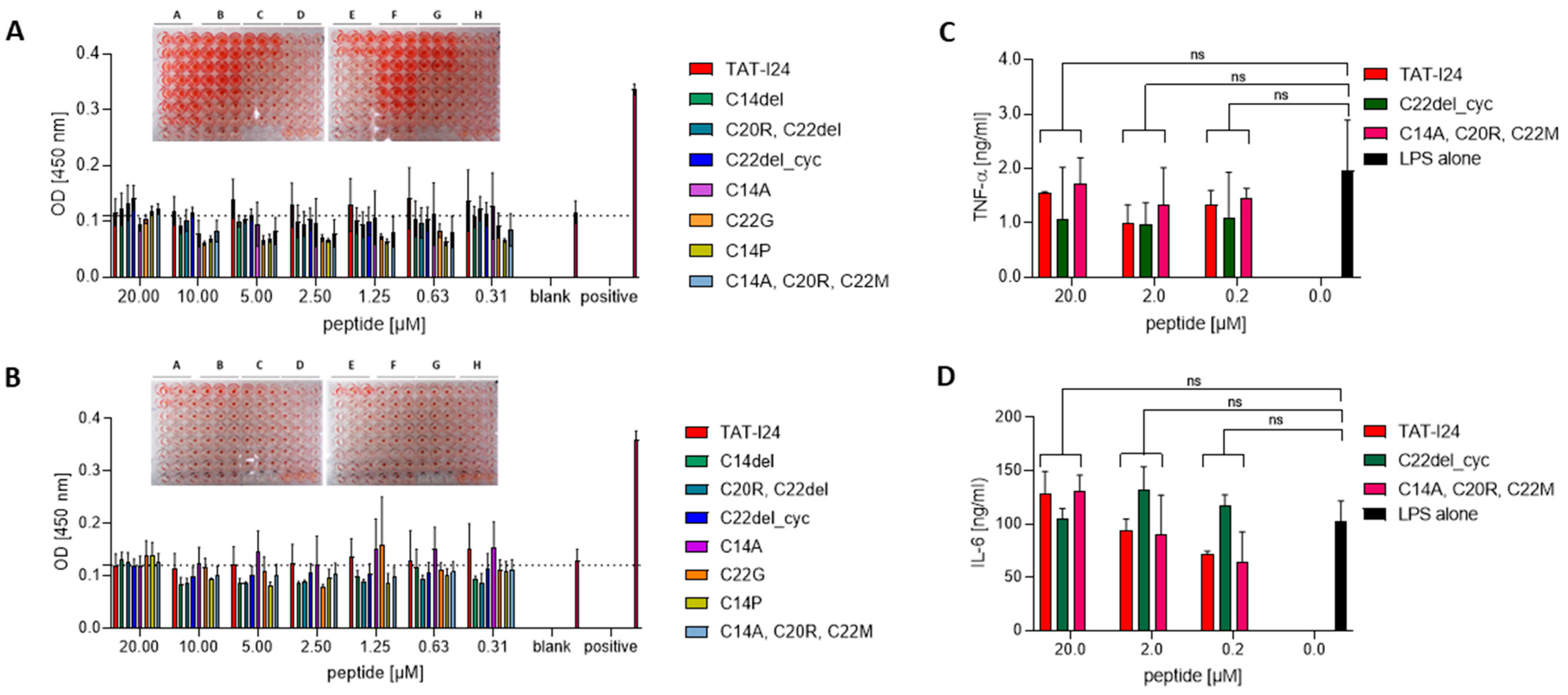
2.4. Effect of TAT-I24 and Its Variants on Blood Cells
2.5. Biochemical Analysis of TAT-I24 and Variant C14del
3. Materials and Methods
3.1. Peptides
3.2. Cell Culture
3.3. Baculovirus
3.4. Mouse Cytomegalovirus
3.5. Staining and Microscopy
3.6. RNA Isolation, cDNA Synthesis and Quantitative Real-Time PCR
3.7. Cell Viability Assay
3.8. DNA Binding Assay
3.9. Isolation of Blood Cells and Hemolysis Assay
3.10. Isolation and Stimulation of PBMCs
3.11. Enzyme-Linked Immunosorbent Assay (ELISA)
3.12. SDS-PAGE and Western Blot Analysis
3.13. Molecular Modeling
3.14. Data Analysis
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Clercq, E.; Field, H.J. Antiviral Prodrugs—The Development of Successful Prodrug Strategies for Antiviral Chemotherapy. Br. J. Pharmacol. 2006, 147, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Littler, E.; Oberg, B. Achievements and Challenges in Antiviral Drug Discovery. Antivir. Chem. Chemother. 2005, 16, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Adamson, C.S.; Chibale, K.; Goss, R.J.M.; Jaspars, M.; Newman, D.J.; Dorrington, R.A. Antiviral Drug Discovery: Preparing for the next Pandemic. Chem. Soc. Rev. 2021, 50, 3647–3655. [Google Scholar] [CrossRef] [PubMed]
- Adalja, A.; Inglesby, T. Broad-Spectrum Antiviral Agents: A Crucial Pandemic Tool. Expert. Rev. Anti-Infect Ther. 2019, 17, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-D.; Meng, W.; Wang, X.-J.; Wang, H.-C.R. Broad-Spectrum Antiviral Agents. Front. Microbiol. 2015, 6, 517. [Google Scholar] [CrossRef]
- Chitalia, V.C.; Munawar, A.H. A Painful Lesson from the COVID-19 Pandemic: The Need for Broad-Spectrum, Host-Directed Antivirals. J. Transl. Med. 2020, 18, 390. [Google Scholar] [CrossRef]
- Mousavi Maleki, M.S.; Sardari, S.; Ghandehari Alavijeh, A.; Madanchi, H. Recent Patents and FDA-Approved Drugs Based on Antiviral Peptides and Other Peptide-Related Antivirals. Int. J. Pept. Res. Ther. 2023, 29, 5. [Google Scholar] [CrossRef]
- Agarwal, G.; Gabrani, R. Antiviral Peptides: Identification and Validation. Int. J. Pept. Res. Ther. 2021, 27, 149–168. [Google Scholar] [CrossRef]
- Gao, B.; Zhao, D.; Li, L.; Cheng, Z.; Guo, Y. Antiviral Peptides with in Vivo Activity: Development and Modes of Action. Chempluschem 2021, 86, 1547–1558. [Google Scholar] [CrossRef]
- Vanzolini, T.; Bruschi, M.; Rinaldi, A.C.; Magnani, M.; Fraternale, A. Multitalented Synthetic Antimicrobial Peptides and Their Antibacterial, Antifungal and Antiviral Mechanisms. Int. J. Mol. Sci. 2022, 23, 545. [Google Scholar] [CrossRef]
