Abstract
The diversity of phage-related sequences (PRSs) and their site-specific integration into the genomes of nonpathogenic, agriculturally valuable, nitrogen-fixing root nodule bacteria, such as Sinorhizobium meliloti, were evaluated in this study. A total of 314 PRSs, ranging in size from 3.24 kb to 88.98 kb, were identified in the genomes of 27 S. meliloti strains. The amount of genetic information foreign to S. meliloti accumulated in all identified PRSs was 6.30 Mb. However, more than 53% of this information was contained in prophages (Phs) and genomic islands (GIs) integrated into genes encoding tRNAs (tRNA genes) located on the chromosomes of the rhizobial strains studied. It was found that phiLM21-like Phs were predominantly abundant in the genomes of S. meliloti strains of distant geographical origin, whereas RR1-A- and 16-3-like Phs were much less common. In addition, GIs predominantly contained fragments of phages infecting bacteria of distant taxa, while rhizobiophage-like sequences were unique. A site-specific integration analysis revealed that not all tRNA genes in S. meliloti are integration sites, but among those in which integration occurred, there were “hot spots” of integration into which either Phs or GIs were predominantly inserted. For the first time, it is shown that at these integration “hot spots”, not only is the homology of attP and attB strictly preserved, but integrases in PRSs similar to those of phages infecting the Proteobacteria genera Azospirillum or Pseudomonas are also present. The data presented greatly expand the understanding of the fate of phage-related sequences in host bacterial genomes and also raise new questions about the role of phages in bacterial–phage coevolution.
1. Introduction
The successful ecological adaptation of bacteria may depend on the ability to expand the bacterial genome with genetic material, which the cell can obtain through horizontal gene transfer processes, including transduction [,,]. Phage DNA is integrated into the bacterial genome through site-specific recombination and can remain there for a long time as part of the replicon that was targeted for integration; alternatively, if its mobility is lost, the “grounding” of the phage DNA sequence in the host bacterial genome occurs [,]. In bacteria, phage-related sequences are present in significant numbers and can account for up to 10–20% of the genome [,,]. For example, up to 18 and 6 of such sequences are found in E. coli and Desulfovibrio vulgaris strains, respectively, and their presence in a genome does not depend on the genetic background [,,].
Once integrated into the host bacterial genome, temperate phages can persist for a long time as prophages that evolve further through intragenomic recombination []. Prophages can be subdivided into three groups as follows: (i) intact prophages (int-Phs), which are sequences capable of excision and existence as part of a viral particle; (ii) incomplete prophages (inc-Phs), which are sequences that have lost some or all structural genes responsible for excision and/or virulence; and (iii) questionable prophages (q-Phs), which cannot be classified as intact or incomplete [,]. All of these sequences (int-Ph, q-Ph, and inc-Ph) have different levels of integrity and can be grouped together as “phage-related sequences” (PRSs). Genomic islands (GIs) containing not only genes of phage origin but also a significant number of genes of bacterial origin can also belong to this group []. This is consistent with the hypothesis that genomic islands originated from prophages or plasmids [,].
The integration or loss of PRSs may contribute to intraspecific variability in bacterial strains, along with mutations and genomic rearrangements [,,,,,]. The presence of genes or gene blocks in phage-related sequences can confer new, previously absent properties to the host bacterium (lysogenic conversion), which has been studied mainly in pathogenic bacteria and single strains of nonpathogenic bacteria. For example, changes in bacterial properties such as pathogenicity [,,,], antibiotic resistance [], immunity [,,], symbiotic activity [,,], and viability and adaptability to environmental conditions [,,,,,] have been observed. However, the expression level of the introduced genetic material depends on the intact genomic background of the bacterium, which may or may not support the expression or function of the recruited genes, consistent with []. Prophages have been shown to affect the activity of recombination processes in the host genome [,,,], which may play a significant role in rapid bacterial evolution. In addition, the presence of prophages can influence the ecology of microbiocenosis. Thus, phage infection results in the immunization of bacterial hosts and protects them from superinfection by other closely related or unrelated phages []. All of these effects contribute to the viability of potential host bacteria, making the study of antiphage defense systems in the host bacterial genome all the more relevant [,,,,,].
The integration of a PRS into the host bacterial genome occurs through site-specific recombination with the participation of enzymes (Figure 1a), which are conserved site-specific tyrosine and serine recombinases belonging to two unrelated families. These recombinases are widely distributed in bacterial and viral genomes and are often referred to as “phage-like integrases”, responsible for the mobility of mobile genetic elements of different types [,].
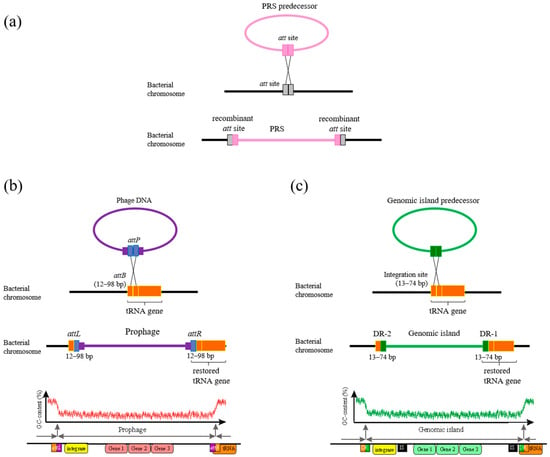
Figure 1.
General scheme of PRS integration into bacteria genome (a) adopted from []. PRS predecessor—the PRS before integration into a bacterial chromosome. att site—attachment site for PRS integration. Scheme of integration of a phage (b) and a genomic island (c) into the tRNA gene, designed according to the data obtained in this work for S. meliloti strains and adopted from [,,]. attP, attB—sites for phage integration (see text); attL, attR—recombinant sites (see text); DR-1 and -2—short direct repeat sequences flanking the GI (see text).
In most cases, the attachment site (attB) of a tyrosine integrase in the bacterial genome corresponds to a region of a gene encoding transfer or transfer–messenger RNA (tRNA or tmRNA gene, respectively) and that fact has the biological importance of preserving the integrity and function of essential tRNA genes after PRS integration (Figure 1) [,,,,]. Attachment sites are usually short 15–30 bp sequences that typically correspond to the sequence at the 3′ end of the tRNA gene and, in some cases, attB sequences may extend beyond the gene boundary. However, it has been suggested that the local genetic context of the attB site may not be crucial for PRS integration [].
The attachment site of a PRS (attP) undergoes recombination with an attB site to generate the recombinant sites attL and attR (left and right sites), which are direct repeats flanking the PRS (DR) integrated into the bacterial genome (Figure 1a). The attP site in the phage genome is much more complex and longer than the attB site and can bind to recombination proteins [,,].
The fact that the site-specific integration of phage sequences occurs precisely in tRNA genes may be due to a number of factors, including the conservation of evolutionarily ancient genes and their small size. This is thought to be responsible for minimizing the size of the host bacterial DNA sequences used in attP, which is necessary to restore gene function in the case of phage sequence integration (Figure 1) []. For some species of bacteria, not all tRNA genes are integration sites, and there may be other sites in the genome where PRS integration is more active [,].
Recently, it was shown that the integration of PRSs into the chromosome of Alphaproteobacteria of the Rhodobacteraceae family depended on the structural organization of the chromosome and, above all, on the location of oriC (replication origin) and terC (replication terminus) [,]. The sequence of the dnaA-encoding gene is among the conserved, taxonomically significant markers used to determine oriC and thus is the starting point for chromosomal gene coordinates [,]. The location of terC is less well studied, and its location is not universal; therefore, techniques based on the analysis of chromosome nucleotide composition asymmetry are used to search for potential terC sequences. Both of these regions have been described for a limited number of bacteria [].
Information on the integration sites and origins of phage sequences, as well as on the structural organization of chromosomes, is mainly known for pathogenic bacterial strains including strains of Pseudomonas, Staphylococcus, Streptococcus, Bacillus, and E. coli [,,,,]. For nonpathogenic species such as Sinorhizobium meliloti, which are nitrogen-fixing symbionts of legume crops, such data are almost nonexistent.
S. meliloti are agriculturally valuable alfalfa symbionts and typical representatives of the soil microbiome. These bacteria have a multicomponent genome that includes a chromosome and two megaplasmids, and they often contain nonsymbiotic plasmids that vary greatly in number and size. There are data on the structure of the chromosome of the model strain S. meliloti 1021 (Rm1021), as well as on the locations of extended chromosomal genomic islands, which are presumably of phage origin [,,]. The predicted functions of the ORFs of S. meliloti 1021 genomic islands showed that these sequences are similar to the ORFs of bacteria belonging to at least 10 different classes in six different phyla, which, together, suggests the active participation of GIs in horizontal transfer processes []. GIs of similar localization, like GIs of Rm1021, were detected in almost half of the S. meliloti strains isolated from soil samples collected at the origin of plant diversity in the Northern Caucasus as well from an alfalfa introgressive hybridization center in Mugodzhary, NW Kazakhstan [,]. We found that some other S. meliloti isolates may have GIs whose localization differs from the above []. Thus, there is some understanding of the structure, location, and functional significance of genomic islands in S. meliloti genomes, but little is known about the origin, diversification, and specificity of GI integration into the rhizobial genome.
The aim of our study was to estimate the abundance of phage-related sequences (PRSs) in S. meliloti bacterial genomes based on analyses of full-genome data of strains independently sampled from geographically distant areas, according to the NCBI database (27 strains). For the first time, data on the site-specific integration of PRSs allowed us to identify integration hot spots on the S. meliloti chromosome. As a result of this study, the mechanisms underlying the integration of phage-originating sequences into bacterial genomes are better understood, allowing us to place a new focus on the evolution of bacterial–phage interactions.
2. Results
2.1. Phage-Related Sequences (PRSs)
Three hundred fourteen phage-related sequences (PRSs) were detected in the genomes of 27 strains of Sinorhizobium meliloti (Table 1), independently sampled from geographically distant areas (see Section 4 Materials and Methods). That is, on average, there could be 11 ± 1.1 PRSs per genome of S. meliloti, as predicted using PHASTER, PHASTEST and Islander (see Section 4 Materials and Methods). We identified seven inc-Phs in the genome of S. meliloti 1021—one on the chromosome, four on pSymA, and two on pSymB—in addition to the three extended genomic islands described previously [].

Table 1.
Evaluation of the occurrence of phage-related sequences (PRSs) in genomes (gray squares) and on chromosomes (white squares) in S. meliloti strains.
PRSs, considered “foreign sequences” in rhizobial genomes, varied significantly in length, ranging from 3.24 to 88.98 kb. On average, there were 233 ± 25 kb of PRSs per genome (or 3.3 ± 0.3% of genome size), with minimum and maximum PRS lengths of 21.28 kb and 576.29 kb (0.8 to 7.9% of genome size), respectively. More than half of the PRSs were represented by inc-Phs (frequency of 0.52), and int-Phs and GIs occurred with equal frequencies (0.17). In addition, q-Phs were detected at a frequency of 0.14 (Table 1). The total length of all 314 PRSs was 6.30 Mb and included 7.4 thousand ORFs.
PRSs could be detected on any replicon in the rhizobial genome, and their distribution among replicons was not random (Χ2 = 22.0, P = 6.5 × 10−5). PRSs were found twice as frequently on chromosomes as on the megaplasmids pSymA and pSymB, on which they occurred at similar frequencies (0.42 and an average frequency of 0.21, respectively). PRSs were also found on 28 cryptic plasmids (frequency of 0.17), which were identified in the genomes of 16 of the 27 strains studied.
