Physiological Features of the Neural Stem Cells Obtained from an Animal Model of Spinal Muscular Atrophy and Their Response to Antioxidant Curcumin
Abstract
1. Introduction
2. Results
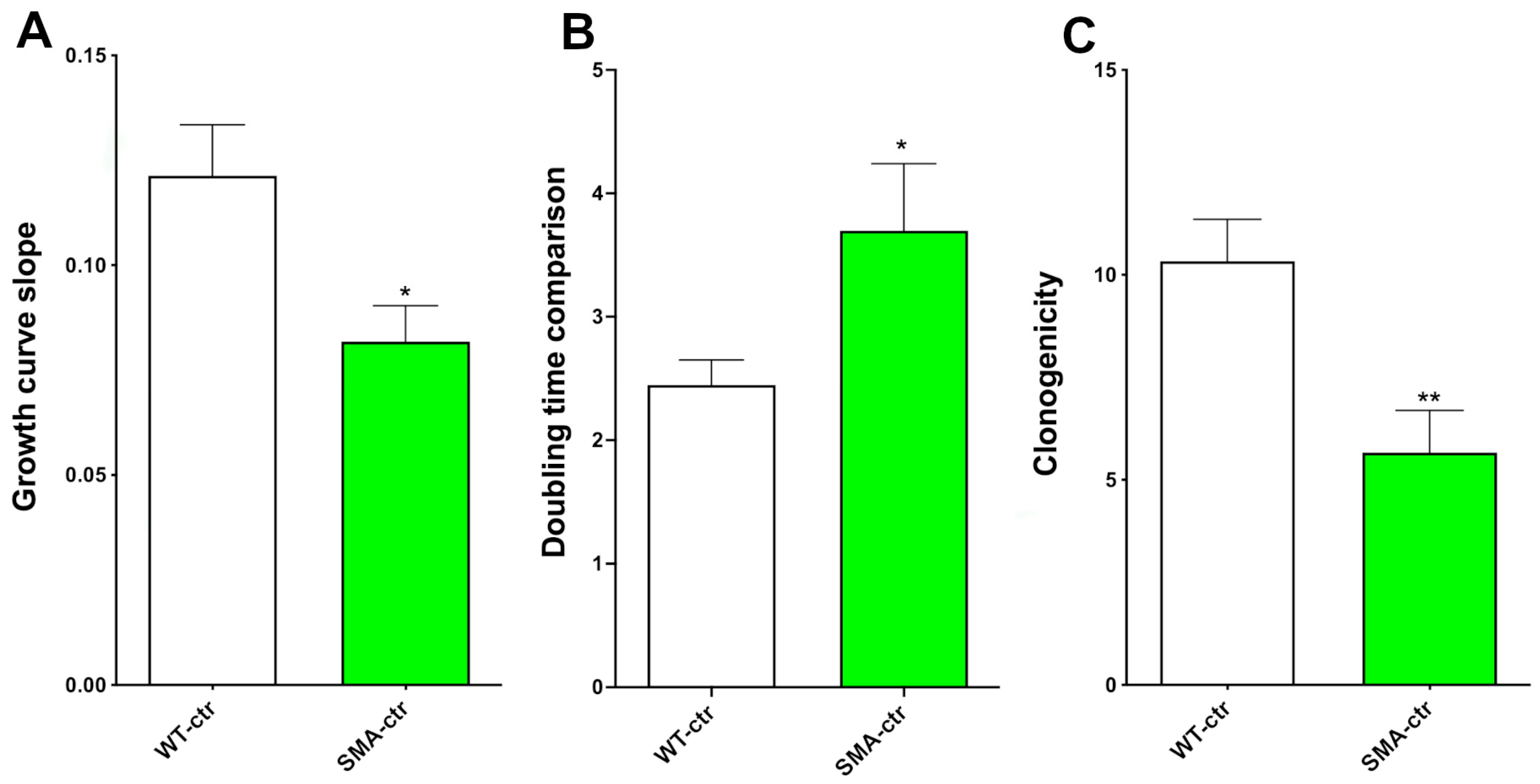
2.1. Proliferation Capability