- Vilas Boas, L.C.P.; Campos, M.L.; Berlanda, R.L.A.; de Carvalho Neves, N.; Franco, O.L. Antiviral Peptides as Promising Therapeutic Drugs. Cell. Mol. Life Sci. 2019, 76, 3525–3542. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Chou, Y.Y.; Chang, T.L. Defensins in Viral Infections. J. Innate Immun. 2009, 1, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.S.; Wiens, M.E.; Smith, J.G. Antiviral Mechanisms of Human Defensins. J. Mol. Biol. 2013, 425, 4965–4980. [Google Scholar] [CrossRef] [PubMed]
- Gwyer Findlay, E.; Currie, S.M.; Davidson, D.J. Cationic Host Defence Peptides: Potential as Antiviral Therapeutics. BioDrugs 2013, 27, 479–493. [Google Scholar] [CrossRef]
- Chessa, C.; Bodet, C.; Jousselin, C.; Wehbe, M.; Lévêque, N.; Garcia, M. Antiviral and Immunomodulatory Properties of Antimicrobial Peptides Produced by Human Keratinocytes. Front. Microbiol. 2020, 11, 1155. [Google Scholar] [CrossRef]
- Park, M.S.; Kim, J., II; Lee, I.; Park, S.; Bae, J.Y.; Park, M.S. Towards the Application of Human Defensins as Antivirals. Biomol. Ther. 2018, 26, 242–254. [Google Scholar] [CrossRef]
- Zupin, L.; Crovella, S. Human Defensins from Antivirals to Vaccine Adjuvants: Rediscovery of the Innate Immunity Arsenal. Protein Pept. Lett. 2021, 29, 121–124. [Google Scholar] [CrossRef]
- Skalickova, S.; Heger, Z.; Krejcova, L.; Pekarik, V.; Bastl, K.; Janda, J.; Kostolansky, F.; Vareckova, E.; Zitka, O.; Adam, V.; et al. Perspective of Use of Antiviral Peptides against Influenza Virus. Viruses 2015, 7, 5428–5442. [Google Scholar] [CrossRef]
- Hoffmann, A.R.; Guha, S.; Wu, E.; Ghimire, J.; Wang, Y.; He, J.; Garry, R.F.; Wimley, W.C. Broad-Spectrum Antiviral Entry Inhibition by Interfacially Active Peptides. J. Virol. 2020, 94, e01682-20. [Google Scholar] [CrossRef]
- Xia, S.; Lan, Q.; Pu, J.; Wang, C.; Liu, Z.; Xu, W.; Wang, Q.; Liu, H.; Jiang, S.; Lu, L. Potent MERS-CoV Fusion Inhibitory Peptides Identified from HR2 Domain in Spike Protein of Bat Coronavirus HKU4. Viruses 2019, 11, 56. [Google Scholar] [CrossRef]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S.; et al. Fusion Mechanism of 2019-NCoV and Fusion Inhibitors Targeting HR1 Domain in Spike Protein. Cell. Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef]
- Xia, S.; Yan, L.; Xu, W.; Agrawal, A.S.; Algaissi, A.; Tseng, C.-T.K.; Wang, Q.; Du, L.; Tan, W.; Wilson, I.A.; et al. A Pan-Coronavirus Fusion Inhibitor Targeting the HR1 Domain of Human Coronavirus Spike. Sci. Adv. 2019, 5, eaav4580. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (Previously 2019-NCoV) Infection by a Highly Potent Pan-Coronavirus Fusion Inhibitor Targeting Its Spike Protein That Harbors a High Capacity to Mediate Membrane Fusion. Cell Res. 2020, 30, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhou, J.; Zhang, K.; Chu, H.; Liu, D.; Poon, V.K.-M.; Chan, C.C.-S.; Leung, H.-C.; Fai, N.; Lin, Y.-P.; et al. A Novel Peptide with Potent and Broad-Spectrum Antiviral Activities against Multiple Respiratory Viruses. Sci. Rep. 2016, 6, 22008. [Google Scholar] [CrossRef] [PubMed]