The analysis of the chromosome structure was a priority. Of the 314 PRSs, 132 PRSs with a total length of 4.09 Mb (i.e., 64.8% of the total amount of detected foreign DNA of phage origin) fell on the chromosomes of the studied strains (Table S1). On average, 152 ± 16 kb of foreign DNA was found per S. meliloti chromosome (the minimum and maximum average sizes were 12.2 and 307.9 kb, respectively). We found that 88 of the 132 PRSs were site-specifically integrated into tRNA genes. These 88 PRSs accounted for 53.5% of the detected foreign DNA of phage origin (Table S1). These PRSs were represented by 51 GIs (frequency of 0.58) and 37 prophages (Phs; Table 1).
Of the 37 prophages, 27 (frequency of 0.73) were similar to only three rhizobiophages (Table 2). The vast majority of these prophages were similar to the Sinorhizobium phage phiLM21 (frequency of 0.67); much less frequently, the phages were similar to Rhizobium phage 16-3 or phage RR1-A (with equal frequencies, 0.17). The other 10 of the 37 phages were similar to phages infecting Mesorhizobium, Brucella, Sulfitobacter, and Enterobacteria (Table 2), which are members of different orders including Hyphomicrobiales (Mesorhizobium and Brucella), Rhodobacterales, and Enterobacteriales, respectively.

Table 2.
Similarity of prophages integrated into tRNA genes of S. meliloti to particular phages.
We found it interesting that prophages similar to phages phiLM21 and RR1-A were only int-Ph (Table 2). In contrast, the following sequences similar to Rhizobium phage 16-3 were found with varying degrees of integrity: int-Phs, inc-Phs, and q-Phs (Table 2). The int-Phs were single sequences similar to phages infecting Mesorhizobium, Brucella, or Sulfitobacter, while sequences similar to phages infecting Enterobacteria occurred only as q-Phs (Table 2 and Table S2).
The analysis of the 51 GI sequences showed that 32 of them contained no sequences of phage origin, with the exception of the gene encoding integrase (see below). The analysis of the other 19 GIs (frequency of 0.37) showed that they included short sequences of phage origin annotated as inc-Phs and q-Phs, according to PHASTER and PHASTEST, but the extent of these sequences was less than 50% of the length of the genomic island.
Rhizobiophage-like sequences were in only 8 of the 19 GIs discussed above. These GIs contained single inc-Ph sequences similar to Sinorhizobium phage phiLM21, Rhizobium phage 16-3, and Sinorhizobium phage ort11, as well as inc-Phs similar to two Sinorhizobium phages, phiN3 and phiM7, and four q-Ph sequences similar to Rhizobium phage vB_RleM_PPF1. However, GIs may contain several distinct prophages at the same time. For example, the GI of strain AK21 has two inc-Phs simultaneously, one similar to Sinorhizobium phage phiLM21 and the other one similar to Sulfitobacter phage NYA_2014a. Similarity was found between phage sequences in GIs and a wide variety of phages infecting bacteria of distant taxa. For example, one of the q-Ph sequences similar to Rhizobium phage vB_RleM_PPF1 (strain T073) was similar to Azospirillum phage Cd and Rhodobacter phage RcapNL (Table S2).
The remaining 11 of the 19 GIs contained sequences similar to phages infecting Brucella, Sulfitobacter, and Enterobacter species, as well as sequences similar to three different Stx-encoding Escherichia phages (F451, C1717, SH2026) and Ralstonia phage RsoM1USA (USDA1106 strain). In addition, a q-Ph similar to a phage infecting Pelagibacter was present in one of these GIs (Table S2).
In summary, S. meliloti chromosomes contain prophages integrated into tRNA genes, which differ in their degree of structural integrity, and GIs with and without phage-like sequences. It was observed that int-Phs did not occur in the GIs: only inc-Phs and q-Phs were present in a ratio of 14:5. These sequences were mostly similar to phages infecting bacteria of taxonomically distant genera (frequency of 0.68). Interestingly, int-Phs were predominantly rhizobiophage-like and were integrated into tRNA genes, whereas rhizobiophage-like sequences were extremely rare in GIs (frequencies of 0.31 and 0.08, respectively).
2.2. Site-Specific Integration of PRSs
All 37 Phs and 51 GIs integrated into tRNA genes were found to be flanked by two short direct repeats (DRs) ranging in size from 12 to 98 bp, which are presumably recombinant sites named attL and attR (see Introduction) (Figure 1b,c). In this work, one of the paired DR sequences corresponding to the sequence at the 3′ or 5′ end of a tRNA gene was labeled attR, whereas the opposite DR sequence was always labeled attL (Figure 1b,c). attR matched the sequence at the 3′ end of the encoding gene in 77 of 88 PRSs (frequency of 0.88); in only nine cases did attR match the sequence at the 5’ end of the tRNA gene (frequency of 0.10) (Table S2). In all of these cases, the integrity of the tRNA genes was always preserved (Figure 1b,c).
Two cases were identified where attR corresponded to the sequence of the central part of the tRNA gene, resulting in a disruption of the tRNA gene structure. In these cases, the integration of int-Phs similar to Mesorhizobium phage vB_MloP_Lo5R7ANS led to the inactivation of the gene encoding tRNALeu(UAG)026 (strains AK21 and RU11/001). However, in both cases, a gene functionally similar to tRNA was present in the int-Ph (see below).
The site-specific integration of PRSs occurred in only 25 out of all 52 tRNA genes on the chromosome. The sequence analysis of the genes encoding the corresponding tRNAs showed that their sequences were identical in each of the 27 strains studied with few exceptions. The sequences of three genes encoding tRNAArg(ACG), tRNALeu(CAA), and tRNAGlu(CTC) (strains 1021, AK170, and USDA1021, respectively) had SNPs. As a result, the average identity of these sequences was 98.7% (coverage 100%) relative to the corresponding sequences among all of the strains studied. In certain S. meliloti strains, the number of tRNA genes could be higher or lower than in the reference strain (1021) because of intrachromosomal rearrangements affecting the regions containing the tRNA genes (strains SM11, KH46, USDA1157, T073, B399). Thus, in strain B399, 60 tRNA genes were detected, which resulted from the duplication of a 223 kb fragment containing 8 tRNA genes (018-025) (Table 3). This is the highest number of tRNA genes among the S. meliloti genomes studied. The lowest number of tRNA genes, namely, 46 and 48, as in strains M270 and USDA1021, respectively, was due to the deletion of extended regions containing the corresponding genes (Table S2).

Table 3.
Essential tRNA genes of the reference strain S. meliloti 1021.
The locations of the above 25 tRNA genes were estimated relative to the structural organization of the chromosomes of the analyzed S. meliloti strains. The sequence of the oriC region was identical to that of the reference strain (477 bp) in 18 out of 27 cases, whereas in the remaining cases, SNPs, deletions, and/or insertions were detected (identity 97.91–99.79%; π = 4.28 × 10−3 (see Section 4 Materials and Methods)). Thus, the first nucleotide of the oriC region was taken as a reference point for the alignment of the chromosomes [,]. The terC region was defined for each studied chromosome, and its distance from oriC was 170° ± 0.6°. The terC region was determined for each chromosome examined, and its distance from oriC was 170° ± 0.6°. An exception in the position of terC relative to oriC was detected only in cases of genomic rearrangements affecting extended sequences. Thus, terC in strains M270, SM11, B399, and USDA1021 had coordinates 185°, 215°, 192°, and 156°, respectively. The length of the terC region was 8.26 kb, and the level of similarity to the reference was 99.8–100% in 10 out of 27 cases. In the remaining 17 cases, the size of the terC region ranged from 6.60 to 11.71 kb. The variability in the size of this region was due to the presence of one to four ORFs encoding transposases or hypothetical proteins.
Determining the oriC-terC axis made it possible to unify the locations of all 52 tRNA genes on the chromosomes and to show that the order of their locations is identical in all studied S. meliloti strains. An analysis of the locations of the above 25 tRNA genes into which PRS insertions occurred showed that they are relatively evenly distributed on the chromosome, with 13 and 12 of these tRNA genes on the plus and minus strands of DNA, respectively. Conversely, the distribution of all 52 tRNA genes between the plus and minus strands of the chromosome was significantly asymmetric (Χ2 = 15.8, P = 1.3 × 10−3) (Figure 2). This fact allowed us to unify the designation of genes encoding tRNAs and to give each gene its own ordinal number in the direction from oriC to terC, namely, 001-052 (further in the text, only the numbers of the corresponding genes will predominantly be used) (Figure 2; Table 3).
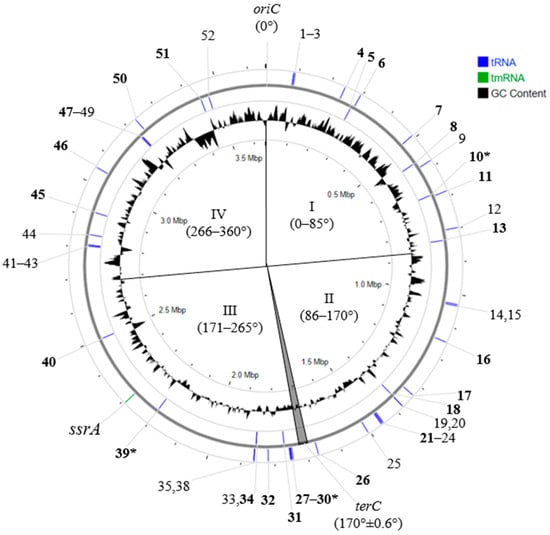
Figure 2.
Localization of essential tRNA genes on S. meliloti 1021 chromosome. Designations: 1-52—ordinal numbers of tRNA genes (in the text 001-052); oriC and terC—origin and terminus of replication, respectively; I–IV—quarters of the chromosome, defined relative to oriC and terC; in bold—ordinal number of tRNA genes with integrated PRSs; *—“hot spots” of PRS integration (genes 030 and 039) and spot of preferential integration (gene 010) (see text).
The frequency of occurrence of PRSs integrated into any of the 25 tRNA genes was significantly different from a discrete uniform distribution, according to the Chi-square test (P = 1.97 × 10−18). We found that the average frequency of occurrence of integrated PRSs in the sequences of 16 out of the 25 tRNA genes was 0.02 ± 2 × 10−3; that in the other six tRNA genes (008, 013, 016, 032, 040, and 045) was almost threefold higher at 0.05 ± 3 × 10−3. The frequency of PRSs integrated into the sequences of the last 3 of the 25 tRNA genes (010, 030, and 039) was significantly higher, equal to 0.11 ± 8 × 10−3 (Figure 3).
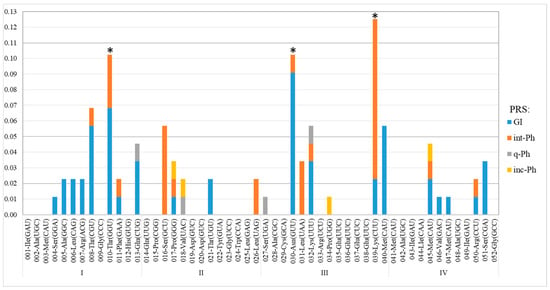
Figure 3.
