H3K27me3 and EZH Are Involved in the Control of the Heat-Stress-Elicited Morphological Changes in Diatoms
Abstract
1. Introduction
2. Results
2.1. Analysis of the Physiological Responses in the Ptezh Mutants under Standard Conditions
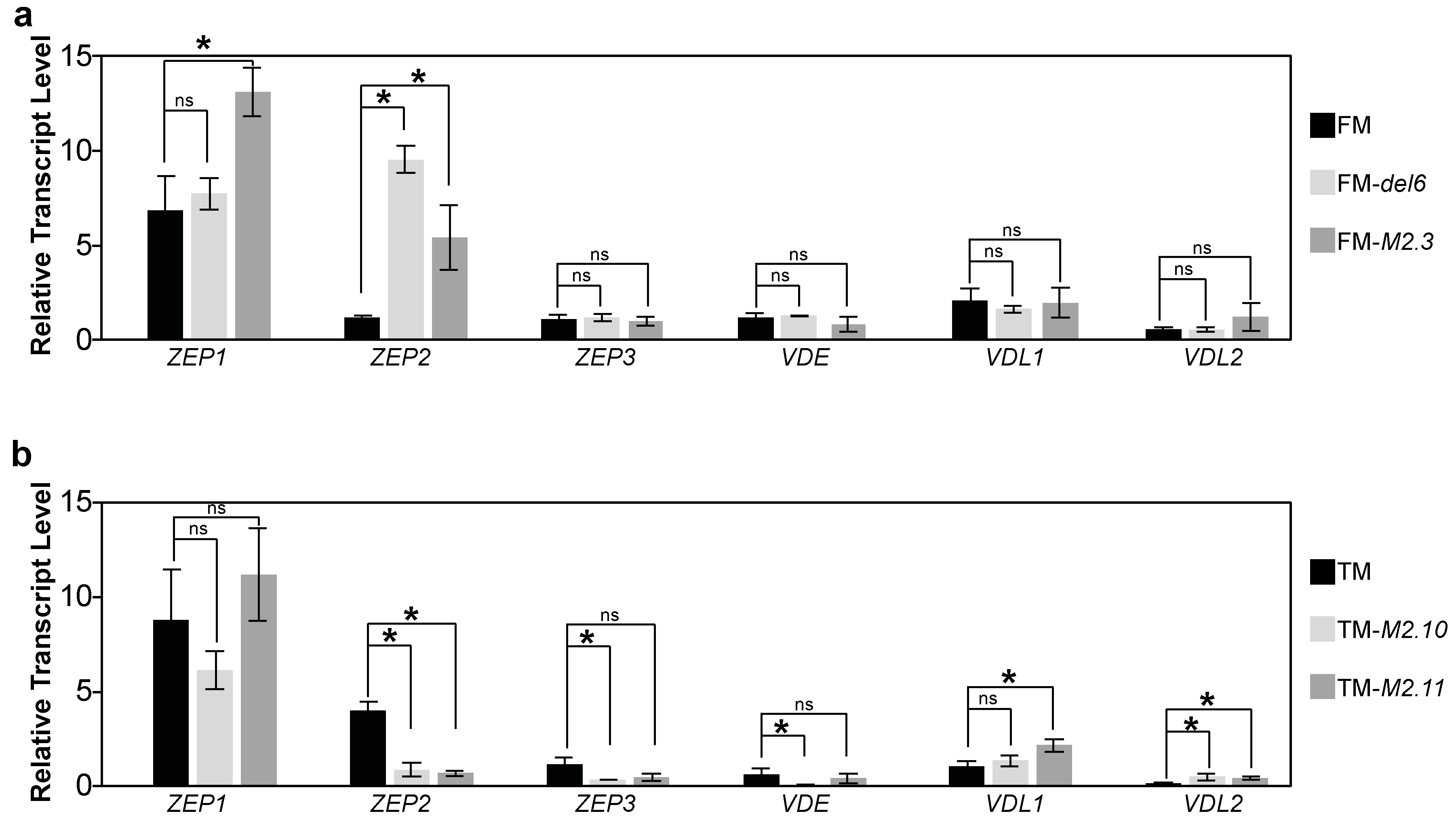
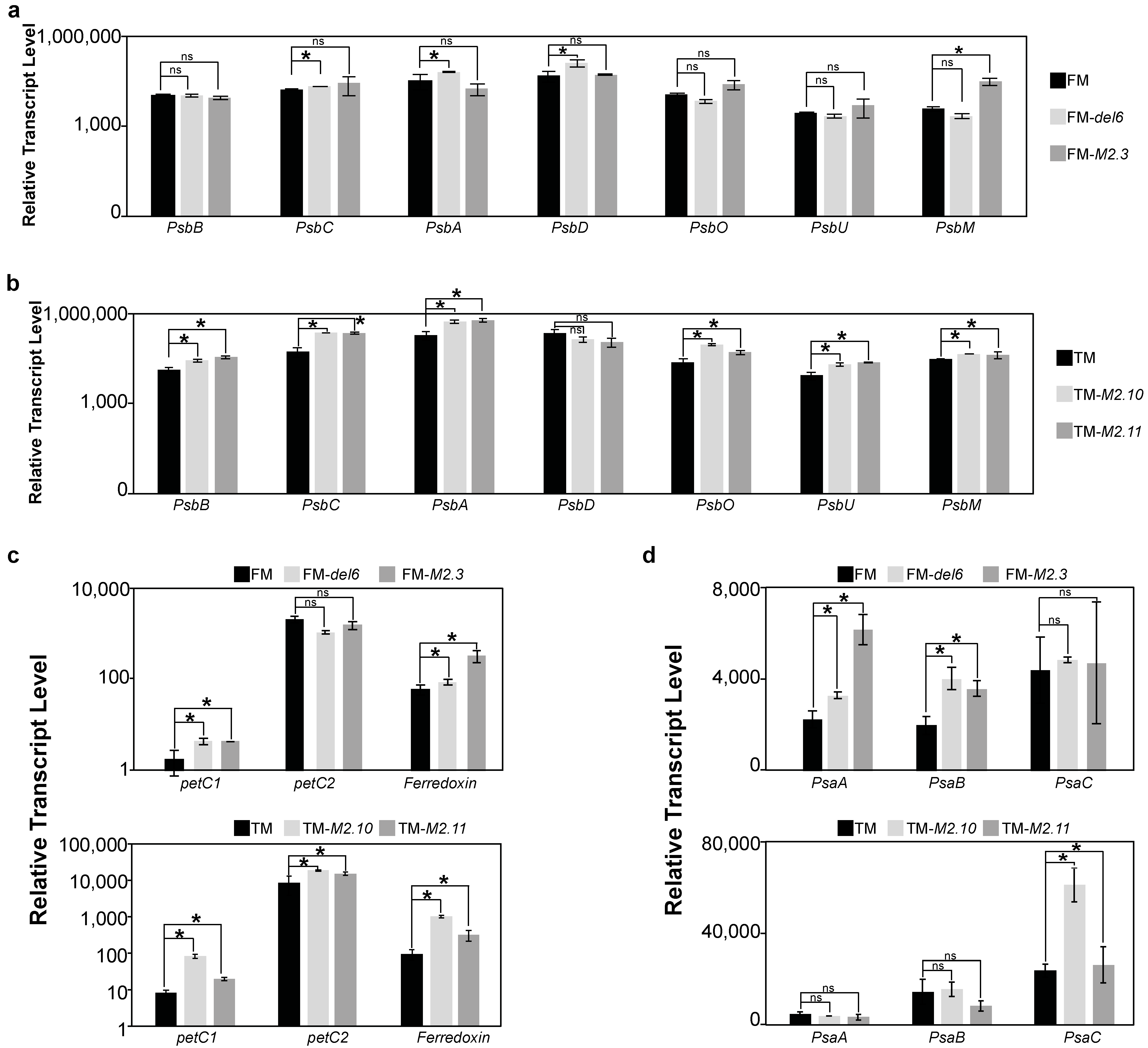
2.2. Gene Expression Analysis of Photosynthesis Proteins in Ptezh Mutants
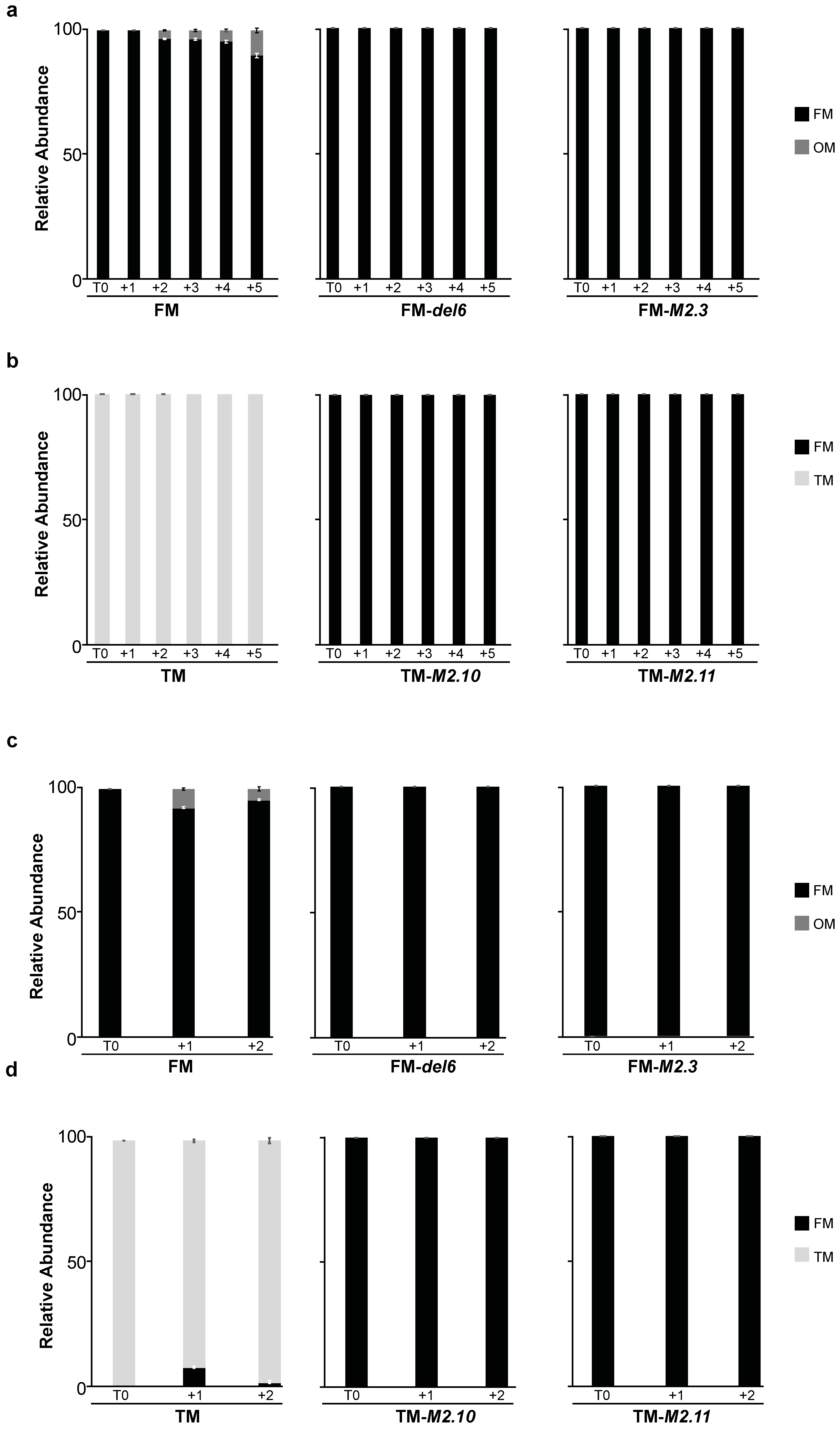
2.3. Morphotype Conversion in Response to Prolonged Heat Stress
2.3.1. Morphotype Conversion in Ecotypes of Pure Morphotypes
2.3.2. Regulation of Morphotype Conversion by PtEZH during a Prolonged Heat Stress
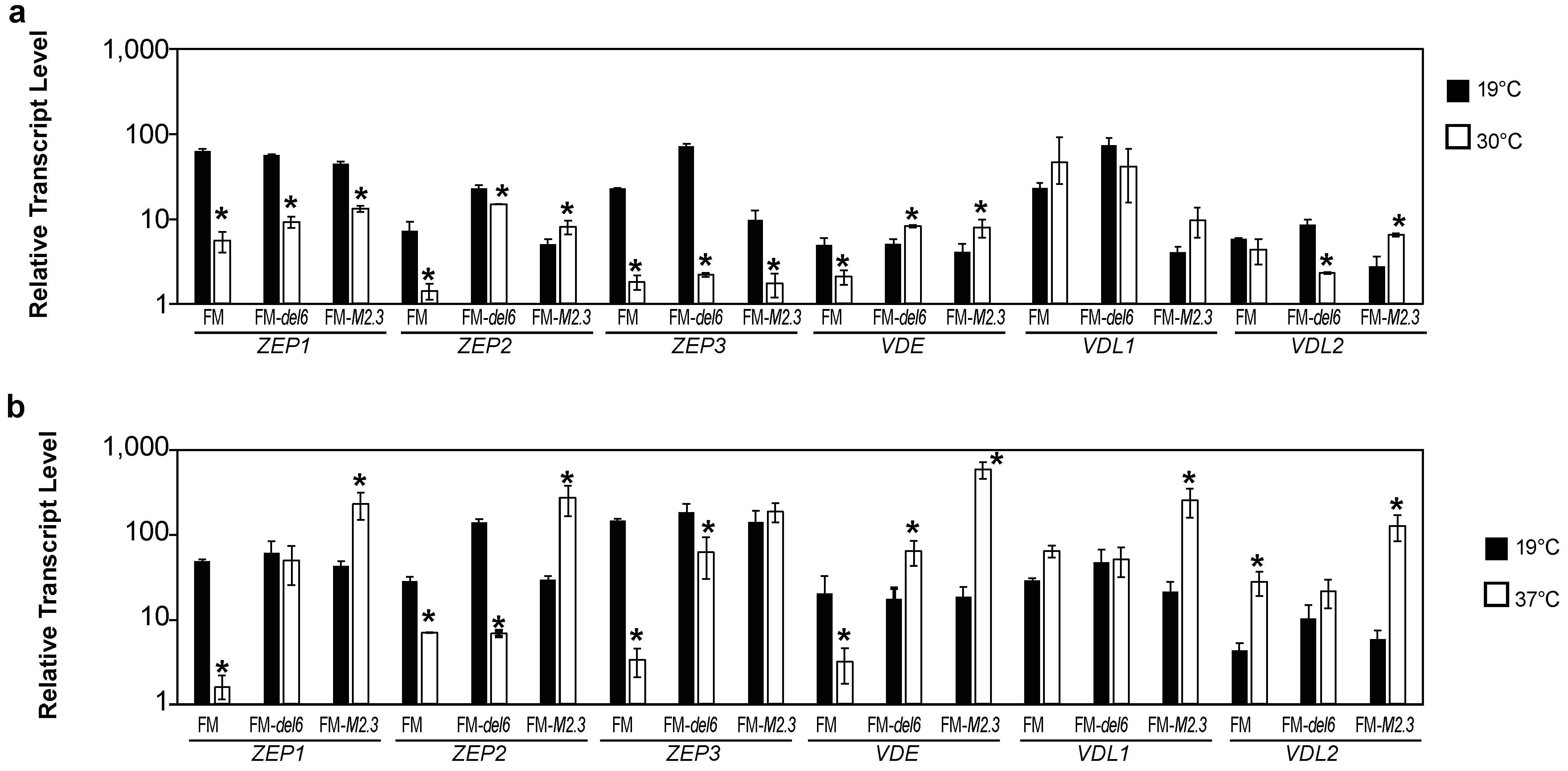
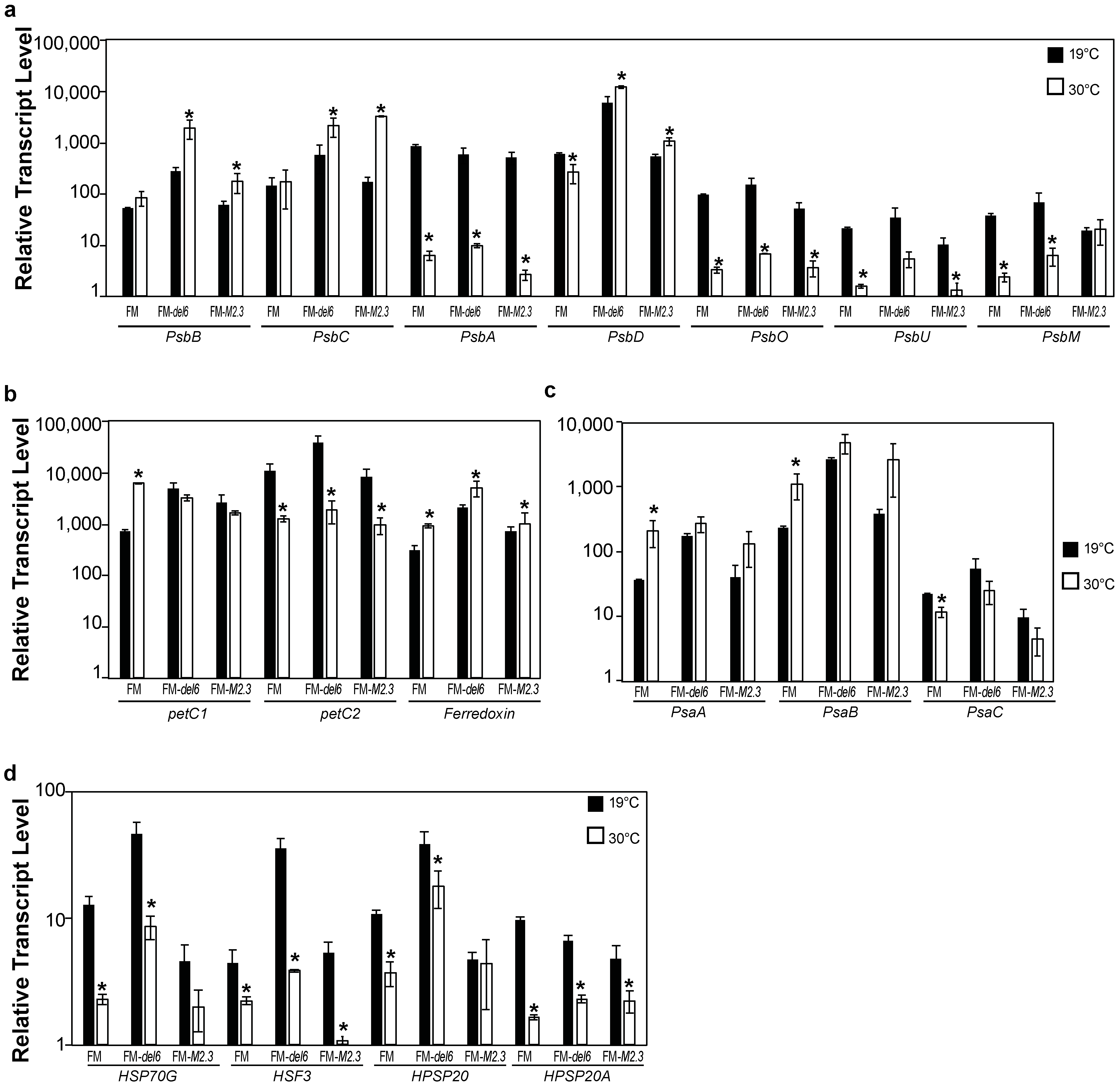
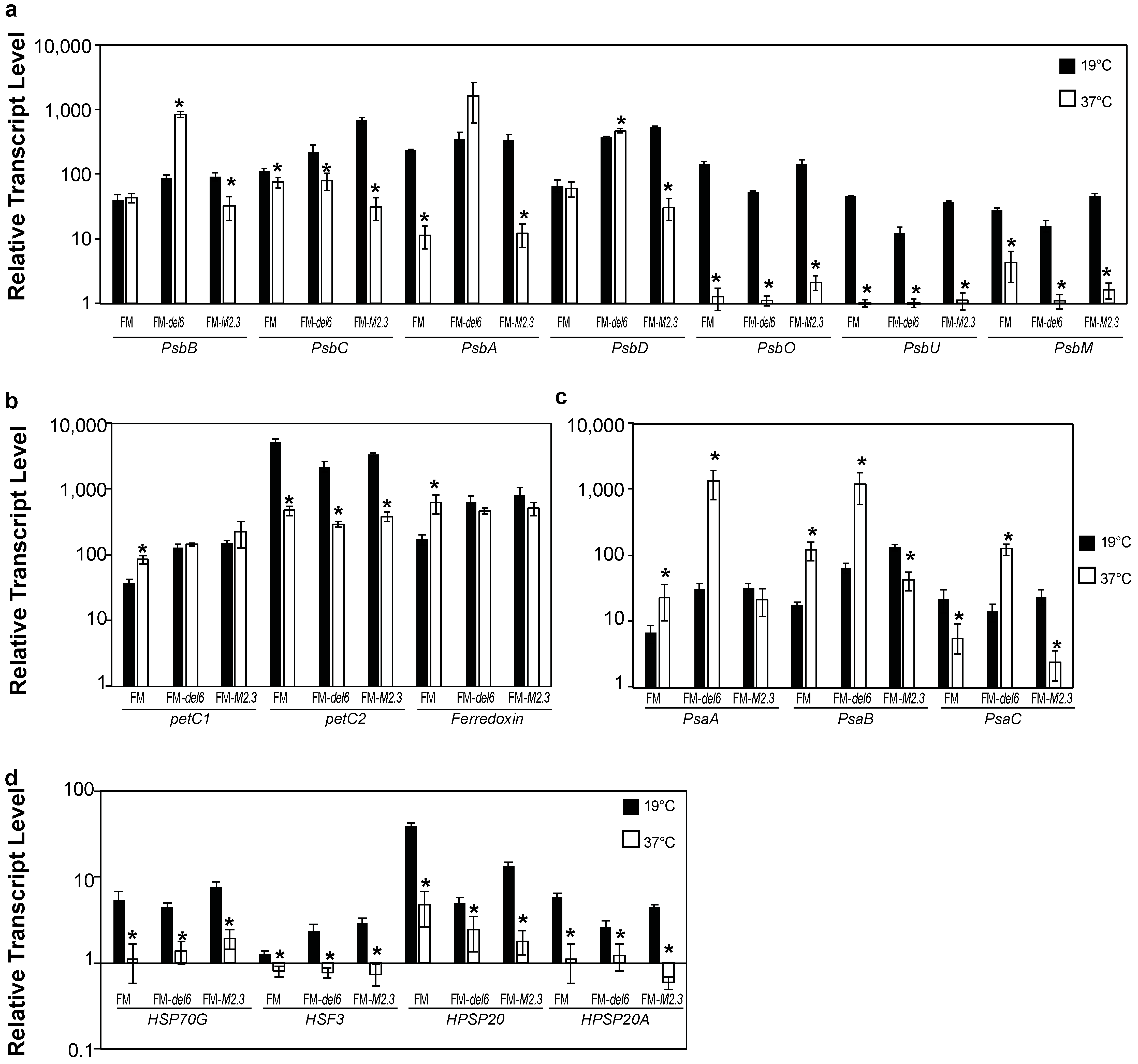
2.4. Analysis of the Effects of Prolonged Heat Stresses on Photosynthesis Gene Expression
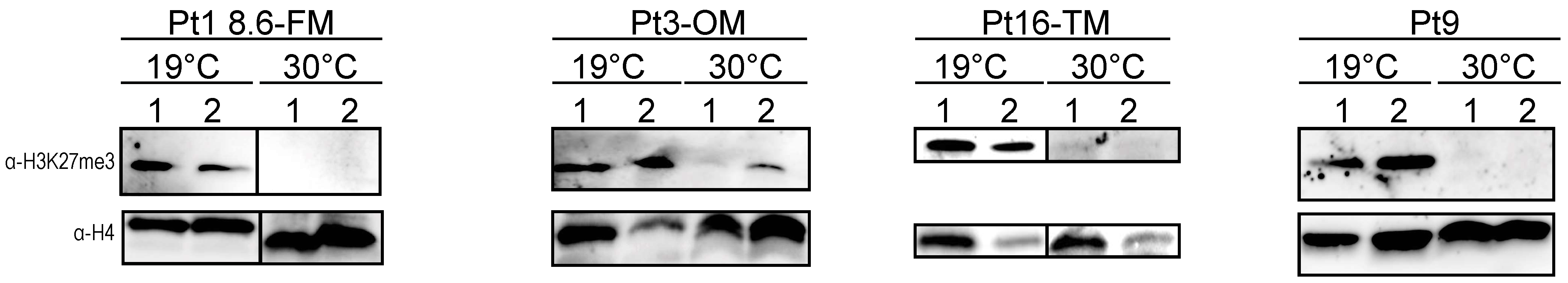
2.5. Analysis of the Effects of Prolonged Heat Stress on H3K27me3 Levels
3. Discussion
4. Materials and Methods
4.1. Used Ecotypes and Genetically-Modified Lines
4.2. Heat Stress Experiments
4.3. Heat Stress Experiments on Synchronised Cells
4.4. PAM Fluorescence Measurements
4.5. Protein Extraction and Immunoblot Analysis
4.6. RNA Extraction and Quantitative RT-PCR
4.7. Protein Sequence Alignments and Structures
