Genome-Wide Identification of NAC Gene Family Members of Tree Peony (Paeonia suffruticosa Andrews) and Their Expression under Heat and Waterlogging Stress
Abstract
1. Introduction
2. Results
2.1. Identification of PsNAC Family Members and Analysis of Physicochemical Properties
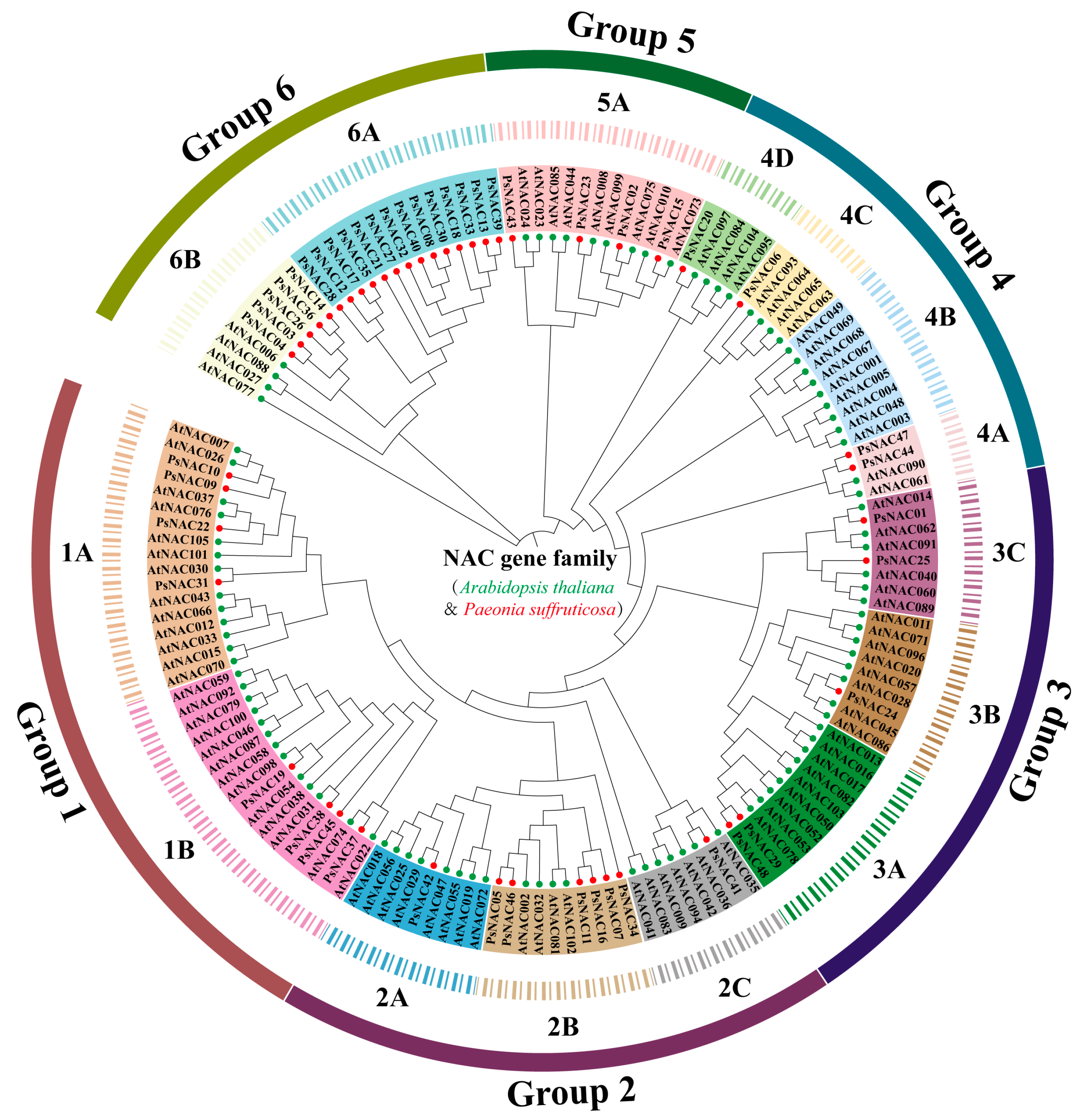
2.2. Phylogenetic Analysis and Classification of PsNAC
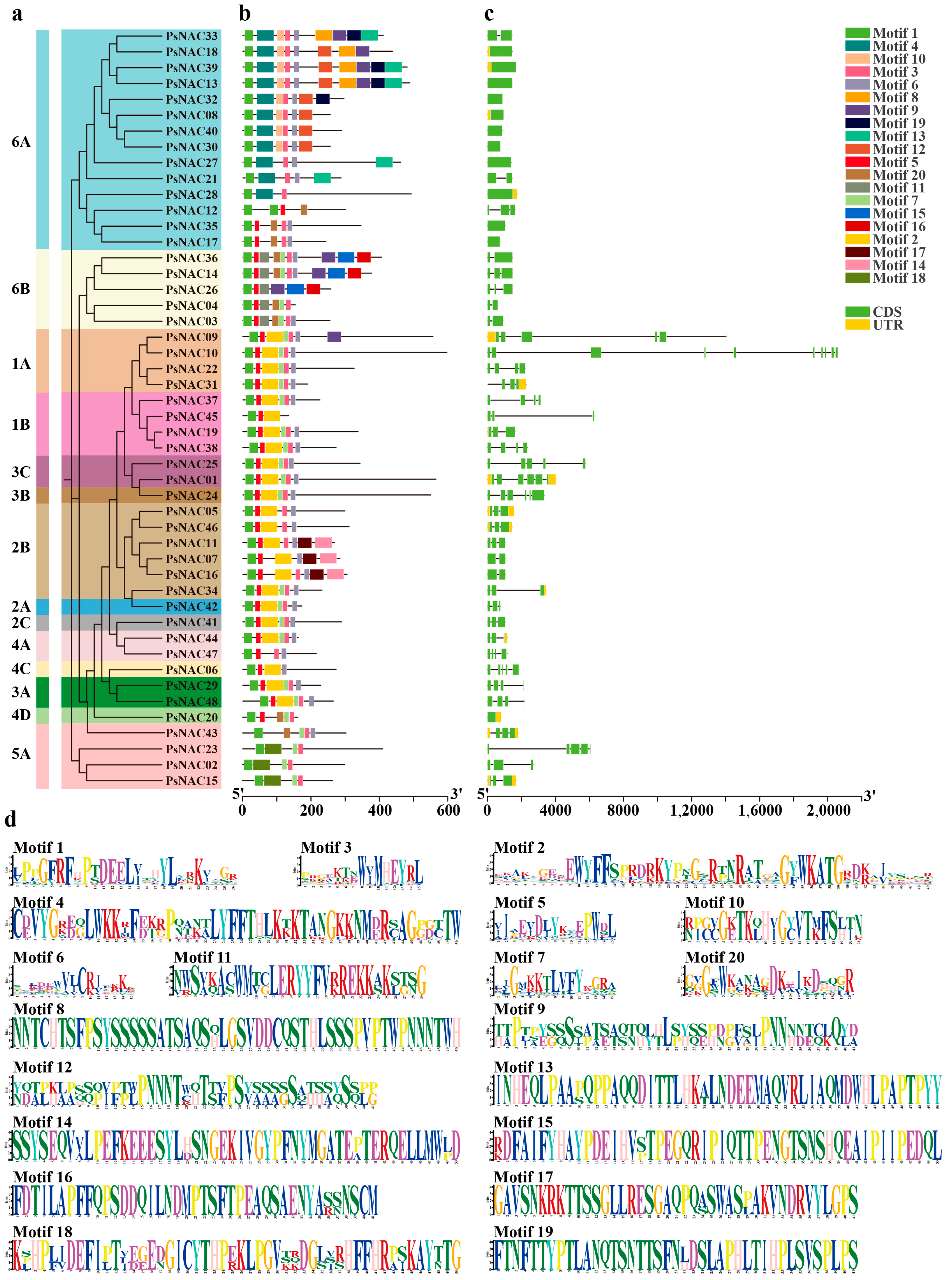
2.3. Protein Conservation Motif and Gene Structure Analysis of PsNAC Family
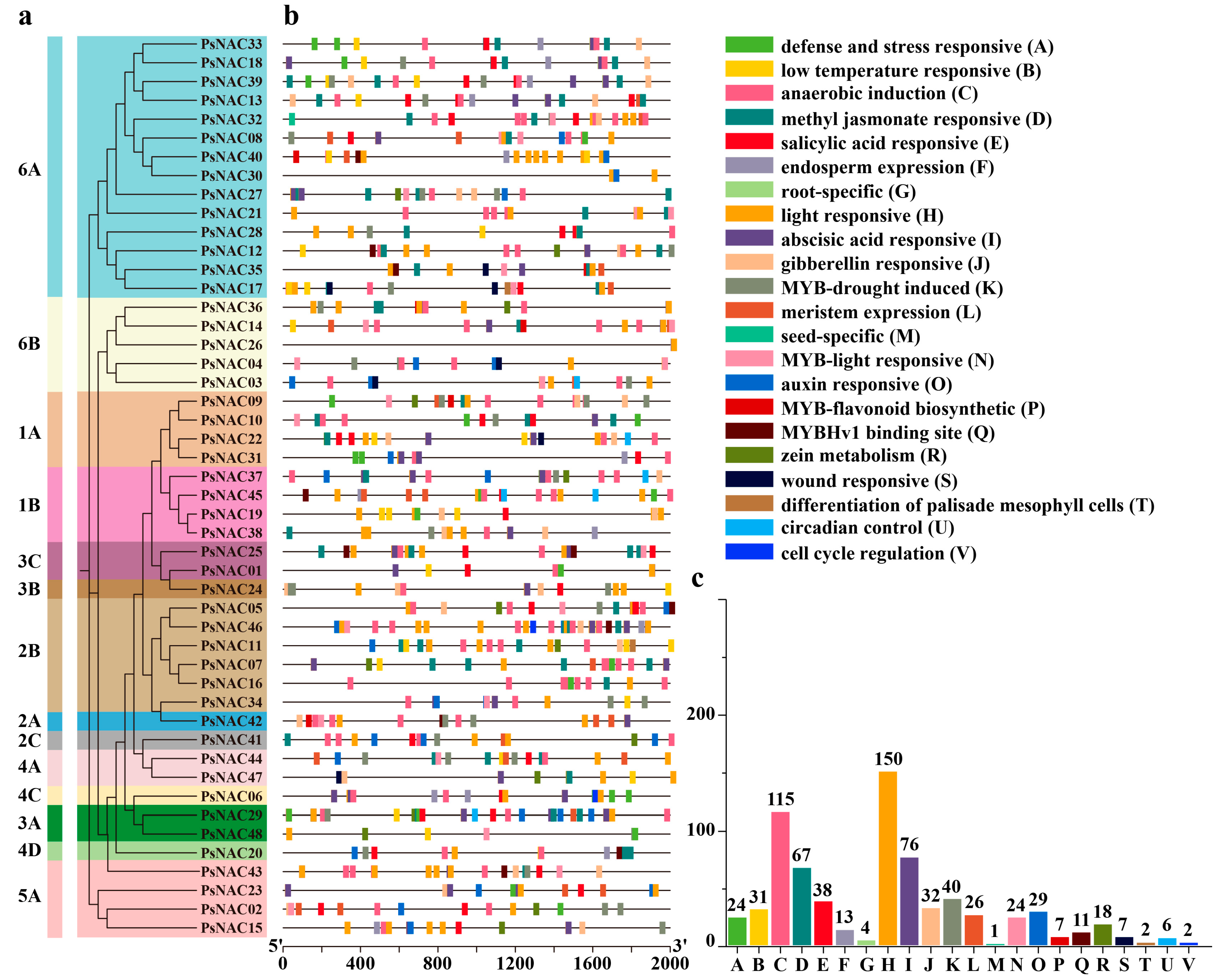
2.4. Analysis of Cis-Acting Elements in the Promoter Region of PsNAC Genes
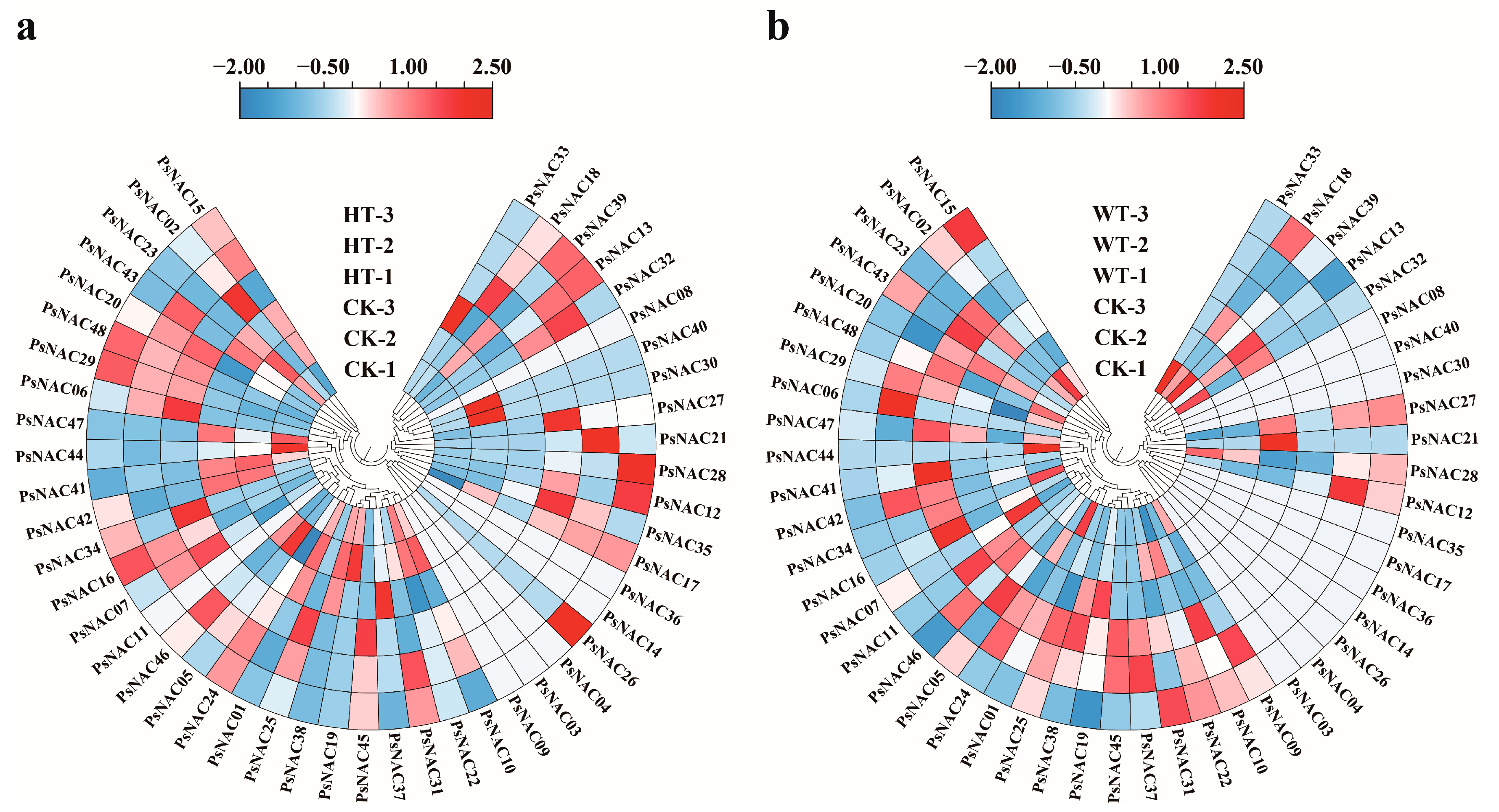
2.5. Expression Pattern Analysis of PsNAC Genes
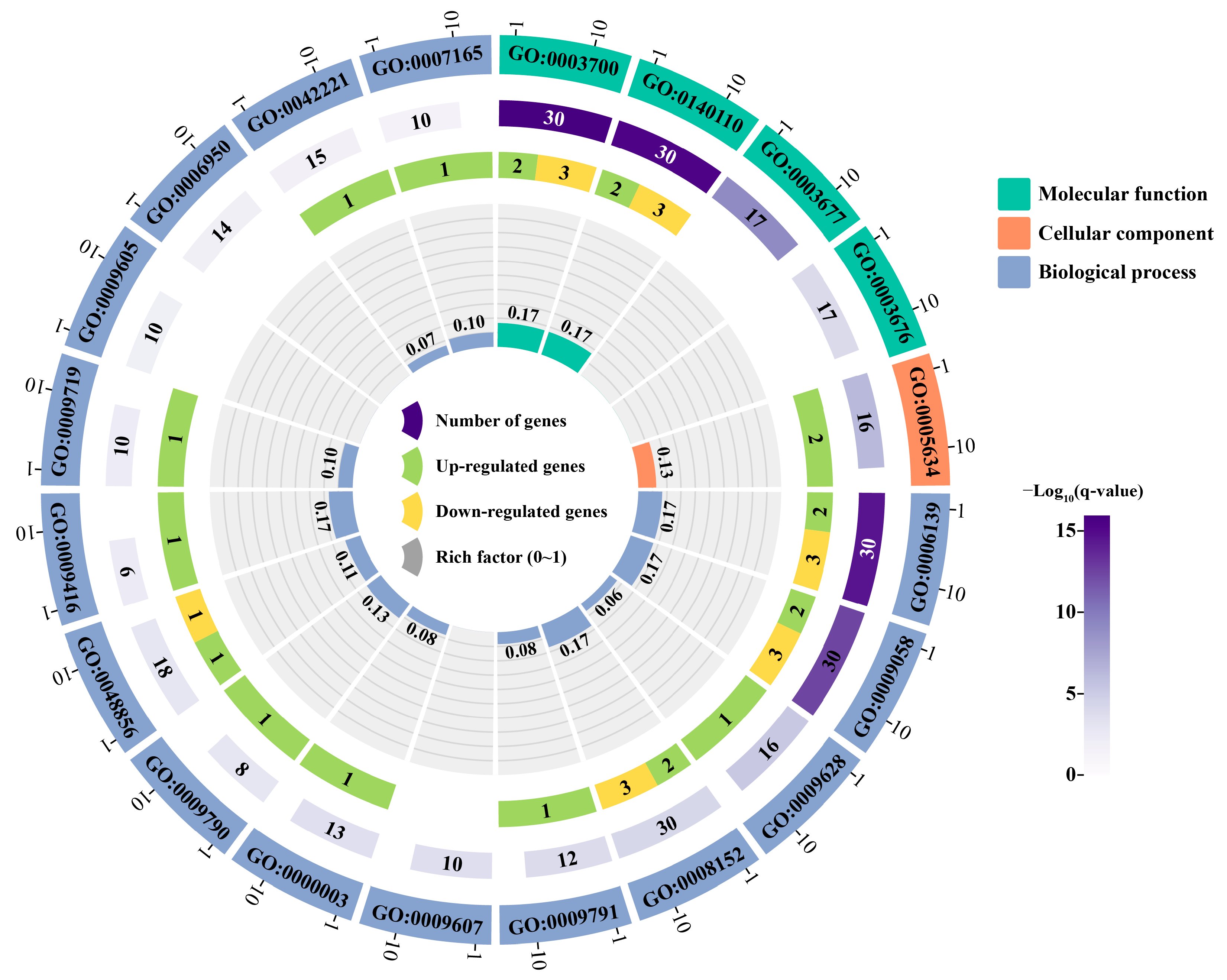
2.6. Annotation and Enrichment Analysis of PsNAC Genes
2.7. qRT-PCR Validation of Differentially Expressed PsNAC Genes
3. Discussion
4. Materials and Methods
4.1. Identification and Testing of Transcription Factors
4.2. Analysis of Physicochemical Properties of Protein Sequences
4.3. Construction of Phylogenetic Tree
4.4. Conserved Sequence Comparison and Motif Analysis
4.5. Analysis of Cis-Acting Elements in the Promoter Region
4.6. Gene Expression Analysis
4.7. Annotation and Enrichment Analysis in GO and KEGG Databases
4.8. Plant Material and Stress Treatments
4.9. qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skirycz, A.; Inzé, D. More from less: Plant growth under limited water. Curr. Opin. Biotechnol. 2010, 21, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Zeng, Y.L.; Yin, F.L.; Wei, R.; Mao, X.F. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in sunflower during salt and drought stress. Sci. Rep. 2021, 11, 19865. [Google Scholar] [CrossRef] [PubMed]