Frequencies of PRS integration in essential tRNA genes of S. meliloti. Designations: along the abscissa axis—the signature combines the ordinal number of tRNA genes, with a corresponding amino acid and anticodon following the dash (see Table 3); along the abscissa axis—frequency of occurrence of PRSs integrated into different tRNA genes; *—tRNA genes at “hot spots” of integration (genes 030 and 039) and spot of preferential integration (gene 010); I–IV—chromosome quarters defined relative to oriC and terC (see Figure 2).
The analysis of PRSs integrated into the three above-mentioned tRNA-encoding genes showed that Phs similar to phage RR1-A were predominantly integrated into the gene encoding tRNAThr(GGU)010. In addition, phiLM21-like int-Phs were significantly more frequently integrated into the gene encoding tRNALys(CUU)039 (X2 = 15.95, P = 2.7 × 10−3), but similar sequences were also detected in gene 016. GIs were significantly more frequently used for integration into the sequences of the gene encoding tRNAAsn(GUU)030 (X2 = 12.8, P = 1.2 × 10−3). GIs were more frequently integrated than Phs in genes 008, 013, and 010. The occurrence of repeated integration into the same gene, namely, gene 039, supports the conclusion that integration “hot spots” are favored. Thus, in the chromosome of strain KH46, a 137.5 kb sequence was detected as integrated into the gene encoding tRNALys(CUU)039, which is flanked by 49 bp direct repeat (DR) identical to the 3′ end of the gene. There are 46 bp identical to the flanking DR just in the middle of this sequence. Apparently, the 137.5 kb sequence was formed by the sequential integration of a GI and then an int-Ph, whose putative sizes were apparently 84.2 and 53.3 kb, according to PHASTER and PHASTEST annotation (Table S2).
In conclusion, tRNA genes 010, 030, and 039 are proposed to be sites of preferential integration of PRSs, or integration “hot spots”, on the Sinorhizobium meliloti chromosome (the preferential insertion of GIs and Phs into corresponding tRNA gene sequences is a statistically reliable fact).
2.3. tRNA Genes in PRSs (tRNAPRS)
Twenty-three tRNA genes were identified in 22 PRSs integrated into the sequences of tRNA genes in 13 out of the 27 studied S. meliloti strains (Table 4). These tRNA genes (hereafter, tRNAPRS) were in addition to the tRNA genes located on the chromosomes of the strains studied. Two tRNA genes were identified in two PRSs that were also located on the chromosome but were not integrated into tRNA genes (Table 4). A total of 25 tRNAPRS genes encoded the following tRNAs: nine tRNAs fMet(CAT), eight tRNAs Met(CAT), four tRNAs Thr(GGT), two tRNAs Leu(TAG), and one tRNA each for Val(CAC) and Ser(GCT).

Table 4.
tRNA genes in PRSs in S. meliloti strains.
As a result, we showed for the first time that PRSs can contain one or two genes encoding tRNAs at the same frequency of 0.38. Cases of three or four tRNA genes in PRSs are rare occurrences (two and one strain of S. meliloti, respectively).
tRNAPRS genes were found to be 4-fold more frequent in Phs than in GIs (0.82 and 0.18, respectively). These genes could be within one or several different PRSs, similar to different rhizobiophages or phages infecting bacteria belonging to different taxa. The four tRNAPRS-encoding genes identified in the genome of strain M270 were represented by two genes encoding tRNAfMet(CAU)PRS within two phiLM21-like int-Phs and two tRNAfMet(CAU)PRS-encoding genes located within inc-Phs: one was similar to Sulfitobacter phage NYA-2014a, and the other was similar to Paracoccus phage Shpa (Table 4).
The sequence similarity in the genes encoding tRNAPRS and the corresponding chromosomal tRNA genes was assessed. As an example, two tRNAPRS-encoding genes in strain AK83 were analyzed. These are the genes encoding tRNAVal(CAC)PRS and tRNASer(GCU)PRS, each located within one of two int-Phs. One was a phiLM21-like int-Ph, and the other int-Ph was similar to both Ruegeria phage DSS3-P1 and Loktanella phage pCB2051-A simultaneously. The similarity level of the gene encoding tRNASer(GCU)PRS with the analogous chromosomal gene encoding tRNASer(GCU)016 did not exceed 96.7% (coverage 100%) but was higher with the gene encoding tRNASer(GCU) detected in the genomes of Stappia indica PHM037 and Jiella pelagia HL-NP1 (GenBank: CP046908.1 and CP114029.1, respectively; identity 98.9%; coverage 100%). For the gene encoding tRNAVal(CAC)PRS, no similar sequences were detected in the genomes of the studied S. meliloti strains or those of other bacterial species (coverage < 45%).
Another example is the analysis of tRNALeu(UAG)PRS genes present in two int-Phs similar to Mesorhizobium phage vB_MloP_Lo5R7ANS (strains AK21 and RU11/001, Table 4). The analysis showed that the similarity in the tRNALeu(UAG)PRS gene to the sequence of the corresponding gene located on the chromosome, tRNALeu(UAG)026, did not exceed 80.6%, but it was significantly higher (97.6%) when compared with the corresponding genes detected in both Octadecabacter arcticus 238 (GenBank: CP003742.1) and O. antarcticus 307 (GenBank: CP003740.1). Thus, the gene encoding tRNALeu(UAG)026 was inactivated by PRS integration (see above) but replaced by the tRNALeu(UAG)PRS gene, similar to that in another species of Alphaproteobacteria, as mentioned above.
The sequences of three genes encoding tRNAThr(GGU)PRS identified in three GIs that had inc-Phs similar to phage RR1-A (strains RU11/001, SM11, USDA1157; Table 4) were shown to be identical to each other. All three genes were identical to the gene encoding tRNAThr(GGU)PRS present in an RR1-A-like int-Ph integrated into gene 050 (strain USDA1021). All four sequences encoding tRNAThr(GGU)PRS differed from the sequence of gene 010 (coverage 59%, identity 95.8%) and had no similarity to the corresponding genes in other bacterial species.
Thus, we showed that PRSs can contribute additional tRNA genes to the host bacterial genome. Such tRNA genes have higher similarity to the corresponding genes of bacteria of other species than to the corresponding tRNA gene in the host bacterium. Such substitutions resulted in the diversification of evolutionarily old genes in host bacterial genomes and could in principle help the codon adaptation of genes harbored by GIs. The presence of identical genes encoding tRNAPRS in prophages and genomic islands is evidence confirming that the origin of GIs are the corresponding prophages.
Phylogenetic analysis was performed on the nucleotide sequences of genes encoding tRNAMet(CAU)PRS and tRNAfMet(CAU)PRS, which occurred most frequently in the genomes of the strains studied (frequency of 0.68). As a result, we analyzed the sequences of 17 genes encoding tRNAMet(CAU)PRS and tRNAfMet(CAU)PRS, as well as the sequences of genes 003, 041, and 047, belonging to the three rrn operons, and gene 045, located on the chromosome (Figure 4). Three clusters, A, B, and C, were identified. Cluster A had only the sequence of gene 045 and was identical in all S. meliloti strains studied (Figure 4). Cluster B included the sequences of eight tRNAfMet(CAU)PRS genes belonging to phiLM21-like phages, as well as the sequence of the tRNAfMet(CAU)PRS gene belonging to an inc-Ph similar to Paracoccus phage Shpa (the bootstrap value was 75%; Figure 4). The third cluster (C) comprised two monophyletic subclusters, C1 and C2 (bootstrap 77%). Subcluster C1 combined the sequences of seven tRNAfMet(CAU)PRS-encoding genes belonging to PRSs similar to different phages, including the rhizobiophages Sinorhizobium phage phiLM21 and Rhizobium phage 16-3, as well as phages Shpa and NYA-2014a, which infect Paracoccus and Sulfitobacter, respectively (Table 4). Subcluster C2 was represented by three tRNAfMet(CAU) genes in rrn operons, as well as the sequence of the tRNAfMet(CAU)PRS gene, which was part of a Ph similar to Rhizobium phage 16-3 and was integrated into gene 045 (strain RU11/001; Figure 4).
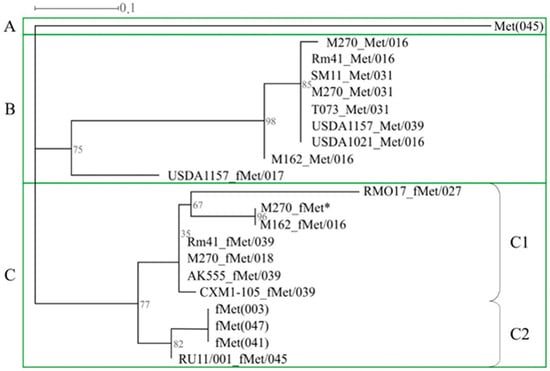
Figure 4.
Phylogenetic analysis of genes encoding essential tRNAMet(CAT) and tRNAfMet(CAT) of S. meliloti strains. Designations: the signatures of tRNA genes are in the form X_Y/N, where X—the name of the strain, Y—amino acid Met or fMet, N—ordinal number of the tRNA gene in which the PRS with an additional tRNA gene is integrated; see also the designations given in Table 3; *—additional tRNAfMet(CAT) located in PRS that is not integrated into the tRNA gene.
In summary, our results clearly show, for the first time, that tRNA genes present in the S. meliloti genome have different phylogenetic origins and, apparently, are linked to horizontal transfer processes involving phages, which is also true for tRNAfMet(CAU)PRS-encoding genes, similar to those from rrn operons. It should be noted that there is no evidence that tRNAPRS genes are used as new potential sites for integration.
2.4. The Synteny of PRSs Integrated into Essential tRNA Genes
Using pairwise sequence alignment (see Section 4 Materials and Methods), we analyzed the sequence synteny of the 88 above PRSs and the 3 PRSs that were also integrated into tRNA genes but located on pSymB (Table S2). This allowed us to identify the top-matching alignment results (hereinafter, synteny blocks or SBs) between all PRSs, amounting to 1057 SBs. SB sizes ranged from 37 to 71,338 bp (identity 73.6–100%; Table S3A), and of these, 983 were less than 8 kb (identity < 90%) and were not considered further (Figure S1). Of interest for analysis were 83 SBs, each with a size larger than 8 kb (identity > 90%; Table S3B). These SBs were identified between 19 GIs and 25 Phs, whereas the other 32 GIs and 12 Phs did not have the above SBs. There were 48 SBs between Ph/Ph pairs, 18 SBs between Ph/GI pairs, 7 SBs between GI/GI pairs that had phage-like sequences, and 10 SBs between GI/GI pairs that were free of such sequences (Figure 5). It should also be noted that the lack of detected SBs was 2.6 times more frequent in GIs than in Phs.
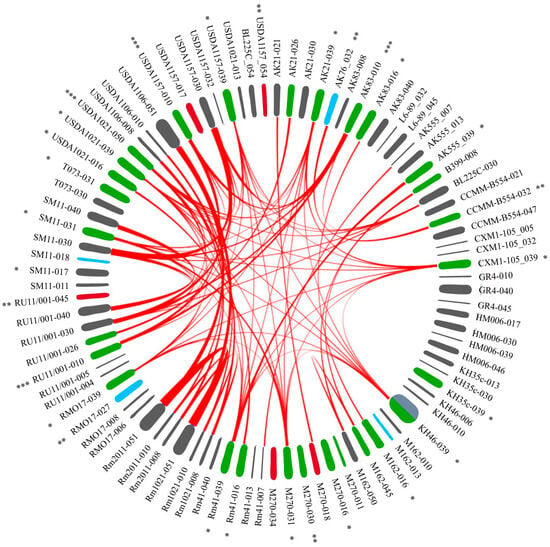
Figure 5.