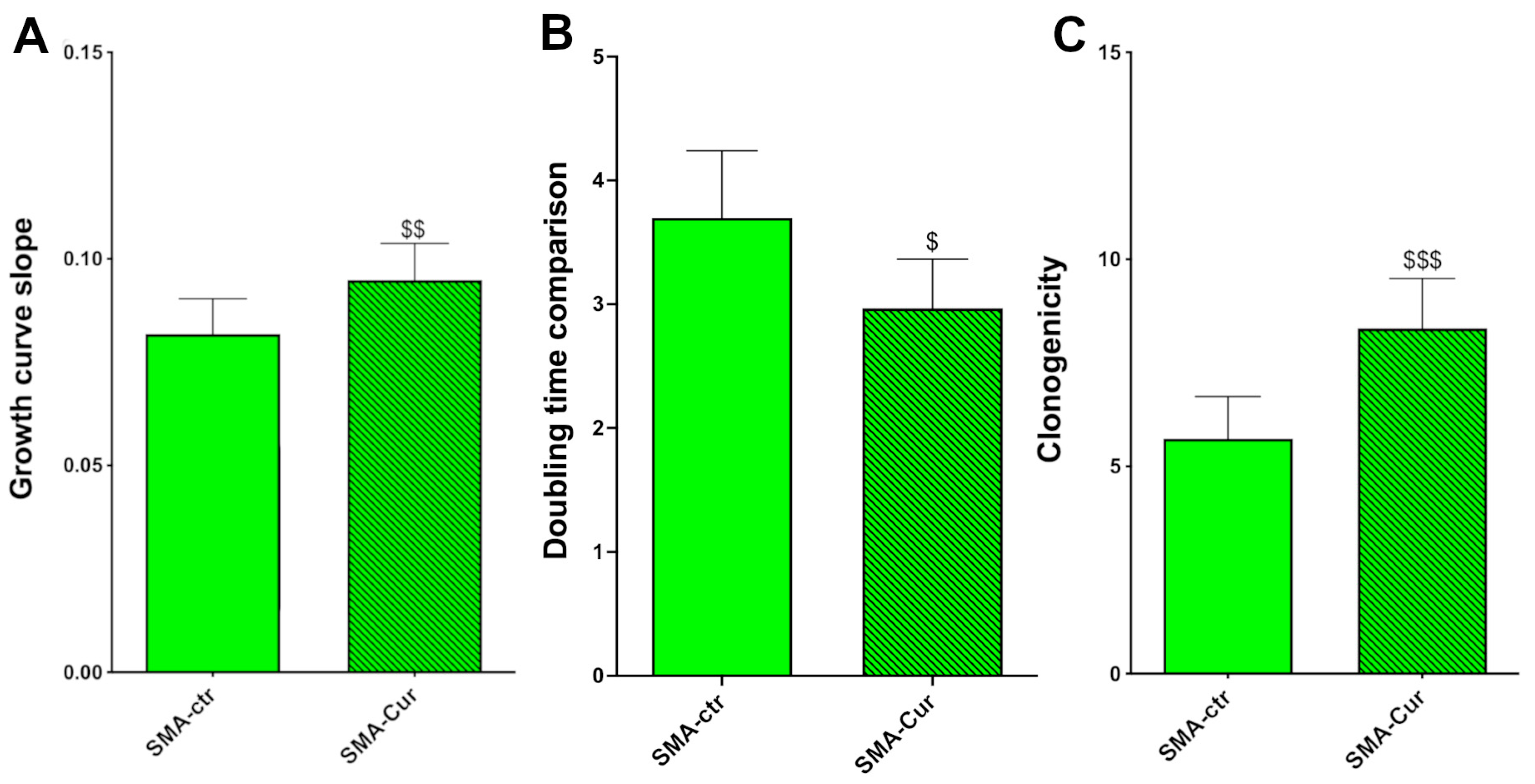
2.2. Clonogenic Capability
2.3. Expression of Stemness Neural Markers