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falkowski, P.G.; Barber, R.T.; Smetacek, V. Biogeochemical Controls and Feedbacks on Ocean Primary Production. Science 1998, 281, 200–206. [Google Scholar] [CrossRef] [PubMed]
- De Martino, A.; Meichenin, A.; Shi, J.; Pan, K.; Bowler, C. Genetic and Phenotypic Characterization of Phaeodactylum tricornutum (Bacillariophyceae) Accessions. J. Phycol. 2007, 43, 992–1009. [Google Scholar] [CrossRef]
- De Martino, A.; Bartual, A.; Willis, A.; Meichenin, A.; Villazán, B.; Maheswari, U.; Bowler, C. Physiological and Molecular Evidence That Environmental Changes Elicit Morphological Interconversion in the Model Diatom Phaeodactylum tricornutum. Protist 2011, 162, 462–481. [Google Scholar] [CrossRef]
- Chaumier, T.; Yang, F.; Manirakiza, E.; Ait-Mohamed, O.; Wu, Y.; Chandola, U.; Jesus, B.; Piganeau, G.; Groisillier, A.; Tirichine, L. Genome-Wide Assessment of Genetic Diversity and Transcript Variations in 17 Accessions of the Model Diatom Phaeodactylum tricornutum. ISME Commun. 2024, 4, ycad008. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, A.; Vieira, F.R.J.; Deton-Cabanillas, A.F.; Veluchamy, A.; Cantrel, C.; Wang, G.; Vanormelingen, P.; Bowler, C.; Piganeau, G.; Hu, H.; et al. A Genomics Approach Reveals the Global Genetic Polymorphism, Structure, and Functional Diversity of Ten Accessions of the Marine Model Diatom Phaeodactylum tricornutum. ISME J. 2020, 14, 347–363. [Google Scholar] [CrossRef]