The synteny blocks of PRSs integrated into essential tRNA genes of S. meliloti. Designations: there are sequences of PRSs around the circle; PRS signature: the strain that contained a PRS, with the ordinal number of the tRNA gene following the dash (see Table S2, Figure 2). Type of PRS indicated by color: gray—GI, green—int-Ph, blue—q-Ph, red—inc-Ph (see text). The red lines inside the circle connect matching sequences between pairs of PRSs (see Section 4 Materials and Methods; Table S3B). Detailed blastn PRS alignment results are in Table S3A. *, **, ***—PRS similar to phages phiLM21, 16-3, and RR1-A, respectively (see text). USDA1157-054, BL225C-054, and USDA1021-013—PRSs localized on the megaplasmid pSymB.
2.4.1. Analysis of 48 SBs between Ph/Ph Pairs
Forty-six synteny blocks were detected between rhizobiophage-like Phs. Of these, 39 SBs were between phiLM21-like Phs integrated into particular tRNA genes (Table 3, Figure 2). The integration points of these PRSs were genes 016, 031, and 039. Six other SBs were between RR1-A-like Phs integrated into genes 010 and 050, and one block was between an int-Ph and a q-Ph similar to Rhizobium phage 16-3 integrated into gene 032 (Figure 5). In addition, a block between pairs of phages similar to Mesorhizobium phage vB_MloP_Lo5R7ANS (integrated into gene 026) and another block between pairs of phages similar to Sulfitobacter phages (integrated into genes 008 and 045) were identified (Figure 5, Table S3B).
2.4.2. Analysis of 18 SBs between Ph/GI Pairs
A Ph/GI-pair analysis revealed 15 SBs between four int-Phs (integrated into genes 010 and 050) and four GIs (integrated into gene 030) containing fragments of Rhizobium phage vB_RleM_PPF1, and 3 SBs were identified between Ph/GI pairs that had sequences similar to those of the Sulfitobacter phage (integrated into genes 008, 018, 045, 021, and 050) (Figure 5; Table S3B).
2.4.3. Analysis of 17 SBs between GI/GI Pairs
Seven SBs were detected between pairs of GIs that had sequences similar to phages. Of these, six blocks were between four GIs integrated into gene 030, each containing a q-Ph similar to Rhizobium phage vB_RleM_PPF1. In one of these GIs, the sequence of interest was identified only through its similarity to the above sequences (using BLASTn; strain USDA1157). Another SB was between pairs of GIs integrated into gene 040, and each harbored sequences similar to both Enterobacter phage HK542 and Bacillus phage BM5 (Figure 5; Table S3B).
Ten SBs were detected between pairs of GIs that did not have sequences similar to phages. Of these, nine SBs (ranging from 14.8 to 71.3 kb) were between pairs of GIs integrated into genes 008, 010, and 051, which were identified in the genomes of closely related strains (1021, 2011, and USDA1106). The GI integrated into gene 051 of strain USDA1106 had a sequence similar to that of Stx-encoding phages, which did not overlap with SBs. Another block was between pairs of GIs integrated into gene 007 (Figure 5; Table S3B).
Thus, the highest number of synteny blocks was detected between pairs of Phs, whereas the number of blocks was 2.7-fold lower between pairs of Ph/GI sequences and even less frequent, namely, 2.6-fold, between pairs of GIs (48, 18, and 7 blocks, respectively). The decrease in the number of SBs identified in the pairwise sequence comparison of these variants is similar to a decreasing geometric progression whose denominator is (2.62 ± 0.05)−1.
A comparative analysis of GIs and Phs showed that sequences similar to rhizobiophages were abundant in prophages, namely, in 27 out of 37 Phs (frequency 7 × 10−1), and were extremely rare in GIs. The sequences of interest were only in 3 out of 51 GIs, i.e., the indicated sequences were an order of magnitude less frequent in GIs (frequency 6 × 10−2). Thus, rhizobiophage-like sequences introduced into the genomes of S. meliloti strains are not maintained (not “fixed”). This appears to be the result of the cell’s defense systems against DNA of phage origin or “foreign DNA”. At the same time, in GIs, there is a “fixation” of phage sequences for which rhizobia are not specific hosts. This may contribute to the formation of bacterial defenses against superinfection, i.e., the formation of immunity to phages belonging to the same immunity group as the “fixed” prophage.
2.4.4. Analysis of SBs between PRSs of Sinorhizobium spp.
A search for sequences similar to the rhizobiophages phiLM21, RR1-A, and 16-3 identified in S. meliloti strains was also carried out in the full-genome sequences of various species of the genus Sinorhizobium available in GenBank (access date June 2024). Extended sequences similar to Rhizobium phage 16-3 were not found at all in the genomes of other members of the genus Sinorhizobium, and these results are in agreement with the data that we obtained previously [].
SBs were detected between phiLM21-like Phs present in the genomes of six S. meliloti strains (AK21, AK555, CXM1-105, USDA1157, RMO17, and Rm41), both strains of S. kummerowiae, and 13 tested S. medicae strains (coverage > 50%, identity > 90%; see Section 4 Materials and Methods; Table 5). The similarity in the indicated SBs was higher in the pairwise comparison of PRSs from S. meliloti/S. kummerowiae than in the pairwise comparison of PRSs from S. meliloti/S. medicae (coverage 61–78%, identity 92.90–99.54% and coverage 53–68%, identity 90.24–98.49%, respectively).

Table 5.
List of analyzed bacterial genomes.
SBs similar to rhizobiophage RR1-A were found in all tested S. medicae strains (coverage 52–100%, identity 92.53–97.98%), except for three strains (SU277, T2, and T10) (see Section 4 Materials and Methods; Table 5). The above-indicated SBs were only in 4 out of 27 S. meliloti strains (USDA1157, RU11/001, AK83, and USDA1021) and were absent in both mentioned S. kummerowiae strains. PRSs similar to rhizobiophage RR1-A identified in S. meliloti strain USDA1021 and S. medicae strains ml42 and ml20 were found to have a high degree of similarity (coverage 100%, identity 97.98%).
Consequently, SBs similar to rhizobiophages phiLM21 and RR1-A were abundant in S. medicae strains (100% and 77% of strains of those tested, respectively), while the above sequences were found in S. meliloti strains almost 5-fold and 7-fold less frequently than in S. medicae strains. Both SBs similar to RR1-A- and phiLM21-like phages were identified in only one S. meliloti strain (USDA1157) and almost all tested S. medicae strains. Thus, it could be concluded that two closely related species, S. meliloti and S. medicae, as well as S. kummerowiae, recently assigned to S. meliloti species [], are significantly distinct in terms of the infectivity of these two rhizobiophages.
2.5. Analysis of Integrases Identified in PRSs
All 88 PRSs (51 GI and 37 Phs) had att sites and were site-specifically integrated into different tRNA genes (Figure 6). All of these sequences had a gene encoding an integrase that is responsible for site-specific integration. All identified integrases were tyrosine integrases similar to phage integrases, except one showed similarity not only to the tyrosine integrase of Mycobacterium phage Dori but also to the serine recombinase of Burkholderia phage KS14 (Figure 6).
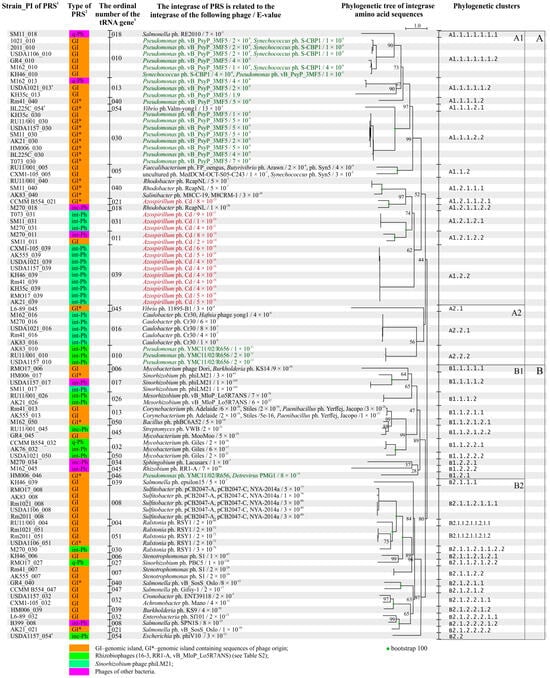
Figure 6.
Phylogenetic tree constructed using the amino acid sequences of integrases of PRS integrated into tRNA genes of S. meliloti. Designations: 1—PI means a point of integration, it combines the strain and an ordinal number of tRNA genes through a dash (see Table 3); 2—GI—genomic island; int-Ph—intact phage; inc-Ph—incomplete phage sequence; q-Ph—questionable phage sequence; 3—ordinal numbers of tRNA genes (see Table 3); 4—PRS localized on pSymB in case of the corresponding strains (see text and Table S2).
Both the nucleotide and amino acid sequences of PRS integrases shared similarities with certain phage integrases in only 14 of the 88 cases. This was the case for three Sinorhizobium phage phiLM21-like integrases, two Mesorhizobium phage vB_MloP_Lo5R7ANS-like integrases, five integrases similar to integrases of different Sulfitobacter phages, and four Ralstonia phage RSY1-like integrases (E-value ≤ 4 × 10−61; according to viruSITE) (Figure 6). In the other 74 cases, similarity to the integrases of different phages was established based on amino acid sequence analysis (E-value ≤ 7 × 10−3; in 1 case, the E-value was 1.9 (Figure 6, Table S4)). In total, similarities to the integrases of 44 different phages were detected, but half of these phages belonged to 15 different families within the class Caudoviricetes and their host bacteria belonged to 25 distant taxa (Table S4). The remaining phages were unclassified Caudoviricetes, and one of them was an unclassified bacterial virus, according to the latest phage classification [,].
The genes encoding integrases identified in each of the 51 GIs were analyzed. Only 1 of the 51 sequences was similar to a rhizobiophage integrase, namely, Sinorhizobium phage phiLM21. All other sequences were predominantly similar to integrases of different phages infecting Sulfitobacter or Pseudomonas, which are representatives of Alphaproteobacteria and Gammaproteobacteria, respectively (frequency of 0.61). Similarity to integrases of phages infecting Betaproteobacteria or bacteria of other classes was also revealed (average frequency of 0.15), while similarity to integrases of phages infecting bacteria from other genera of Alphaproteobacteria was extremely rare (frequency of 0.08; Table S4). A significant difference was observed between the group of GIs that did not have sequences of phage origin (except genes encoding integrases) and the group of GIs that had such sequences. This difference was evidenced by the occurrence of integrases similar to those of a larger number of different phages in the former group (Shannon indices H = 1.55 and 1.08, respectively), in which integrases of different Sulfitobacter phages and Pseudomonas phage vB_PsyP_3MF5 (Χ2 = 9.3, P = 2.5 × 10−2) were abundant (Figure 6; Table S4).