- Ridyard, K.E.; Overhage, J. The Potential of Human Peptide Ll-37 as an Antimicrobial and Anti-Biofilm Agent. Antibiotics 2021, 10, 650. [Google Scholar] [CrossRef]
- Ahmed, A.; Siman-Tov, G.; Keck, F.; Kortchak, S.; Bakovic, A.; Risner, K.; Lu, T.K.; Bhalla, N.; de la Fuente-Nunez, C.; Narayanan, A. Human Cathelicidin Peptide LL-37 as a Therapeutic Antiviral Targeting Venezuelan Equine Encephalitis Virus Infections. Antivir. Res. 2019, 164, 61–69. [Google Scholar] [CrossRef]
- Gentilucci, L.; De Marco, R.; Cerisoli, L. Chemical Modifications Designed to Improve Peptide Stability: Incorporation of Non-Natural Amino Acids, Pseudo-Peptide Bonds, and Cyclization. Curr. Pharm. Des. 2010, 16, 3185–3203. [Google Scholar] [CrossRef]
- Chew, M.-F.; Poh, K.-S.; Poh, C.-L. Peptides as Therapeutic Agents for Dengue Virus. Int. J. Med. Sci. 2017, 14, 1342–1359. [Google Scholar] [CrossRef]
- Matthews, T.; Salgo, M.; Greenberg, M.; Chung, J.; DeMasi, R.; Bolognesi, D. Enfuvirtide: The First Therapy to Inhibit the Entry of HIV-1 into Host CD4 Lymphocytes. Nat. Rev. Drug Discov. 2004, 3, 215–225. [Google Scholar] [CrossRef]
- Blank, A.; Markert, C.; Hohmann, N.; Carls, A.; Mikus, G.; Lehr, T.; Alexandrov, A.; Haag, M.; Schwab, M.; Urban, S.; et al. First-in-Human Application of the Novel Hepatitis B and Hepatitis D Virus Entry Inhibitor Myrcludex B. J. Hepatol. 2016, 65, 483–489. [Google Scholar] [CrossRef]
- Ruzsics, Z.; Hoffmann, K.; Riedl, A.; Krawczyk, A.; Widera, M.; Sertznig, H.; Schipper, L.; Kapper-Falcone, V.; Debreczeny, M.; Ernst, W.; et al. A Novel, Broad-Acting Peptide Inhibitor of Double-Stranded DNA Virus Gene Expression and Replication. Front. Microbiol. 2020, 11, 2934. [Google Scholar] [CrossRef] [PubMed]
- Harant, H.; Höfinger, S.; Kricek, F.; Ruf, C.; Ruzsics, Z.; Hengel, H.; Lindley, I.J.D. The Peptide TAT-I24 with Antiviral Activity against DNA Viruses Binds Double-Stranded DNA with High Affinity. Biologics 2021, 1, 41–60. [Google Scholar] [CrossRef]
- Vivès, E.; Brodin, P.; Lebleu, B. A Truncated HIV-1 Tat Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef]
- Green, M.; Loewenstein, P.M. Autonomous Functional Domains of Chemically Synthesized Human Immunodeficiency Virus Tat Trans-Activator Protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef]
- Frankel, A.D.; Pabo, C.O. Cellular Uptake of the Tat Protein from Human Immunodeficiency Virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Ignatovich, I.A.; Dizhe, E.B.; Pavlotskaya, A.V.; Akifiev, B.N.; Burov, S.V.; Orlov, S.V.; Perevozchikov, A.P. Complexes of Plasmid DNA with Basic Domain 47-57 of the HIV-1 Tat Protein Are Transferred to Mammalian Cells by Endocytosis-Mediated Pathways. J. Biol. Chem. 2003, 278, 42625–42636. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.; Seelig, J. High Affinity of the Cell-Penetrating Peptide HIV-1 Tat-PTD for DNA. Biochemistry 2007, 46, 8138–8145. [Google Scholar] [CrossRef]