- Feng, Y.; Li, Z.; Li, X.; Shen, L.; Liu, X.; Zhou, C.; Zhang, J.; Sang, M.; Han, G.; Yang, W.; et al. Structure of a Diatom Photosystem II Supercomplex Containing a Member of Lhcx Family and Dimeric FCPII. Sci. Adv. 2023, 9, eadi8446. [Google Scholar] [CrossRef]
- Grouneva, I.; Rokka, A.; Aro, E.M. The Thylakoid Membrane Proteome of Two Marine Diatoms Outlines Both Diatom-Specific and Species-Specific Features of the Photosynthetic Machinery. J. Proteome Res. 2011, 10, 5338–5353. [Google Scholar] [CrossRef]
- Sandmann, G.; Reck, H.; Kessler, E.; Böger, P. Distribution of Plastocyanin and Soluble Plastidic Cytochrome c in Various Classes of Algae. Arch. Microbiol. 1983, 134, 23–27. [Google Scholar] [CrossRef]
- Nagao, R.; Kato, K.; Ifuku, K.; Suzuki, T.; Kumazawa, M.; Uchiyama, I.; Kashino, Y.; Dohmae, N.; Akimoto, S.; Shen, J.-R.; et al. Structural Basis for Assembly and Function of a Diatom Photosystem I-Light-Harvesting Supercomplex. Nat. Commun. 2020, 11, 2481. [Google Scholar] [CrossRef]
- Coesel, S.; Oborník, M.; Varela, J.; Falciatore, A.; Bowler, C. Evolutionary Origins and Functions of the Carotenoid Biosynthetic Pathway in Marine Diatoms. PLoS ONE 2008, 3, e2896. [Google Scholar] [CrossRef]