- Rummukainen, M. Changes in climate and weather extremes in the 21st century. Wiley Interdiscip. Rev. Clim. Chang. 2012, 3, 115–129. [Google Scholar] [CrossRef]
- Walter, J.; Harter, D.E.V.; Beierkuhnlein, C.; Jentsch, A. Transgenerational effects of extreme weather: Perennial plant offspring show modified germination, growth and stoichiometry. J. Ecol. 2016, 104, 1032–1040. [Google Scholar] [CrossRef]
- Zhang, X.P.; Xu, Z.D.; Yu, X.Y.; Zhao, L.Y.; Zhao, M.Y.; Han, X.; Qi, S. Identification of two novel R2R3-MYB transcription factors, PsMYB114L and PsMYB12L, related to anthocyanin biosynthesis in Paeonia suffruticosa. Int. J. Mol. Sci. 2019, 20, 1055. [Google Scholar] [CrossRef]
- Lv, S.Z.; Cheng, S.; Wang, Z.Y.; Li, S.M.; Jin, X.; Lan, L.; Yang, B.; Yu, K.; Ni, X.M.; Li, N.; et al. Draft genome of the famous ornamental plant Paeonia suffruticosa. Ecol. Evol. 2020, 10, 4518–4530. [Google Scholar] [CrossRef]
- Wang, S.L.; Ren, X.X.; Xue, J.Q.; Xue, Y.Q.; Cheng, X.D.; Hou, X.G.; Zhang, X.X. Molecular characterization and expression analysis of the Squamosa Promoter Binding Protein-Like gene family in Paeonia suffruticosa. Plant Cell Rep. 2020, 39, 1425–1441. [Google Scholar] [CrossRef]
- Geng, D.D.; Song, S.F.; Li, Y.; Li, T.T.; Shu, Q.Y.; Hao, Q. A novel transcription factor PsMYBM enhances the biosynthesis of anthocyanins in response to light in tree peony. Ind. Crops Prod. 2023, 200, 116800. [Google Scholar] [CrossRef]