- Richard, J.P.; Melikov, K.; Brooks, H.; Prevot, P.; Lebleu, B.; Chernomordik, L.V. Cellular Uptake of Unconjugated TAT Peptide Involves Clathrin-Dependent Endocytosis and Heparan Sulfate Receptors. J. Biol. Chem. 2005, 280, 15300–15306. [Google Scholar] [CrossRef]
- Christianson, H.C.; Belting, M. Heparan Sulfate Proteoglycan as a Cell-Surface Endocytosis Receptor. Matrix Biol. 2014, 35, 51–55. [Google Scholar] [CrossRef]
- Ziegler, A.; Seelig, J. Interaction of the Protein Transduction Domain of HIV-1 TAT with Heparan Sulfate: Binding Mechanism and Thermodynamic Parameters. Biophys. J. 2004, 86, 254–263. [Google Scholar] [CrossRef]
- Vivès, R.R.; Lortat-Jacob, H.; Chroboczek, J.; Fender, P. Heparan Sulfate Proteoglycan Mediates the Selective Attachment and Internalization of Serotype 3 Human Adenovirus Dodecahedron. Virology 2004, 321, 332–340. [Google Scholar] [CrossRef]
- Dechecchi, M.C.; Tamanini, A.; Bonizzato, A.; Cabrini, G. Heparan Sulfate Glycosaminoglycans Are Involved in Adenovirus Type 5 and 2-Host Cell Interactions. Virology 2000, 268, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chopra, P.; Li, X.; Bouwman, K.M.; Tompkins, S.M.; Wolfert, M.A.; De Vries, R.P.; Boons, G.J. Heparan Sulfate Proteoglycans as Attachment Factor for SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Duisit, G.; Saleun, S.; Douthe, S.; Barsoum, J.; Chadeuf, G.; Moullier, P. Baculovirus Vector Requires Electrostatic Interactions Including Heparan Sulfate for Efficient Gene Transfer in Mammalian Cells. J. Gene Med. 1999, 1, 93–102. [Google Scholar] [CrossRef]
- Chowdhury, A.S.; Reehl, S.M.; Kehn-Hall, K.; Bishop, B.; Webb-Robertson, B.J.M. Better Understanding and Prediction of Antiviral Peptides through Primary and Secondary Structure Feature Importance. Sci. Rep. 2020, 10, 19260. [Google Scholar] [CrossRef]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic Peptides: Current Applications and Future Directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef]
- Haney, E.F.; Wu, B.; Lee, K.; Hilchie, A.L.; Hancock, R.E.W. Aggregation and Its Influence on the Immunomodulatory Activity of Synthetic Innate Defense Regulator Peptides. Cell Chem. Biol. 2017, 24, 969–980.e4. [Google Scholar] [CrossRef]
- Zapadka, K.L.; Becher, F.J.; Gomes dos Santos, A.L.; Jackson, S.E. Factors Affecting the Physical Stability (Aggregation) of Peptide Therapeutics. Interface Focus. 2017, 7, 20170030. [Google Scholar] [CrossRef]