The genes encoding integrases identified in each of the 37 Phs were also analyzed. These integrases were predominantly similar to integrases of phages infecting different representatives of Alphaproteobacteria (frequency of 0.70), according to amino acid sequence analysis. Half of these sequences were similar to phage Cd integrase; this phage infects associative nitrogen-fixing soil bacteria of the genus Azospirillum (Figure 6). About 30% of the other sequences were similar to integrases of phages infecting the following bacteria of distant taxa: Betaproteobacteria (Ralstonia), Gammaproteobacteria (Salmonella, Pseudomonas), and Actinomycetia (Mycobacterium, Streptomyces) (Shannon index H = 0.88; Table S4).
It is noteworthy that the origin of the integrase (according to viruSITE) rarely matched the origin of the PRS (according to PHASTER and PHASTEST). There are only three cases, namely, a phiLM21-like Ph and two Phs similar to the Mesorhizobium phage vB_MloP_Lo5R7ANS, where the integrase and PRS had the same origin. In all other cases (34 of 37), where the PRS integrases were similar to integrases of other phages, according to amino acid sequence analysis. For example, in 18 phiLM21-like Phs, an integrase corresponding to the phiLM21 phage was found only once, whereas in other cases, the similarity was predominantly to the integrase of Azospirillum phage Cd or, with half the frequency, to the integrase of Caulobacter phage Cr30 (frequencies of 0.67 and 0.28, correspondingly) (Table S4).
Thus, the integrases detected in Phs and GIs (with and without sequences of phage origin) originated from different groups of phages infecting predominantly Alpha- and Gamma-/Betaproteobacteria, respectively, and the detected difference was significant (Χ2 = 20.5 and 23.7, p < 0.05).
Integrases similar to Azospirillum phage Cd and various Pseudomonas phages constituted the most representative groups, 15 and 21 sequences, respectively, of the 88 sequences tested (frequencies of 0.17 and 0.24, respectively). Therefore, they were used for a phylogenetic analysis conducted jointly considering PRS integration sites.
The 15 sequences of integrases similar to Azospirillum phage Cd integrase formed four subclusters in cluster A1.2. It turned out that the sequences of integrases that clustered in subclusters also coincided with the integration sites of the corresponding PRSs, namely, gene 039, 031, 021, or 011. The frequencies of occurrence of the integrase sequences in the corresponding subclusters were 0.60, 0.20, 0.07, and 0.13, respectively (Figure 6).
We analyzed the similarities between the amino acid sequences of PRS integrases and Azospirillum phage Cd integrase. Both integrase sequences had a higher degree of similarity when PRS integration occurred at the “hot spot”, gene 039 (subcluster A1.2.2 (bootstrap 62)), compared with when the PRS had integrated into another gene, such as gene 031 (subcluster A1.2.1.2.1 (bootstrap 100); E-value 4 × 10−16–6 × 10−16 and 4 × 10−11–1 × 10−10, respectively; Figure 6). In the cases where PRSs integrated into genes 021 and 011, the corresponding integrases had a lower degree of similarity to the integrase sequence of Azospirillum phage Cd and belonged to the following subclusters: A1.2.1.1.2.1 (bootstrap 100) and A1.2.1.2.2 (bootstrap 100; Figure 6, Table S4), respectively. Two GIs also appeared to be integrated into the same gene 039 in different strains. However, the integrase of one of the GIs was similar to that of Burkholderia phage KS9, and that of the second GI was similar to that of Salmonella phage epsilon15. These integrases belonged to different clusters (clusters B2.1.2.2.1.2 (bootstrap 100) and B2.1.1.1 (bootstrap 84)) (Figure 6).
The 21 integrase sequences similar to Pseudomonas phage integrases were identified in GIs (with or without phage genes) and prophages (8:9:4 ratio, respectively) and were mainly integrated into genes 010 and 030 (frequencies of 0.43 and 0.38, respectively) but were also detected in genes 013, 040, and 046 (frequencies of 0.09, 0.05, and 0.05, respectively) (Figure 6, Table S4). Out of the 21 integrase sequences, 17 belonged to four subclusters of cluster A1.1.1 (bootstrap 99) (Figure 6). Sequences grouped into subclusters also coincided with the integration sites of the corresponding PRSs (Figure 6).
We analyzed the amino acid sequences of integrases from this group. The integrases of GIs identified in “hot spot” gene 030 had a higher degree of similarity to the Pseudomonas phage vB_PsyP_3MF5 integrase (subcluster A1.1.1.2.2 (bootstrap 97); E-value 4 × 10−9–1 × 10−8) compared with the sequences of integrases of GIs identified in gene 010 (subcluster A1.1.1.1.1.1.2 (bootstrap 90); E-value 4 × 10−8—1 × 10−6) (Figure 6). Moreover, five of the six integrase sequences in the last cluster were also similar to those of Synechococcus phage S-CBP1 integrases (Figure 6). Three int-Phs appeared to be integrated into the same gene (010), but their integrases were similar to the integrase of Pseudomonas YMC11/02/R656 phage, and these sequences belonged to another cluster A2.2.2 (bootstrap 56; Figure 6, Table S4).
Therefore, “hot spot”-integrating PRSs containing integrases that show a high degree of similarity to the integrase of the corresponding original phage may have a preferential ability to insert into the host bacterial genome.
2.6. Analysis of PRS Attachment Sites (att)
Using two integration “hot spots”, genes 039 and 030, and gene 010, we analyzed the complementarity in the sequences of the attP and attB sites corresponding to PRSs and the sequence of the corresponding tRNA gene in S. meliloti. The gene sequences of the corresponding tRNAs of other bacterial species that were potential hosts for the aforementioned phage groups were used as a comparison group.
The analysis of the attP sites of PRSs integrated into “hot spot” gene 039 showed that these sequences were identical and 46 bp long in 10 cases, and another attP site was 3 bp longer (strain KH46). Both 46 and 49 bp long attP sites were found to be identical to attB sites located at the 3′ end of gene 039 in S. meliloti (Figure 7).
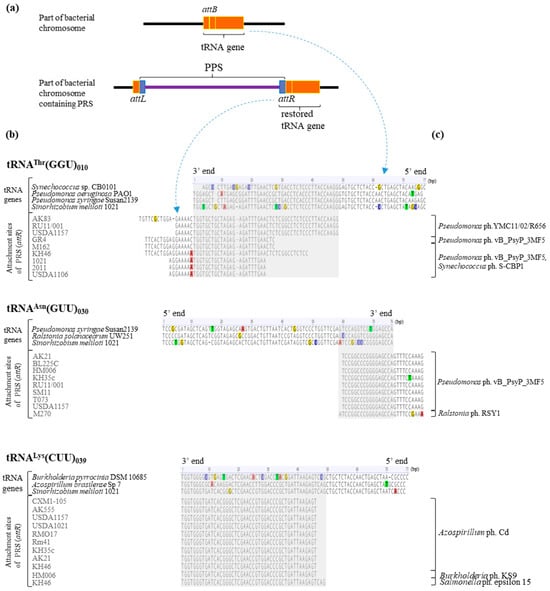
Figure 7.
The alignment of the 3′ ends of essential tRNA genes and PRS attachment sequences (attR sites). Designations: (a)—principal scheme of PRSs integrated into tRNA genes; (b)—the alignment of the 3′ ends of tRNA genes of bacteria and attachment sites of PRS detected in S. meliloti strains; and (c)—phages whose integrases were similar to those of the PRSs. The similarity in phage integrases and PRS integrases was determined by analyzing their amino acid sequences (see text).
The sequence similarity between gene 039 of S. meliloti 1021 and its counterparts encoding tRNALys(CUU) in potential bacteria hosts of the corresponding phages mentioned above was analyzed. The similarity to the corresponding gene in Azospirillum brasilience Sp7 was 94.7%, whereas the similarity to the same gene in Burkholderia pyrrocinia DSM 10685 was lower, at 83.1% (coverage 100% in both cases). The attB site corresponding to the sequence at the 3′ end of the gene encoding tRNALys(CUU) had two and nine SNPs in Azospirillum brasilience Sp7 and Burkholderia pyrrocinia DSM 10685, respectively (Figure 7). In Salmonella enterica LT2, a gene encoding tRNALys(CUU) was not detected at all.
The attP sites of PRSs integrated into “hot spot” gene 030 were 27 bp long in eight cases, and another attP site had one nucleotide more (Figure 7). These attP sites had 17 or 18 bp (respectively; see above) corresponding to the sequence at the 3′ end of gene 030 and 10 bp in the intergenic spacer adjacent to the 3′ end of the gene (Figure 7). In the case of two attP sites, the intergenic spacers included transitions, namely, C/T and G/A in one and A/G in the other sequences (strains KH35c and M270, respectively; Figure 7).
The sequence similarity between gene 030 in S. meliloti 1021 and its counterparts encoding tRNAAsn(GUU) in potential bacterial hosts of the corresponding phages was analyzed. The similarity to the corresponding genes in Pseudomonas syringae Susan2139 and Ralstonia solanacearum UW251 was 85.5% and 89.5%, respectively (coverage 100%), because of the presence of four SNPs in each of the two sequences (Figure 7).
The analysis of the attP sites of the nine PRSs integrated into gene 010 showed that their sequences varied significantly, from 31 to 64 bp. However, only 23 to 47 bp corresponded to the sequence at the 3′ end of gene 010, and the rest of the attP site (6 to 17 bp) was in the sequence of the intergenic spacer adjacent to the tRNA gene (Figure 7). SNPs, namely, a C/A nucleotide transversion adjacent to the tRNA gene and a G/A nucleotide transversion, were detected in the intergenic spacer regions in the case of five attP sites, and one other attP had a single-nucleotide deletion (Figure 7). Consequently, the attP sites of PRSs integrated into gene 010 are more variable in sequence length than the attP sites presented above in cases of “hot spots”.
We analyzed the sequence similarity in gene 010 in S. meliloti 1021 to its counterparts encoding tRNAThr(GGU) in potential bacterial hosts of the above phages. The similarity did not exceed 85% (coverage from 96.1% to 100%; identity from 76.6% to 84.4%) because of the presence of SNPs in the attB site (Figure 7).
Thus, in the “hot spots” of integration (genes 030 and 039) and the spot of preferential integration (gene 010) in S. meliloti, single-nucleotide substitutions never occurred at an att site corresponding to the sequence at the 3′ end of a particular tRNA gene.
Finally, it is intriguing that the attB sequences of the above tRNA genes in S. meliloti are recognized by PRS integrases that have a high level of similarity to phage integrases specific to bacteria of the genera Azospirillum and Pseudomonas.
3. Discussion
In this study, we conducted phage-related sequences (PRSs) analysis on the genomes of nonpathogenic, agriculturally valuable nitrogen-fixing root nodule bacteria. We focused on Sinorhizobium meliloti strains, which form symbiotic relationships with leguminous grasses of the genera Medicago, Melilotus, and Trigonella.
Our findings revealed a significant enrichment of PRSs within the genomes of these strains. The total information capacity of these “foreign DNA” sequences (314 PRSs) is comparable to the genome size of the reference S. meliloti 1021 strain (6.93 Mb). Surprisingly, over 64% of this “foreign DNA” is located on the chromosomes of these strains, despite chromosomes typically being considered highly conserved [,,]. Therefore, we can assume that these “foreign DNA sequences” become genetic objects that are inherited through vertical evolutionary pathways, which may contribute to active processes of genetic diversification and microbial genome evolution, in agreement with [].