- Gu, Z.Y.; Zhu, J.; Hao, Q.; Yuan, Y.W.; Duan, Y.W.; Men, S.Q.; Wang, Q.Y.; Hou, Q.Z.; Liu, Z.A.; Shu, Q.Y.; et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 2019, 60, 599–611. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Q.; Wei, L.L.; Zhao, Y.; Zhang, G.Z.; Wang, J.; Gu, C.H. Responses of the tree peony (Paeonia suffruticosa, Paeoniaceae) cultivar ’Yu Hong’ to heat stress revealed by iTRAQ-based quantitative proteomics. Proteome Sci. 2022, 20, 18. [Google Scholar] [CrossRef]
- Ma, J.; Wang, J.; Wang, Q.; Shang, L.X.; Zhao, Y.; Zhang, G.Z.; Ma, Q.Q.; Hong, S.D.; Gu, C.H. Physiological and transcriptional responses to heat stress and functional analyses of PsHSPs in tree peony (Paeonia suffruticosa). Front. Plant Sci. 2022, 13, 926900. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.T.; Shi, H.J.; Li, X.Q.; Jin, S.H. Salicylic acid induces physiological and biochemical changes in peony under waterlogging stress. Acta Sci. Pol. Hortorum Cultus 2020, 19, 41–52. [Google Scholar] [CrossRef]
- Zhang, X.X.; Liu, X.; Zhou, M.H.; Hu, Y.H.; Yuan, J.H. PacBio full-length sequencing integrated with RNA-seq reveals the molecular mechanism of waterlogging and its recovery in Paeonia ostii. Front. Plant Sci. 2022, 13, 1030584. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, Y.H.; Li, H.X.; Wang, T.T.; Zhang, J.H.; Ouyang, B.; Ye, Z.B. Molecular and functional characterization of ShNAC1, an NAC transcription factor from Solanum habrochaites. Plant Sci. 2018, 271, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozakiy, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.S.; Stewart, C.N. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, S.J.; Guan, C.J.; Kong, X.; Zhang, Y.N. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020, 149, 96–110. [Google Scholar] [CrossRef]