- Maxian, T.; Gerlitz, L.; Riedl, S.; Rinner, B.; Zweytick, D. Effect of L- to D-amino Acid Substitution on Stability and Activity of Antitumor Peptide Rdp215 against Human Melanoma and Glioblastoma. Int. J. Mol. Sci. 2021, 22, 8469. [Google Scholar] [CrossRef]
- Najjar, K.; Erazo-Oliveras, A.; Brock, D.J.; Wang, T.Y.; Pellois, J.P. An L- to D-Amino Acid Conversion in an Endosomolytic Analog of the Cell-Penetrating Peptide TAT Influences Proteolytic Stability, Endocytic Uptake, and Endosomal Escape. J. Biol. Chem. 2017, 292, 847–861. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorg Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, C. A Review of Methodologies for the Detection, Quantitation, and Localization of Free Cysteine in Recombinant Proteins: A Focus on Therapeutic Monoclonal Antibodies. Front. Mol. Biosci. 2022, 9, 886417. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.B.; Braun, G.B.; She, Z.G.; Kotamraju, V.R.; Sugahara, K.N.; Teesalu, T.; Ruoslahti, E. A Free Cysteine Prolongs the Half-Life of a Homing Peptide and Improves Its Tumor-Penetrating Activity. J. Control. Release 2014, 175, 48–53. [Google Scholar] [CrossRef]
- Aubry, S.; Burlina, F.; Dupont, E.; Delaroche, D.; Joliot, A.; Lavielle, S.; Chassaing, G.; Sagan, S. Cell-surface Thiols Affect Cell Entry of Disulfide-conjugated Peptides. FASEB J. 2009, 23, 2956–2967. [Google Scholar] [CrossRef]
- Bultmann, H.; Teuton, J.; Brandt, C.R. Addition of a C-Terminal Cysteine Improves the Anti-Herpes Simplex Virus Activity of a Peptide Containing the Human Immunodeficiency Virus Type 1 TAT Protein Transduction Domain. Antimicrob. Agents Chemother. 2007, 51, 1596–1607. [Google Scholar] [CrossRef]
- Kim, W.J.; Kim, G.R.; Cho, H.J.; Choi, J.M. The Cysteine-Containing Cell-Penetrating Peptide AP Enables Efficient Macromolecule Delivery to T Cells and Controls Autoimmune Encephalomyelitis. Pharmaceutics 2021, 13, 1134. [Google Scholar] [CrossRef]
- Jha, D.; Mishra, R.; Gottschalk, S.; Wiesmüller, K.H.; Ugurbil, K.; Maier, M.E.; Engelmann, J. CyLoP-1: A Novel Cysteine-Rich Cell-Penetrating Peptide for Cytosolic Delivery of Cargoes. Bioconjug. Chem. 2011, 22, 319–328. [Google Scholar] [CrossRef]
- Trilling, M.; Le, V.T.K.; Fiedler, M.; Zimmermann, A.; Bleifuß, E.; Hengel, H. Identification of DNA-Damage DNA-Binding Protein 1 as a Conditional Essential Factor for Cytomegalovirus Replication in Interferon-γ-Stimulated Cells. PLoS Pathog. 2011, 7, e1002069. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.V.; Salguero, C.; Khan, S.N.; Meagher, J.L.; Brown, W.C.; Humbert, N.; de Rocquigny, H.; Smith, J.L.; D’Souza, V.M. HIV-1 Tat Interactions with Cellular 7SK and Viral TAR RNAs Identifies Dual Structural Mimicry. Nat. Commun. 2018, 9, 4266. [Google Scholar] [CrossRef]