The fact that the Phs of geographically distant S. meliloti strains are similar to a limited number of phages and mainly to Sinorhizobium phage phiLM21 was unexpected. This result leads us to suggest that S. meliloti has not yet properly developed defense systems against infection with this phage and that phiLM21-like phages are probably the most active participants in rhizobia lysogenization in modern ecosystems. This is confirmed by the fact that similar phiLM21-like sequences were also detected in strains of the closely related species S. medicae, as well as in S. kummerowiae.
The analysis of GIs, whose number and size varied considerably among S. meliloti strains, showed, for the first time, that they predominantly contained sequences similar to fragments of phages infecting bacteria of distant taxa (according to PHASTER and PHASTEST annotation). This similarity may indicate the presence of “gene flow” between phylogenetically distant bacteria involving phages, which is in agreement with []. In addition, the presence of such sequences in the genomes of bacterial hosts may contribute to the development of defense systems against superinfections, according to [,].
The analysis of S. meliloti GIs showed, for the first time, that there are GIs with and without sequences of phage origin, although both types of GI sequences retain genomic island features, according to the Islander algorithm []. Since each of the GIs studied had an integrase-encoding gene and was flanked by direct repeats, attL and attR sites, these sequences were most likely derived from a site-specific recombination mechanism similar to Lambda phage integration. The obtained data are consistent with the previously proposed theory of the “life cycle” of GIs [], as well as with the theory of the “grounding” of prophages in the bacterial genome []. However, the presented data also demonstrate an active “washout” of phage genes from bacterial genomes, which is apparently associated with the action of host bacterial defense systems.
Of the identified PRSs, 28% were inserted into chromosomal tRNA genes, and these sequences were of most significance for this analysis. Of great interest is the finding that these integrated sequences represented more than half of the total amount of foreign DNA (53.5%) detected in the studied S. meliloti strains. Here, for the first time, we present data showing that PRS integration does not occur randomly and does not affect all tRNA genes located on the chromosome, but only half of the 52 tRNA genes identified in the genome of S. meliloti species. The attB sites of the three genes encoding the isoacceptor tRNAThr(GGU), tRNAAsn(GUU), and tRNALys(CUU) are proposed to be “hot spots (sites)” of integration. The fact that phage sequences integrate into certain tRNAs is confirmed by recently obtained data for Helicobacter pylori phages [].
The most curious and still poorly described phenomenon is that the tRNA gene that is inactivated through PRS integration is replaced by a tRNA gene that is a part of the PRS but encodes a similar isoacceptor tRNA. However, genes encoding tRNAs introduced by PRSs have greater similarity to similar genes present in the genomes of other phylogenetically unrelated Alphaproteobacteria species. This result suggests that a tRNA gene specific to the host bacterium is substituted by a tRNA gene that is similar to genes in taxonomically unrelated bacterial species. Such cases do not seem to be unique: a similar case was recently described for Mycobacterium, and the authors described it as tRNA-dependent lysogeny []. Therefore, there is a process for the diversification of tRNA genes, which are assumed to be conserved and evolutionarily ancient.
Thus, as a result of phage infection, bacterial genomes can acquire an additional tRNA gene, which can lead to an increase in the number of tRNAs per genome, as shown in our study. The genes encoding tRNAfMet and tRNAMet, which are required for efficient protein synthesis in most groups of bacteria, were the most frequently introduced into S. meliloti genomes by PRSs. The resulting increase in the number of tRNA genes per genome may have a positive effect on the metabolic characteristics of bacteria, according to [,,]. At the same time, phage-encoded tRNA was found to replace the depleted bacterial host tRNA, which occurs, for example, via the action of a specific bacterial anticodon tRNA-nuclease as an early response to phage infection []. Thus, the presence of phage genes encoding tRNAs may be significant in resisting bacterial antiphage defense systems and thus assist in preventing the inhibition of phage propagation [,,].
All site-specifically integrated PRSs harbored a gene encoding an integrase similar to the tyrosine integrase of a phage. The fact that PRS integrases were not similar to any integrases of different rhizobiophages (viruSITE database), except in rare cases, was entirely unexpected. Notably, GIs and Phs contain integrases of different phylogenetically distant origins, a distinction we showed to be reliable.
GIs preferentially contained integrases similar to those of Pseudomonas phages, whereas Phs most often carried integrases of Azospirillum phages. Moreover, PRSs carrying integrases of Azospirillum or Pseudomonas phages contained att sites identical to fragments of tRNA genes in S. meliloti. However, these att sites were not similar to the corresponding sites of tRNA genes in bacteria that were potential hosts of the corresponding phages. Thus, the results presented testify to the fact of the targeted and obviously effective insertion of “foreign sequences” into the genome of a typical nonpathogenic saprophytic soil bacterium recipient, S. meliloti.
Our findings provide valuable insights into the molecular genetic mechanisms underlying phage integration into bacterial genomes and may contribute to the development of new methodologies for gene construct creation and targeted insertion vectors. Additionally, these results shed light on the role of phages in the evolution of bacteria, particularly phylogenetically distant classes like Gammaproteobacteria and Alphaproteobacteria.
4. Materials and Methods
4.1. Bacterial Genomes Analyzed in This Study
Complete genomes of 27 Sinorhizobium meliloti strains, 13 S. medicae strains, and 2 S. kummerowiae strains, as well as 10 strains of other genera, deposited in GenBank (accessed on 1 June 2024), were used in this study. The list of the genomes is presented in Table 5.
4.2. List and Purpose of the Programs Used in This Study
A search for phage-related sequences (PRSs). PHASTER (accessed on 25 June 2023), PHASTEST (accessed on 13 May 2024) [,,] and Islander tools from IslandViewer4 (https://www.pathogenomics.sfu.ca/islandviewer; accessed on 5 June 2023) [,] were applied, respectively, to search prophages (Phs) and genomic islands (GIs), i.e., PRSs. The Islander algorithm was also used to identify the tRNA genes into which the GI integration occurred, as well as the identification of direct repeats flanking the GIs. In the case of Phs, the search for such tRNA genes was performed by aligning the flanking Ph sequences and sequences of the tRNA gene of S. meliloti using the BLASTn tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 10 January 2024) [,].
The search and annotation of tRNA genes were carried out using Aragorn [] and tRNAscan-SE v. 2.0 tools (https://trna.ucsc.edu/tRNAscan-SE/; accessed on 8 May 2024), which allowed us to identify the tRNA isotype and its anticodon [,]. The localization of tRNA genes in the chromosomes of S. meliloti strains was determined relative to oriC and terC.
Analysis of the structural organization of chromosomes. Sequences corresponding to the origin of replication (oriC) were carried out by similarity to the oriC (SMc04880) of the reference strain S. meliloti 1021 (477 bp) according to [] using BLASTn [,]. DnaSP v. 5.10.01 software (University of Barcelona, Barcelona, Spain) [] was used to determine nucleotide diversity index (π) for oriC sequences. The first nucleotide of the sequence of the oriC was assumed as an origin (start) point of each of the studied chromosomes. The method of standardization of coordinates of chromosomal sequences of S. meliloti relative to oriC was described in [,]. The origin point was assumed to be 0°, and the opposite was 360°, consistent with []. Coordinates in degrees were determined by the following formula: C° = Cbp/(Lbp/360), where C° is the coordinate in degrees, Cbp is the coordinate in bp, and Lbp is the length of the chromosome in bp. The potential terminus location (terC) was determined using a method based on an asymmetrically distributed octomeric sequence (GGGCAGGG) on both strands in the genome of the Alphaproteobacteria. The abundance of this octomeric sequence on each strand around the terC region is maximal []. This method is specific for Alphaproteobacteria and does not depend on the settings of the octomeric sequence search program. The localization of the octomeric sequence in S. meliloti chromosomes was determined using BLASTn. To verify the results obtained, the classical method of determining the potential terC sequence based on GC-skew was used. GC-skew was determined using GenSkew (https://genskew.csb.univie.ac.at/; accessed on 2 November 2023) []; the Stepsize and Windowsize were equal to 0.1% of the chromosome length.
The analysis of the synteny of the nucleotide sequences of PRSs was carried out using the BLASTn tool. Based on the pairwise alignment of all sequences, a table that contained the results of only the best of all the resulting alignments was created (Table S3A; Figure S1). Using the Python JSON library, this table was converted into a document with data in JSON format. A graphical representation of the obtained synteny data was obtained using the dash bio module (https://dash.plotly.com/dash-bio; accessed on 23 October 2023). For the graphic image, only the results of an alignment of more than 8 kb with a length and identity of more than 90% were used.
The viruSITE tool (accessed on 15 March 2024) [] was used to analyze amino acid and nucleotide sequence encoded integrases and to determine the degree of similarity between integrase sequences of different phages.
Phylogenetic trees were reconstructed using MUSCLE (https://www.ebi.ac.uk/jdispatcher/msa/muscle; accessed on 15 June 2024) [] and IQTree tools (http://iqtree.cibiv.univie.ac.at/; accessed on 15 June 2024) [].
The comparison analysis of different groups was performed according to the Chi-square test (significance level, α = 0.05) using PAST v. 4.03 software (Oslo, Norway) [].
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms251910421/s1.
Author Contributions
Conceptualization, M.L.R.; methodology, M.L.R., V.S.M. and M.E.V.; formal analysis, M.E.V., A.S.S. and V.S.M.; validation, M.E.V.; investigation, M.E.V., S.P.G. and V.S.M.; data curation, M.L.R. and M.E.V.; writing—original draft, M.L.R. and M.E.V.; writing—review and editing, M.L.R., A.S.S., V.S.M., S.P.G., A.M. and M.E.V.; project administration, M.L.R. All authors have read and agreed to the published version of the manuscript.
Funding
This paper was produced with support from the Ministry of Science and Higher Education of the Russian Federation in accordance with agreement no. 075-15-2022-320, dated 20 April 2022, on providing grants in the form of subsidies from the federal budget of the Russian Federation. The grant was provided for state support for the creation and development of a World-class Scientific Center, “Agrotechnologies for the Future”.