- Yue, Y.Z.; Li, L.; Li, Y.L.; Li, H.Y.; Wang, L.G. Genome-wide analysis of NAC transcription factors and characterization of the cold stress response in sweet osmanthus. Plant Mol. Biol. Rep. 2020, 38, 314–330. [Google Scholar] [CrossRef]
- Zhuo, X.K.; Zheng, T.C.; Zhang, Z.Y.; Zhang, Y.C.; Jiang, L.B.; Ahmad, S.; Sun, L.D.; Wang, J.; Cheng, T.R.; Zhang, Q.X. Genome-wide analysis of the NAC transcription factor gene family reveals differential expression patterns and cold-stress responses in the woody plant Prunus mume. Genes 2018, 9, 494. [Google Scholar] [CrossRef] [PubMed]
- Geng, L.F.; Su, L.; Fu, L.F.; Lin, S.; Zhang, J.M.; Liu, Q.H.; Jiang, X.Q. Genome-wide analysis of the rose (Rosa chinensis) NAC family and characterization of RcNAC091. Plant Mol. Biol. Rep. 2022, 108, 605–619. [Google Scholar] [CrossRef]
- Wang, H.; Li, T.; Li, W.; Wang, W.; Zhao, H.E. Identification and analysis of Chrysanthemum nankingense NAC transcription factors and an expression analysis of OsNAC7 subfamily members. PeerJ 2021, 9, e11505. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Ni, L.J.; Liu, D.N.; Fu, Z.K.; Hua, J.F.; Lu, Z.G.; Liu, L.Q.; Yin, Y.L.; Li, H.G.; Gu, C.S. Genome-wide identification and characterization of NAC family in Hibiscus hamabo Sieb. et Zucc. under various abiotic stresses. Int. J. Mol. Sci. 2022, 23, 3055. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Guan, Q.M.; Yue, X.L.; Zeng, H.T.; Zhu, J.H. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 2014, 26, 438–453. [Google Scholar] [CrossRef]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal. Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef]
- Alshareef, N.O.; Otterbach, S.L.; Allu, A.D.; Woo, Y.H.; de Werk, T.; Kamranfar, I.; Mueller-Roeber, B.; Tester, M.; Balazadeh, S.; Schmoeckel, S.M. NAC transcription factors ATAF1 and ANAC055 affect the heat stress response in Arabidopsis. Sci. Rep. 2022, 12, 11264. [Google Scholar] [CrossRef]