- Doney, S.C.; Ruckelshaus, M.; Emmett Duffy, J.; Barry, J.P.; Chan, F.; English, C.A.; Galindo, H.M.; Grebmeier, J.M.; Hollowed, A.B.; Knowlton, N.; et al. Climate Change Impacts on Marine Ecosystems. Ann. Rev. Mar. Sci. 2012, 4, 11–37. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.-M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef]
- Glancy, E.; Ciferri, C.; Bracken, A.P. Structural Basis for PRC2 Engagement with Chromatin. Curr. Opin. Struct. Biol. 2021, 67, 135–144. [Google Scholar] [CrossRef]
- Zhao, X.; Rastogi, A.; Deton Cabanillas, A.F.; Ait Mohamed, O.; Cantrel, C.; Lombard, B.; Murik, O.; Genovesio, A.; Bowler, C.; Bouyer, D.; et al. Genome Wide Natural Variation of H3K27me3 Selectively Marks Genes Predicted to Be Important for Cell Differentiation in Phaeodactylum tricornutum. New Phytol. 2021, 229, 3208–3220. [Google Scholar] [CrossRef]
- De Lucia, F.; Crevillen, P.; Jones, A.M.E.; Greb, T.; Dean, C. A PHD-Polycomb Repressive Complex 2 Triggers the Epigenetic Silencing of FLC during Vernalization. Proc. Natl. Acad. Sci. USA 2008, 105, 16831–16836. [Google Scholar] [CrossRef]
- Kim, J.; Bordiya, Y.; Xi, Y.; Zhao, B.; Kim, D.H.; Pyo, Y.; Zong, W.; Ricci, W.A.; Sung, S. Warm Temperature-Triggered Developmental Reprogramming Requires VIL1-Mediated, Genome-Wide H3K27me3 Accumulation in Arabidopsis. Development 2023, 150, dev201343. [Google Scholar] [CrossRef] [PubMed]