Data Availability Statement
The original contributions presented in this study are included in this article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
Acknowledgments
We would like to thank J.P.W. Young (University of York) for fruitful discussions of the results of this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Faruque, S.M.; Mekalanos, J.J. Phage-Bacterial Interactions in the Evolution of Toxigenic Vibrio cholerae. Virulence 2012, 3, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal Gene Transfer: Building the Web of Life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Azarian, T.; Huang, I.-T.; Hanage, W.P. Structure and Dynamics of Bacterial Populations: Pangenome Ecology. In The Pangenome; Tettelin, H., Medini, D., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 115–128. ISBN 978-3-030-38280-3. [Google Scholar]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic Islands in Pathogenic and Environmental Microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Ramisetty, B.C.M.; Sudhakari, P.A. Bacterial ‘Grounded’ Prophages: Hotspots for Genetic Renovation and Innovation. Front. Genet. 2019, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.; Hendrix, R.W. Bacteriophages and the Bacterial Genome. In The Bacterial Chromosome; Higgins, N.P., Ed.; ASM Press: Washington, DC, USA, 2014; pp. 39–52. ISBN 978-1-68367-204-3. [Google Scholar]
- Johnson, G.; Banerjee, S.; Putonti, C. Diversity of Pseudomonas aeruginosa Temperate Phages. mSphere 2022, 7, e01015-21. [Google Scholar] [CrossRef]
- Zhou, W.; Li, Y.; Xu, X.; Rao, S.; Wen, H.; Han, Y.; Deng, A.; Zhang, Z.; Yang, Z.; Zhu, G. Whole-Genome Analysis Showed the Promotion of Genetic Diversity and Coevolution in Staphylococcus aureus Lytic Bacteriophages and Their Hosts Mediated by Prophages via Worldwide Recombination Events. Front. Microbiol. 2023, 14, 1088125. [Google Scholar] [CrossRef]
- Asadulghani, M.; Ogura, Y.; Ooka, T.; Itoh, T.; Sawaguchi, A.; Iguchi, A.; Nakayama, K.; Hayashi, T. The Defective Prophage Pool of Escherichia coli O157: Prophage–Prophage Interactions Potentiate Horizontal Transfer of Virulence Determinants. PLoS Pathog. 2009, 5, e1000408. [Google Scholar] [CrossRef]
- Castillo, A.; Tello, M.; Ringwald, K.; Acuña, L.G.; Quatrini, R.; Orellana, O. A DNA Segment Encoding the Anticodon Stem/Loop of tRNA Determines the Specific Recombination of Integrative-Conjugative Elements in Acidithiobacillus Species. RNA Biol. 2018, 15, 492–499. [Google Scholar] [CrossRef]
- Orellana, R.; Arancibia, A.; Badilla, L.; Acosta, J.; Arancibia, G.; Escar, R.; Ferrada, G.; Seeger, M. Ecophysiological Features Shape the Distribution of Prophages and CRISPR in Sulfate Reducing Prokaryotes. Microorganisms 2021, 9, 931. [Google Scholar] [CrossRef]
- Rocha, E.P.C.; Bikard, D. Microbial Defenses against Mobile Genetic Elements and Viruses: Who Defends Whom from What? PLoS Biol. 2022, 20, e3001514. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Roumiantseva, M.L.; Muntyan, V.S.; Cherkasova, M.E.; Saksaganskaya, A.S.; Andronov, E.E.; Simarov, B.V. Genomic Islands in Sinorhizobium meliloti Rm1021, Nitrogen-Fixing Symbiont of Alfalfa. Russ. J. Genet. 2018, 54, 759–769. [Google Scholar] [CrossRef]
- Finlay, B.B.; Falkow, S. Common Themes in Microbial Pathogenicity Revisited. Microbiol. Mol. Biol. Rev. 1997, 61, 136–169. [Google Scholar] [CrossRef]
- Escobar-Páramo, P.; Clermont, O.; Blanc-Potard, A.-B.; Bui, H.; Le Bouguénec, C.; Denamur, E. A Specific Genetic Background Is Required for Acquisition and Expression of Virulence Factors in Escherichia coli. Mol. Biol. Evol. 2004, 21, 1085–1094. [Google Scholar] [CrossRef]
- Middendorf, B.; Hochhut, B.; Leipold, K.; Dobrindt, U.; Blum-Oehler, G.; Hacker, J. Instability of Pathogenicity Islands in Uropathogenic Escherichia coli 536. J. Bacteriol. 2004, 186, 3086–3096. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shestakov, S.V. How Does the Horizontal Gene Transfer in Bacteria Occur and than Is It Tied Up. Ecol. genet. 2007, 5, 12–24. [Google Scholar] [CrossRef][Green Version]
- Cherkasova, M.E.; Muntyan, V.S.; Saksaganskaia, A.S.; Simarov, B.V.; Roumiantseva, M.L. Sinorhizobium meliloti: Chromosomal Types and Genomic Islands. Ecol. Genet. 2019, 17, 23–38. [Google Scholar] [CrossRef]
- Roumiantseva, M.L.; Vladimirova, M.E.; Saksaganskaia, A.S.; Muntyan, V.S.; Kozlova, A.P.; Afonin, A.M.; Baturina, O.A.; Simarov, B.V. Ensifer meliloti L6-AK89, an Effective Inoculant of Medicago lupulina Varieties: Phenotypic and Deep-Genome Screening. Agronomy 2022, 12, 766. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Finlay, B.B. Pathogenicity Islands: A Molecular Toolbox for Bacterial Virulence. Cell Microbiol. 2006, 8, 1707–1719. [Google Scholar] [CrossRef]
- Sekizuka, T.; Yamamoto, A.; Komiya, T.; Kenri, T.; Takeuchi, F.; Shibayama, K.; Takahashi, M.; Kuroda, M.; Iwaki, M. Corynebacterium ulcerans 0102 Carries the Gene Encoding Diphtheria Toxin on a Prophage Different from the C. diphtheriae NCTC 13129 Prophage. BMC Microbiol. 2012, 12, 72. [Google Scholar] [CrossRef] [PubMed]
- Ingmer, H.; Gerlach, D.; Wolz, C. Temperate Phages of Staphylococcus aureus. Microbiol. Spectrum 2019, 7, 7.5.1. [Google Scholar] [CrossRef]
- Kondo, K.; Kawano, M.; Sugai, M. Distribution of Antimicrobial Resistance and Virulence Genes within the Prophage-Associated Regions in Nosocomial Pathogens. mSphere 2021, 6, e00452-21. [Google Scholar] [CrossRef] [PubMed]
- Mestre, M.R.; Gao, L.A.; Shah, S.A.; López-Beltrán, A.; González-Delgado, A.; Martínez-Abarca, F.; Iranzo, J.; Redrejo-Rodríguez, M.; Zhang, F.; Toro, N. UG/Abi: A Highly Diverse Family of Prokaryotic Reverse Transcriptases Associated with Defense Functions. Nucleic Acids Res. 2022, 50, 6084–6101. [Google Scholar] [CrossRef] [PubMed]
- Millman, A.; Melamed, S.; Leavitt, A.; Doron, S.; Bernheim, A.; Hör, J.; Garb, J.; Bechon, N.; Brandis, A.; Lopatina, A.; et al. An Expanded Arsenal of Immune Systems That Protect Bacteria from Phages. Cell Host Microbe 2022, 30, 1556–1569.e5. [Google Scholar] [CrossRef]
- Rousset, F.; Depardieu, F.; Miele, S.; Dowding, J.; Laval, A.-L.; Lieberman, E.; Garry, D.; Rocha, E.P.C.; Bernheim, A.; Bikard, D. Phages and Their Satellites Encode Hotspots of Antiviral Systems. Cell Host Microbe 2022, 30, 740–753.e5. [Google Scholar] [CrossRef]
- Kaneko, T. Complete Genomic Sequence of Nitrogen-Fixing Symbiotic Bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002, 9, 189–197. [Google Scholar] [CrossRef]
- Itakura, M.; Saeki, K.; Omori, H.; Yokoyama, T.; Kaneko, T.; Tabata, S.; Ohwada, T.; Tajima, S.; Uchiumi, T.; Honnma, K.; et al. Genomic Comparison of Bradyrhizobium japonicum Strains with Different Symbiotic Nitrogen-Fixing Capabilities and Other Bradyrhizobiaceae Members. ISME J. 2009, 3, 326–339. [Google Scholar] [CrossRef]
- Stiffler, A.K.; Hesketh-Best, P.J.; Varona, N.S.; Zagame, A.; Wallace, B.A.; Lapointe, B.E.; Silveira, C.B. Genomic and Induction Evidence for Bacteriophage Contributions to Sargassum-Bacteria Symbioses. Microbiome 2024, 12, 143. [Google Scholar] [CrossRef]
- Cheetham, B.F.; Katz, M.E. A Role for Bacteriophages in the Evolution and Transfer of Bacterial Virulence Determinants. Mol. Microbiol. 1995, 18, 201–208. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Aravind, L. Horizontal Gene Transfer in Prokaryotes: Quantification and Classification. Annu. Rev. Microbiol. 2001, 55, 709–742. [Google Scholar] [CrossRef] [PubMed]
- Jain, R. Horizontal Gene Transfer Accelerates Genome Innovation and Evolution. Mol. Biol. Evol. 2003, 20, 1598–1602. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Itoh, T.; Matsuda, H.; Gojobori, T. Biased Biological Functions of Horizontally Transferred Genes in Prokaryotic Genomes. Nat. Genet. 2004, 36, 760–766. [Google Scholar] [CrossRef]
- Pantůček, R.; Sedláček, I.; Indráková, A.; Vrbovská, V.; Mašlaňová, I.; Kovařovic, V.; Švec, P.; Králová, S.; Krištofová, L.; Kekláková, J.; et al. Staphylococcus edaphicus sp. Nov., Isolated in Antarctica, Harbors the mecC Gene and Genomic Islands with a Suspected Role in Adaptation to Extreme Environments. Appl. Environ. Microbiol. 2018, 84, e01746-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jiao, J.; Tian, C.-F. Adaptive Evolution of Rhizobial Symbiosis beyond Horizontal Gene Transfer: From Genome Innovation to Regulation Reconstruction. Genes 2023, 14, 274. [Google Scholar] [CrossRef]
- Nadeem, A.; Wahl, L.M. Prophage as a Genetic Reservoir: Promoting Diversity and Driving Innovation in the Host Community: BRIEF COMMUNICATION. Evolution 2017, 71, 2080–2089. [Google Scholar] [CrossRef]
- Braga, L.P.P.; Soucy, S.M.; Amgarten, D.E.; Da Silva, A.M.; Setubal, J.C. Bacterial Diversification in the Light of the Interactions with Phages: The Genetic Symbionts and Their Role in Ecological Speciation. Front. Ecol. Evol. 2018, 6, 6. [Google Scholar] [CrossRef]
- Martín-Galiano, A.J.; García, E. Streptococcus pneumoniae: A Plethora of Temperate Bacteriophages With a Role in Host Genome Rearrangement. Front. Cell. Infect. Microbiol. 2021, 11, 775402. [Google Scholar] [CrossRef]
- Mavrich, T.N.; Hatfull, G.F. Evolution of Superinfection Immunity in Cluster A Mycobacteriophages. mBio 2019, 10, e00971-19. [Google Scholar] [CrossRef]
- Gentile, G.M.; Wetzel, K.S.; Dedrick, R.M.; Montgomery, M.T.; Garlena, R.A.; Jacobs-Sera, D.; Hatfull, G.F. More Evidence of Collusion: A New Prophage-Mediated Viral Defense System Encoded by Mycobacteriophage Sbash. mBio 2019, 10, e00196-19. [Google Scholar] [CrossRef]
- Meaden, S.; Fineran, P.C. Bacterial Defense Islands Limit Viral Attack. Science 2021, 374, 399–400. [Google Scholar] [CrossRef] [PubMed]
- Owen, S.V.; Wenner, N.; Dulberger, C.L.; Rodwell, E.V.; Bowers-Barnard, A.; Quinones-Olvera, N.; Rigden, D.J.; Rubin, E.J.; Garner, E.C.; Baym, M.; et al. Prophages Encode Phage-Defense Systems with Cognate Self-Immunity. Cell Host Microbe 2021, 29, 1620–1633.e8. [Google Scholar] [CrossRef]
- Kozlova, A.P.; Muntyan, V.S.; Vladimirova, M.E.; Saksaganskaia, A.S.; Kabilov, M.R.; Gorbunova, M.K.; Gorshkov, A.N.; Grudinin, M.P.; Simarov, B.V.; Roumiantseva, M.L. Soil Giant Phage: Genome and Biological Characteristics of Sinorhizobium Jumbo Phage. IJMS 2024, 25, 7388. [Google Scholar] [CrossRef]
- Grainge, I.; Jayaram, M. The Integrase Family of Recombinases: Organization and Function of the Active Site. Mol. Microbiol. 1999, 33, 449–456. [Google Scholar] [CrossRef]
- Van Houdt, R.; Leplae, R.; Lima-Mendez, G.; Mergeay, M.; Toussaint, A. Towards a More Accurate Annotation of Tyrosine-Based Site-Specific Recombinases in Bacterial Genomes. Mobile DNA 2012, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M.; Van Der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic Islands: Tools of Bacterial Horizontal Gene Transfer and Evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef]
- Guerrero-Bustamante, C.A.; Hatfull, G.F. Bacteriophage tRNA-Dependent Lysogeny: Requirement of Phage-Encoded tRNA Genes for Establishment of Lysogeny. mBio 2024, 15, e03260-23. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.P. Integration Sites for Genetic Elements in Prokaryotic tRNA and tmRNA Genes: Sublocation Preference of Integrase Subfamilies. Nucleic Acids Res. 2002, 30, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Grindley, N.D.F.; Whiteson, K.L.; Rice, P.A. Mechanisms of Site-Specific Recombination. Annu. Rev. Biochem. 2006, 75, 567–605. [Google Scholar] [CrossRef]
- Lysogeny: The λ Paradigm and the Role of Lysogenic Conversion in Bacterial Pathogenesis. In Molecular Genetics of Bacteria; ASM Press: Washington, DC, USA, 2007; Volume 8, pp. 343–376. ISBN 978-1-55581-399-4.