- Rattay, S.; Trilling, M.; Megger, D.A.; Sitek, B.; Meyer, H.E.; Hengel, H.; Le-Trilling, V.T.K. The Canonical Immediate Early 3 Gene Product PIE611 of Mouse Cytomegalovirus Is Dispensable for Viral Replication but Mediates Transcriptional and Posttranscriptional Regulation of Viral Gene Products. J. Virol. 2015, 89, 8590–8598. [Google Scholar] [CrossRef]
- Harant, H. Selective Inhibition of Murine Cytomegalovirus Viral Gene Expression by the Antiviral Peptide TAT-I24. Int. J. Mol. Sci. 2022, 23, 7246. [Google Scholar] [CrossRef] [PubMed]
- Marcinowski, L.; Lidschreiber, M.; Windhager, L.; Rieder, M.; Bosse, J.B.; Rädle, B.; Bonfert, T.; Györy, I.; de Graaf, M.; da Costa, O.P.; et al. Real-Time Transcriptional Profiling of Cellular and Viral Gene Expression during Lytic Cytomegalovirus Infection. PLoS Pathog. 2012, 8, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; To, K.K.W.; Sze, K.-H.; Yung, T.T.-M.; Bian, M.; Lam, H.; Yeung, M.L.; Li, C.; Chu, H.; Yuen, K.-Y. A Broad-Spectrum Virus- and Host-Targeting Peptide against Respiratory Viruses Including Influenza Virus and SARS-CoV-2. Nat. Commun. 2020, 11, 4252. [Google Scholar] [CrossRef]
- Zhang, L.-J.; Wang, S.; Xia, L.; Lv, C.; Tang, H.-W.; Liang, Z.; Xiao, G.; Pang, D.-W. Lipid-Specific Labeling of Enveloped Viruses with Quantum Dots for Single-Virus Tracking. mBio 2020, 11, e00135-20. [Google Scholar] [CrossRef]
- Liu, S.L.; Wang, Z.G.; Xie, H.Y.; Liu, A.A.; Lamb, D.C.; Pang, D.W. Single-Virus Tracking: From Imaging Methodologies to Virological Applications. Chem. Rev. 2020, 120, 1936–1979. [Google Scholar] [CrossRef] [PubMed]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Yuan, L.; Chu, Q.; Yang, B.; Zhang, W.; Sun, Q.; Gao, R. Purification and Identification of Anti-Inflammatory Peptides from Sturgeon (Acipenser Schrenckii) Cartilage. Food Sci. Hum. Wellness 2023, 12, 2175–2183. [Google Scholar] [CrossRef]
- Pahar, B.; Madonna, S.; Das, A.; Albanesi, C.; Girolomoni, G. Immunomodulatory Role of the Antimicrobial Ll-37 Peptide in Autoimmune Diseases and Viral Infections. Vaccines 2020, 8, 517. [Google Scholar] [CrossRef]
- Wu, D.; Fu, L.; Wen, W.; Dong, N. The Dual Antimicrobial and Immunomodulatory Roles of Host Defense Peptides and Their Applications in Animal Production. J. Anim. Sci. Biotechnol. 2022, 13, 141. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, C.F.; Liao, Y. Di Fetal Bovine Serum Albumin Inhibits Antimicrobial Peptide Activity and Binds Drug Only in Complex with A1-Antitrypsin. Sci. Rep. 2021, 11, 1267. [Google Scholar] [CrossRef]
- Sæbø, I.P.; Bjørås, M.; Franzyk, H.; Helgesen, E.; Booth, J.A. Optimization of the Hemolysis Assay for the Assessment of Cytotoxicity. Int. J. Mol. Sci. 2023, 24, 2914. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://amberhub.chpc.utah.edu/detect-aminoacid-interactions (accessed on 25 September 2024).