- Goss, R.; Lepetit, B. Biodiversity of NPQ. J. Plant Physiol. 2015, 172, 13–32. [Google Scholar] [CrossRef] [PubMed]
- Huysman, M.J.; Martens, C.; Vandepoele, K.; Gillard, J.; Rayko, E.; Heijde, M.; Bowler, C.; Inzé, D.; Van De Peer, Y.; De Veylder, L.; et al. Genome-Wide Analysis of the Diatom Cell Cycle Unveils a Novel Type of Cyclins Involved in Environmental Signaling. Genome Biol. 2010, 11, R17. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Tirichine, L.; Bowler, C. Protocol: Chromatin Immunoprecipitation (ChIP) Methodology to Investigate Histone Modifications in Two Model Diatom Species. Plant Methods 2012, 8, 48. [Google Scholar] [CrossRef]
- Hattich, G.S.I.; Jokinen, S.; Sildever, S.; Gareis, M.; Heikkinen, J.; Junghardt, N.; Segovia, M.; Machado, M.; Sjöqvist, C. Temperature Optima of a Natural Diatom Population Increases as Global Warming Proceeds. Nat. Clim. Chang. 2024, 14, 518–525. [Google Scholar] [CrossRef]
- Kléparski, L.; Beaugrand, G.; Edwards, M.; Schmitt, F.G.; Kirby, R.R.; Breton, E.; Gevaert, F.; Maniez, E. Morphological Traits, Niche-Environment Interaction and Temporal Changes in Diatoms. Prog. Oceanogr. 2022, 201, 102747. [Google Scholar] [CrossRef]