- Xi, Y.; Ling, Q.Q.; Zhou, Y.; Liu, X.; Qian, Y.X. ZmNAC074, a maize stress-responsive NAC transcription factor, confers heat stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 2022, 13, 986628. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Xiang, J.; Teng, R.D.; Zhang, D.H.; Teng, N.J. A lily membrane-associated NAC transcription factor LlNAC014 is involved in thermotolerance via activation of the DREB2-HSFA3 module. J. Exp. Bot. 2023, 74, 945–963. [Google Scholar] [CrossRef]
- Ren, Y.; Huang, Z.Q.; Jiang, H.; Wang, Z.; Wu, F.S.; Xiong, Y.F.; Yao, J.L. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J. Exp. Bot. 2021, 72, 2947–2964. [Google Scholar] [CrossRef] [PubMed]
- Eysholdt-Derzso, E.; Renziehausen, T.; Frings, S.; Frohn, S.; von Bongartz, K.; Igisch, C.P.; Mann, J.; Haeger, L.; Macholl, J.; Leisse, D.; et al. Endoplasmic reticulum-bound ANAC013 factor is cleaved by RHOMBOID-LIKE 2 during the initial response to hypoxia in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2023, 120, e2221308120. [Google Scholar] [CrossRef] [PubMed]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Li, C.F.; Zhang, J.Y.; Zhang, Q.Q.; Dong, A.; Wu, Q.H.; Zhu, X.Y.; Zhu, X.L. Genome-wide identification and analysis of the NAC transcription factor gene family in garden asparagus (Asparagus officinalis). Genes 2022, 13, 976. [Google Scholar] [CrossRef]
- Liu, M.Y.; Ma, Z.T.; Sun, W.J.; Huang, L.; Wu, Q.; Tang, Z.Z.; Bu, T.L.; Li, C.L.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef]