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence | HEK293 IC50 [µM] | HEK293 CC50 [µM] | SI * | NIH/3T3 IC50 [µM] | DNA Binding IC50 [µM] |
|---|---|---|---|---|---|---|
| TAT-I24 | GRKKRRQRRRPPQCLAFYACFC | 0.05 | 124 | 2480 | 0.91 | 1.01 |
| C14A | GRKKRRQRRRPPQALAFYACFC | 0.08 | 67 | 838 | 0.92 | 2.11 |
| C14L | GRKKRRQRRRPPQLLAFYACFC | 0.05 | 53 | 1060 | 0.57 | 2.19 |
| C14G | GRKKRRQRRRPPQGLAFYACFC | 0.10 | 50 | 500 | 1.07 | 2.36 |
| C14R | GRKKRRQRRRPPQRLAFYACFC | 0.10 | 106 | 1060 | 2.95 | 1.44 |
| C14P | GRKKRRQRRRPPQPLAFYACFC | 0.09 | 92 | 1022 | 3.51 | 2.54 |
| C14del | GRKKRRQRRRPPQ-LAFYACFC | 0.10 | 37 | 370 | 0.92 | 1.26 |
| C22G | GRKKRRQRRRPPQCLAFYACFG | 0.05 | 71 | 1420 | 0.55 | 1.61 |
| C22S | GRKKRRQRRRPPQCLAFYACFS | 0.05 | 99 | 1980 | 1.19 | 1.35 |
| C22del | GRKKRRQRRRPPQCLAFYACF- | 0.08 | n.d. ** | n.d. | 1.65 | 1.38 |
| C22del_cyc *** | GRKKRRQRRRPPQCLAFYACF- | 1.41 | n.d. | n.d. | 12.17 | 3.77 |
| C20R, C22del | GRKKRRQRRRPPQCLAFYARF- | 0.05 | 66 | 1320 | 1.91 | 2.47 |
| C14A, C20R, C22M | GRKKRRQRRRPPQALAFYARFM | 0.58 | 63 | 109 | 3.59 | 2.35 |
| TAT | GRKKRRQRRRPPQ | [31] | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziu, T.; Sambur, E.; Ruzsics, Z.; Hengel, H.; Grabherr, R.; Höfinger, S.; Harant, H. In Vitro Profiling of the Antiviral Peptide TAT-I24. Int. J. Mol. Sci. 2024, 25, 10463. https://doi.org/10.3390/ijms251910463
Ziu T, Sambur E, Ruzsics Z, Hengel H, Grabherr R, Höfinger S, Harant H. In Vitro Profiling of the Antiviral Peptide TAT-I24. International Journal of Molecular Sciences. 2024; 25(19):10463. https://doi.org/10.3390/ijms251910463
Chicago/Turabian StyleZiu, Theodhora, Ezgi Sambur, Zsolt Ruzsics, Hartmut Hengel, Reingard Grabherr, Siegfried Höfinger, and Hanna Harant. 2024. "In Vitro Profiling of the Antiviral Peptide TAT-I24" International Journal of Molecular Sciences 25, no. 19: 10463. https://doi.org/10.3390/ijms251910463
APA StyleZiu, T., Sambur, E., Ruzsics, Z., Hengel, H., Grabherr, R., Höfinger, S., & Harant, H. (2024). In Vitro Profiling of the Antiviral Peptide TAT-I24. International Journal of Molecular Sciences, 25(19), 10463. https://doi.org/10.3390/ijms251910463