- Allakhverdiev, S.I.; Kreslavski, V.D.; Klimov, V.V.; Los, D.A.; Carpentier, R.; Mohanty, P. Heat Stress: An Overview of Molecular Responses in Photosynthesis. Photosynth. Res. 2008, 98, 541–550. [Google Scholar] [CrossRef]
- Wan, J.; Zhou, Y.; Beardall, J.; Raven, J.A.; Lin, J.; Huang, J.; Lu, Y.; Liang, S.; Ye, M.; Xiao, M.; et al. DNA Methylation and Gene Transcription Act Cooperatively in Driving the Adaptation of a Marine Diatom to Global Change. J. Exp. Bot. 2023, 74, 4259–4276. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Gao, S.; Bao, M.; Li, X.; Liu, Z.; Wang, G. HSP70A Promotes the Photosynthetic Activity of Marine Diatom Phaeodactylum tricornutum under High Temperature. Plant J. 2024, 118, 2085–2093. [Google Scholar] [CrossRef]
- Murray, K.O.; Clanton, T.L.; Horowitz, M. Epigenetic Responses to Heat: From Adaptation to Maladaptation. Exp. Physiol. 2022, 107, 1144–1158. [Google Scholar] [CrossRef] [PubMed]
- Debit, A.; Charton, F.; Pierre-Elies, P.; Bowler, C.; Cruz de Carvalho, H. Differential Expression Patterns of Long Noncoding RNAs in a Pleiomorphic Diatom and Relation to Hyposalinity. Sci. Rep. 2023, 13, 2440. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, A.; Lin, X.; Lombard, B.; Loew, D.; Tirichine, L. Probing the Evolutionary History of Epigenetic Mechanisms: What Can We Learn from Marine Diatoms. AIMS Genet. 2015, 02, 173–191. [Google Scholar] [CrossRef]