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S. IslandViewer 4: Expanded Prediction of Genomic Islands for Larger-Scale Datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- de Assis, J.C.S.; Gonçalves, O.S.; Fernandes, A.S.; de Queiroz, M.V.; Bazzolli, D.M.S.; Santana, M.F. Genomic Analysis Reveals the Role of Integrative and Conjugative Elements in Plant Pathogenic Bacteria. Mobile DNA 2022, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Germon, P.; Roche, D.; Melo, S.; Mignon-Grasteau, S.; Dobrindt, U.; Hacker, J.; Schouler, C.; Moulin-Schouleur, M. tDNA Locus Polymorphism and Ecto-Chromosomal DNA Insertion Hot-Spots Are Related to the Phylogenetic Group of Escherichia coli Strains. Microbiology 2007, 153, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, P.H.; Touchon, M.; Cury, J.; Rocha, E.P.C. The Chromosomal Organization of Horizontal Gene Transfer in Bacteria. Nat. Commun. 2017, 8, 841. [Google Scholar] [CrossRef] [PubMed]
- Kopejtka, K.; Lin, Y.; Jakubovičová, M.; Koblížek, M.; Tomasch, J. Clustered Core- and Pan-Genome Content on Rhodobacteraceae Chromosomes. Genome Biol. Evol. 2019, 11, 2208–2217. [Google Scholar] [CrossRef]
- Leonard, A.C.; Grimwade, J.E. Building a Bacterial Orisome: Emergence of New Regulatory Features for Replication Origin Unwinding. Mol. Microbiol. 2005, 55, 978–985. [Google Scholar] [CrossRef]
- Kaguni, J.M. DnaA: Controlling the Initiation of Bacterial DNA Replication and More. Annu. Rev. Microbiol. 2006, 60, 351–371. [Google Scholar] [CrossRef]
- Mulcair, M.D.; Schaeffer, P.M.; Oakley, A.J.; Cross, H.F.; Neylon, C.; Hill, T.M.; Dixon, N.E. A Molecular Mousetrap Determines Polarity of Termination of DNA Replication in E. coli. Cell 2006, 125, 1309–1319. [Google Scholar] [CrossRef]
- Sibley, C.D.; MacLellan, S.R.; Finan, T. The Sinorhizobium Meliloti Chromosomal Origin of Replication. Microbiology 2006, 152, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Roumiantseva, M.L.; Muntyan, V.S.; Cherkasova, M.E.; Andronov, E.E.; Saksaganskaya, A.S.; Dzyubenko, E.A.; Dzyubenko, N.I.; Simarov, B.V. A Comparative Analysis of Genomic Characters of Reference Sinorhizobium meliloti Strains, the Alfalfa Symbionts (Review). S-kh. Biol. 2017, 52, 928–939. [Google Scholar] [CrossRef]
- Muntyan, V.S.; Cherkasova, M.E.; Andronov, E.E.; Simarov, B.V.; Roumiantseva, M.L. Occurrence of Islands in Genomes of Sinorhizobium Meliloti Native Isolates. Russ. J. Genet. 2016, 52, 1015–1022. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In Gene Prediction; Kollmar, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 1962, pp. 1–14. ISBN 978-1-4939-9172-3. [Google Scholar]
- Vladimirova, M.; Muntyan, V.; Kozlova, A.; Grudinin, M.; Roumiantseva, M. CRISPR/Cas in Genomes of Sinorhizobium meliloti. In Proceedings of the 20th International Multidisciplinary Scientific GeoConference Proceedings SGEM 2020; STEF92 Technology, Albena, Bulgaria, 18 August 2020; Volume 20, pp. 215–222. [Google Scholar]
- Vladimirova, M.; Afonin, A.; Muntyan, V.; Simarov, B.; Roumiantseva, M. Evaluation of Sinorhizobium meliloti Genomic Islands Inserted into the tRNA-Thr. In Proceedings of the 2020 Cognitive Sciences, Genomics and Bioinformatics (CSGB), IEEE, Novosibirsk, Russia, 6–10 July 2020; pp. 118–121. [Google Scholar]
- Kozlova, A.P.; Saksaganskaia, A.S.; Afonin, A.M.; Muntyan, V.S.; Vladimirova, M.E.; Dzyubenko, E.A.; Roumiantseva, M.L. A Temperate Sinorhizobium Phage, AP-16-3, Closely Related to Phage 16-3: Mosaic Genome and Prophage Analysis. Viruses 2023, 15, 1701. [Google Scholar] [CrossRef] [PubMed]
- Menéndez, E.; Flores-Félix, J.D.; Kaur, S.; diCenzo, G.C.; Young, J.P.W.; Peix, A.; Velázquez, E. Reclassification of Type Strains of Rhizobium indigoferae and Sinorhizobium kummerowiae into the Species Rhizobium leguminosarum and Sinorhizobium meliloti, Respectively. Int. J. Syst. Evol. Microbiol. 2024, 74. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; Van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of Morphology-Based Taxa and Change to Binomial Species Names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Mihara, T.; Nishimura, Y.; Shimizu, Y.; Nishiyama, H.; Yoshikawa, G.; Uehara, H.; Hingamp, P.; Goto, S.; Ogata, H. Linking Virus Genomes with Host Taxonomy. Viruses 2016, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Galardini, M.; Mengoni, A.; Brilli, M.; Pini, F.; Fioravanti, A.; Lucas, S.; Lapidus, A.; Cheng, J.-F.; Goodwin, L.; Pitluck, S.; et al. Exploring the Symbiotic Pangenome of the Nitrogen-Fixing Bacterium Sinorhizobium meliloti. BMC Genomics 2011, 12, 235. [Google Scholar] [CrossRef]
- Galardini, M.; Pini, F.; Bazzicalupo, M.; Biondi, E.G.; Mengoni, A. Replicon-Dependent Bacterial Genome Evolution: The Case of Sinorhizobium meliloti. Genome Biol. Evol. 2013, 5, 542–558. [Google Scholar] [CrossRef]
- diCenzo, G.C.; Finan, T.M. The Divided Bacterial Genome: Structure, Function, and Evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef]
- Bobay, L.-M. The Prokaryotic Species Concept and Challenges. In The Pangenome; Tettelin, H., Medini, D., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 21–49. ISBN 978-3-030-38280-3. [Google Scholar]
- Csiszovszki, Z.; Buzás, Z.; Semsey, S.; Ponyi, T.; Papp, P.P.; Orosz, L. immX Immunity Region of Rhizobium Phage 16-3 : Two Overlapping Cistrons of Repressor Function. J. Bacteriol. 2003, 185, 4382–4392. [Google Scholar] [CrossRef]
- Hudson, C.M.; Lau, B.Y.; Williams, K.P. Islander: A Database of Precisely Mapped Genomic Islands in tRNA and tmRNA Genes. Nucleic Acids Res. 2015, 43, D48–D53. [Google Scholar] [CrossRef]
- Vale, F.F.; HpGP Research Network; Roberts, R.J.; Kobayashi, I.; Camargo, M.C.; Rabkin, C.S. Gene Content, Phage Cycle Regulation Model and Prophage Inactivation Disclosed by Prophage Genomics in the Helicobacter pylori Genome Project. Gut Microbes 2024, 16, 2379440. [Google Scholar] [CrossRef]
- Rocha, E.P.C. Codon Usage Bias from tRNA’s Point of View: Redundancy, Specialization, and Efficient Decoding for Translation Optimization. Genome Res. 2004, 14, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Ran, W.; Higgs, P.G. The Influence of Anticodon-Codon Interactions and Modified Bases on Codon Usage Bias in Bacteria. Mol. Biol. Evol. 2010, 27, 2129–2140. [Google Scholar] [CrossRef] [PubMed]
- Ayan, G.B.; Park, H.J.; Gallie, J. The Birth of a Bacterial tRNA Gene by Large-Scale, Tandem Duplication Events. eLife 2020, 9, e57947. [Google Scholar] [CrossRef] [PubMed]
- Azam, A.H.; Chihara, K.; Kondo, K.; Nakamura, T.; Ojima, S.; Tamura, A.; Yamashita, W.; Cui, L.; Takahashi, Y.; Watashi, K.; et al. Viruses Encode tRNA and Anti-Retron to Evade Bacterial Immunity 2023. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yang, J.Y.; Fang, W.; Miranda-Sanchez, F.; Brown, J.M.; Kauffman, K.M.; Acevero, C.M.; Bartel, D.P.; Polz, M.F.; Kelly, L. Degradation of Host Translational Machinery Drives tRNA Acquisition in Viruses. Cell Syst. 2021, 12, 771–779.e5. [Google Scholar] [CrossRef]
- Van Den Berg, D.F.; Van Der Steen, B.A.; Costa, A.R.; Brouns, S.J. Phage tRNAs Evade tRNA-Targeting Host Defenses through Anticodon Loop Mutations. eLife 2023, 12, e85183. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, Better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Laslett, D. ARAGORN, a Program to Detect tRNA Genes and tmRNA Genes in Nucleotide Sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-Line: Integrating Search and Context for Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Kono, N.; Arakawa, K.; Sato, M.; Yoshikawa, H.; Tomita, M.; Itaya, M. Undesigned Selection for Replication Termination of Bacterial Chromosomes. J. Mol. Biol. 2014, 426, 2918–2927. [Google Scholar] [CrossRef]
- Lawrence, J.G.; Hendrickson, H. Lateral Gene Transfer: When Will Adolescence End? Mol. Microbiol. 2003, 50, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Salzberg, S.L. SkewIT: The Skew Index Test for Large-Scale GC Skew Analysis of Bacterial Genomes. PLoS Comput. Biol. 2020, 16, e1008439. [Google Scholar] [CrossRef] [PubMed]
- Stano, M.; Beke, G.; Klucar, L. viruSITE—Integrated Database for Viral Genomics. Database 2016, 2016, baw162. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.; Ryan, P. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).