- Sun, M.M.; Liu, X.; Huang, X.J.; Yang, J.J.; Qin, P.T.; Zhou, H.; Jiang, M.G.; Liao, H.Z. Genome-wide identification and expression analysis of the NAC gene family in Kandelia obovata, a typical mangrove plant. Curr. Issues Mol. Biol. 2022, 44, 5622–5637. [Google Scholar] [CrossRef]
- Xia, F.; Liang, X.Y.; Tan, L.N.; Sun, W.; Dai, X.G.; Yan, H.W. Genome-wide identification, evolution and expression profile analysis of NAC transcription factor in Simmondsia chinensis. Curr. Issues Mol. Biol. 2023, 45, 5422–5436. [Google Scholar] [CrossRef]
- Li, Y.; Han, H.; Fu, M.; Zhou, X.; Ye, J.; Xu, F.; Zhang, W.; Liao, Y.; Yang, X. Genome-wide identification and expression analysis of NAC family genes in Ginkgo biloba L. Plant Biol. 2022, 25, 107–118. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, H.; Su, C.L.; Qi, Y.X.; Liu, X.M.; Pu, J.J. Genome-wide identification and expression profile analysis of the NAC transcription factor family during abiotic and biotic stress in woodland strawberry. PLoS ONE 2018, 13, e0197892. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhou, L.; Han, L.L.; Zou, H.Z.; Wang, Y. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony (Paeonia suffruticosa). Plant Physiol. Biochem. 2020, 154, 396–408. [Google Scholar] [CrossRef]
- Ohta, T. Evolution of gene families. Gene 2000, 259, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, C.L.; Yang, S.; Song, J.S.; Li, X.F.; Dai, X.Z.; Wang, F.; Juntawong, N.; Tan, F.J.; Zhang, X.L.; et al. iTRAQ-based quantitative proteomic analysis of heat stress-induced mechanisms in pepper seedlings. PeerJ 2020, 9, e11509. [Google Scholar] [CrossRef]
- An, G. Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiol. 1986, 81, 86–91. [Google Scholar] [CrossRef]