- Zhao, P.; Gu, W.; Wu, S.; Huang, A.; He, L.; Xie, X.; Gao, S.; Zhang, B.; Niu, J.; Peng Lin, A.; et al. Silicon Enhances the Growth of Phaeodactylum tricornutum Bohlin under Green Light and Low Temperature. Sci. Rep. 2014, 4, 3958. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, M.; Desclés, J.; Quinet, M.; Douady, S.; Lopez, P.J. Plasticity and Robustness of Pattern Formation in the Model Diatom Phaeodactylum tricornutum. New Phytol. 2009, 182, 429–442. [Google Scholar] [CrossRef]
- Jauffrais, T.; Jesus, B.; Méléder, V.; Turpin, V.; Russo, A.D.P.G.; Raimbault, P.; Jézéquel, V.M. Physiological and Photophysiological Responses of the Benthic Diatom Entomoneis Paludosa (Bacillariophyceae) to Dissolved Inorganic and Organic Nitrogen in Culture. Mar. Biol. 2016, 163, 115. [Google Scholar] [CrossRef]
- Platt, T.; Gallegos, C.; Harrison, W. Photoinhibition of Photosynthesis in Natural Assemblages of Marine Phytoplankton. J. Mar. Res. 1980, 38, 687–701. [Google Scholar]
- Serôdio, J.; Lavaud, J. A Model for Describing the Light Response of the Nonphotochemical Quenching of Chlorophyll Fluorescence. Photosynth. Res. 2011, 108, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Siaut, M.; Heijde, M.; Mangogna, M.; Montsant, A.; Coesel, S.; Allen, A.; Manfredonia, A.; Falciatore, A.; Bowler, C. Molecular Toolbox for Studying Diatom Biology in Phaeodactylum tricornutum. Gene 2007, 406, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarif, M.; Rousselot, E.; Jesus, B.; Tirichine, L.; Duc, C. H3K27me3 and EZH Are Involved in the Control of the Heat-Stress-Elicited Morphological Changes in Diatoms. Int. J. Mol. Sci. 2024, 25, 8373. https://doi.org/10.3390/ijms25158373
Zarif M, Rousselot E, Jesus B, Tirichine L, Duc C. H3K27me3 and EZH Are Involved in the Control of the Heat-Stress-Elicited Morphological Changes in Diatoms. International Journal of Molecular Sciences. 2024; 25(15):8373. https://doi.org/10.3390/ijms25158373
Chicago/Turabian StyleZarif, Mhammad, Ellyn Rousselot, Bruno Jesus, Leïla Tirichine, and Céline Duc. 2024. "H3K27me3 and EZH Are Involved in the Control of the Heat-Stress-Elicited Morphological Changes in Diatoms" International Journal of Molecular Sciences 25, no. 15: 8373. https://doi.org/10.3390/ijms25158373
APA StyleZarif, M., Rousselot, E., Jesus, B., Tirichine, L., & Duc, C. (2024). H3K27me3 and EZH Are Involved in the Control of the Heat-Stress-Elicited Morphological Changes in Diatoms. International Journal of Molecular Sciences, 25(15), 8373. https://doi.org/10.3390/ijms25158373