- Lyu, M.L.; Yu, Y.J.; Jiang, J.J.; Song, L.M.; Liang, Y.; Ma, Z.M.; Xiong, X.P.; Cao, J.S. BcMF26a and BcMF26b Are Duplicated Polygalacturonase Genes with Divergent Expression Patterns and Functions in Pollen Development and Pollen Tube Formation in Brassica campestris. PLoS ONE 2015, 10, e0131173. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.R.; Zhang, D.W.; Zhou, H.P.; Zheng, T.; Yin, Y.H.; Lin, H.H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018, 14, e1007336. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Li, S.; Jing, X.L.; Tan, Q.P.; Wen, B.B.; Fu, X.L.; Li, D.M.; Chen, X.D.; Xiao, W.; Li, L. The NAC transcription factor MdNAC29 negatively regulates drought tolerance in apple. Front. Plant Sci. 2023, 14, 1173107. [Google Scholar] [CrossRef]
- Lu, P.L.; Chen, N.Z.; An, R.; Su, Z.; Qi, B.S.; Ren, F.; Chen, J.; Wang, X.C. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. Rep. 2007, 63, 289–305. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.B.; Zhang, H.J.; Li, D.Y.; Song, F.M. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.L.; Eddy, S.R.; Durbin, R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins 1997, 28, 405–420. [Google Scholar] [CrossRef]
- Lu, S.N.; Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Marco, B.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Tiziano, G.C.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.R.; Ye, Z.; Stanton, R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 2020, 26, 903–909. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.Y.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
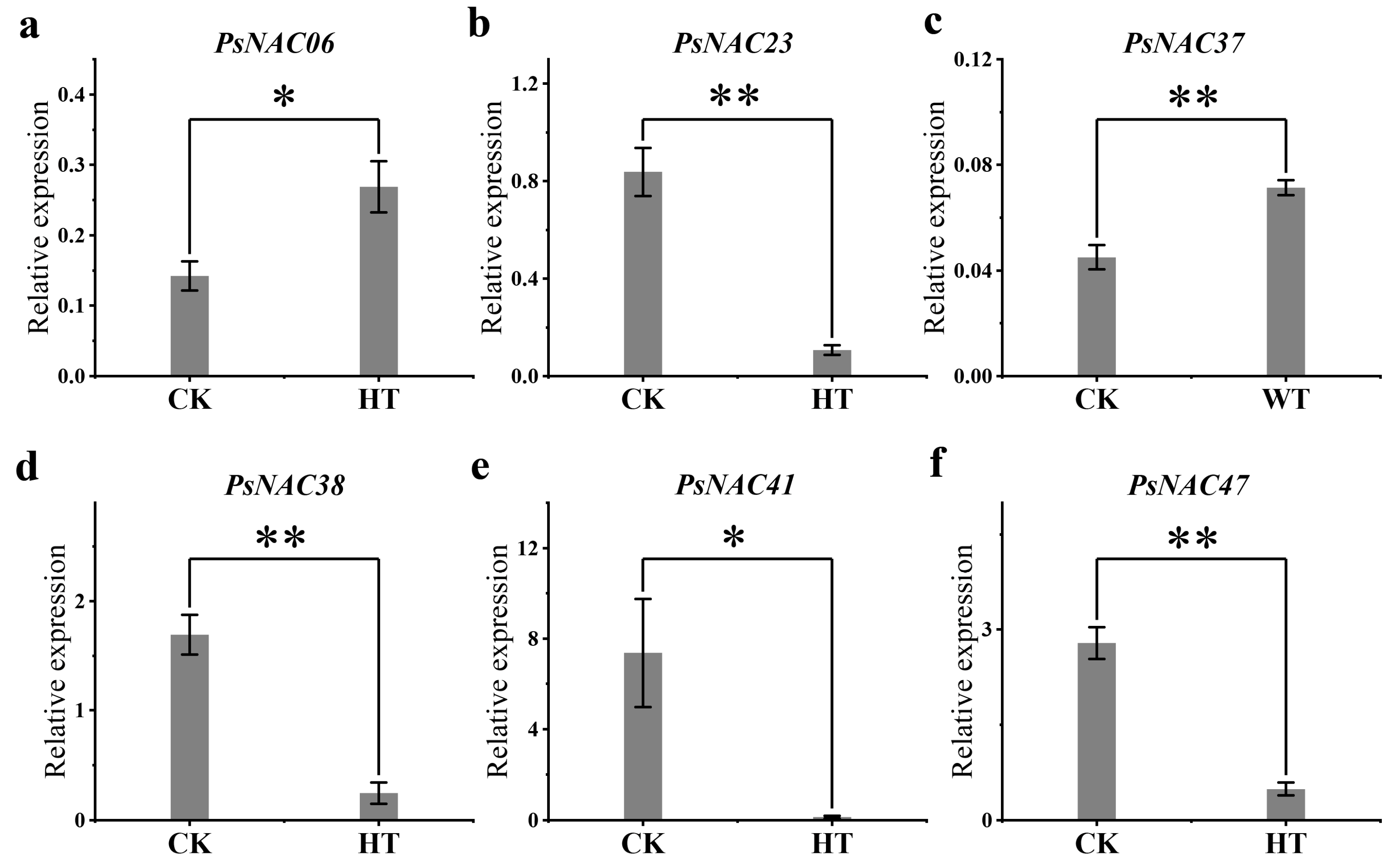
| Treatment | Gene Name | Expression Level | log2 (Fold Change) | q-Value |
|---|---|---|---|---|
| Heat stress | PsNAC06 | up | 3.80155351 | 0.000157825 |
| PsNAC23 | down | −1.142697267 | 3.12 × 10−9 | |
| PsNAC38 | down | −6.598682031 | 0.0000937 | |
| PsNAC41 | down | −2.098132113 | 5.41 × 10−9 | |
| PsNAC47 | down | −2.611193255 | 0.0000686 | |
| Waterlogging stress | PsNAC37 | up | 4.289772524 | 3.41 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhou, L.; Yuan, M.; Peng, F.; Zhu, X.; Wang, Y. Genome-Wide Identification of NAC Gene Family Members of Tree Peony (Paeonia suffruticosa Andrews) and Their Expression under Heat and Waterlogging Stress. Int. J. Mol. Sci. 2024, 25, 9312. https://doi.org/10.3390/ijms25179312
Wang Q, Zhou L, Yuan M, Peng F, Zhu X, Wang Y. Genome-Wide Identification of NAC Gene Family Members of Tree Peony (Paeonia suffruticosa Andrews) and Their Expression under Heat and Waterlogging Stress. International Journal of Molecular Sciences. 2024; 25(17):9312. https://doi.org/10.3390/ijms25179312
Chicago/Turabian StyleWang, Qun, Lin Zhou, Meng Yuan, Fucheng Peng, Xiangtao Zhu, and Yan Wang. 2024. "Genome-Wide Identification of NAC Gene Family Members of Tree Peony (Paeonia suffruticosa Andrews) and Their Expression under Heat and Waterlogging Stress" International Journal of Molecular Sciences 25, no. 17: 9312. https://doi.org/10.3390/ijms25179312
APA StyleWang, Q., Zhou, L., Yuan, M., Peng, F., Zhu, X., & Wang, Y. (2024). Genome-Wide Identification of NAC Gene Family Members of Tree Peony (Paeonia suffruticosa Andrews) and Their Expression under Heat and Waterlogging Stress. International Journal of Molecular Sciences, 25(17), 9312. https://doi.org/10.3390/ijms25179312