2.4. Metabolic Activity
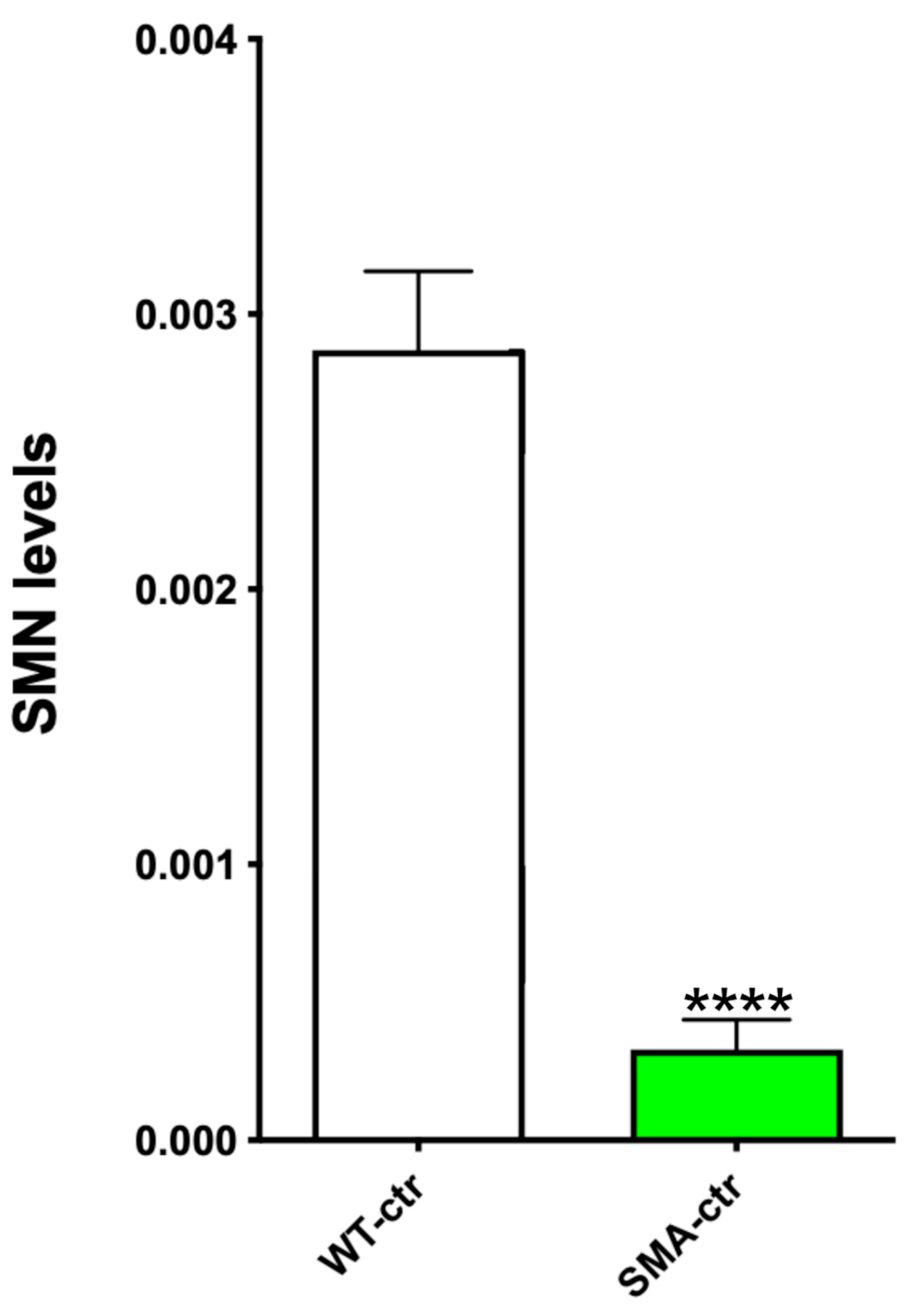
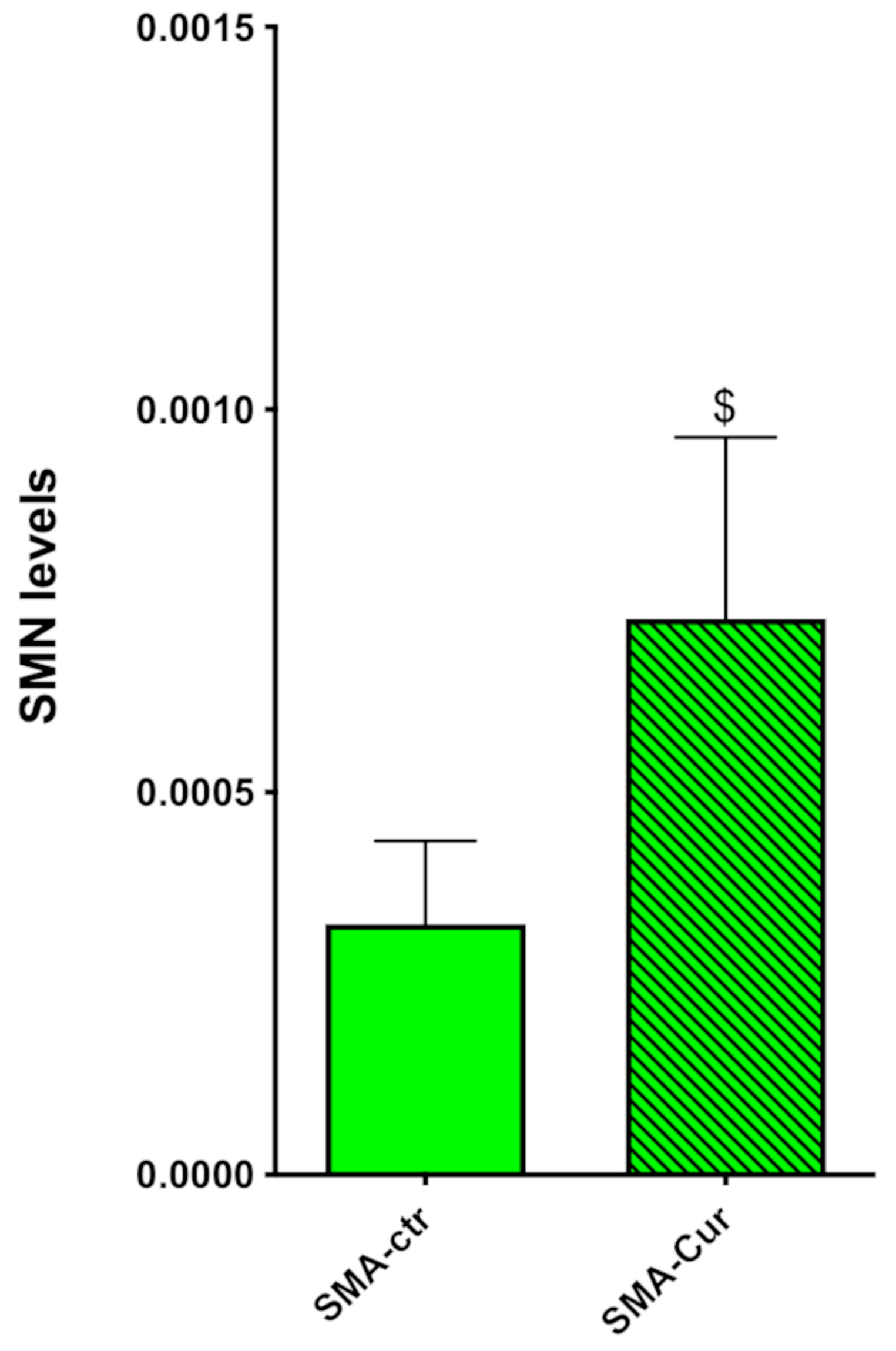
2.5. SMN Protein Levels
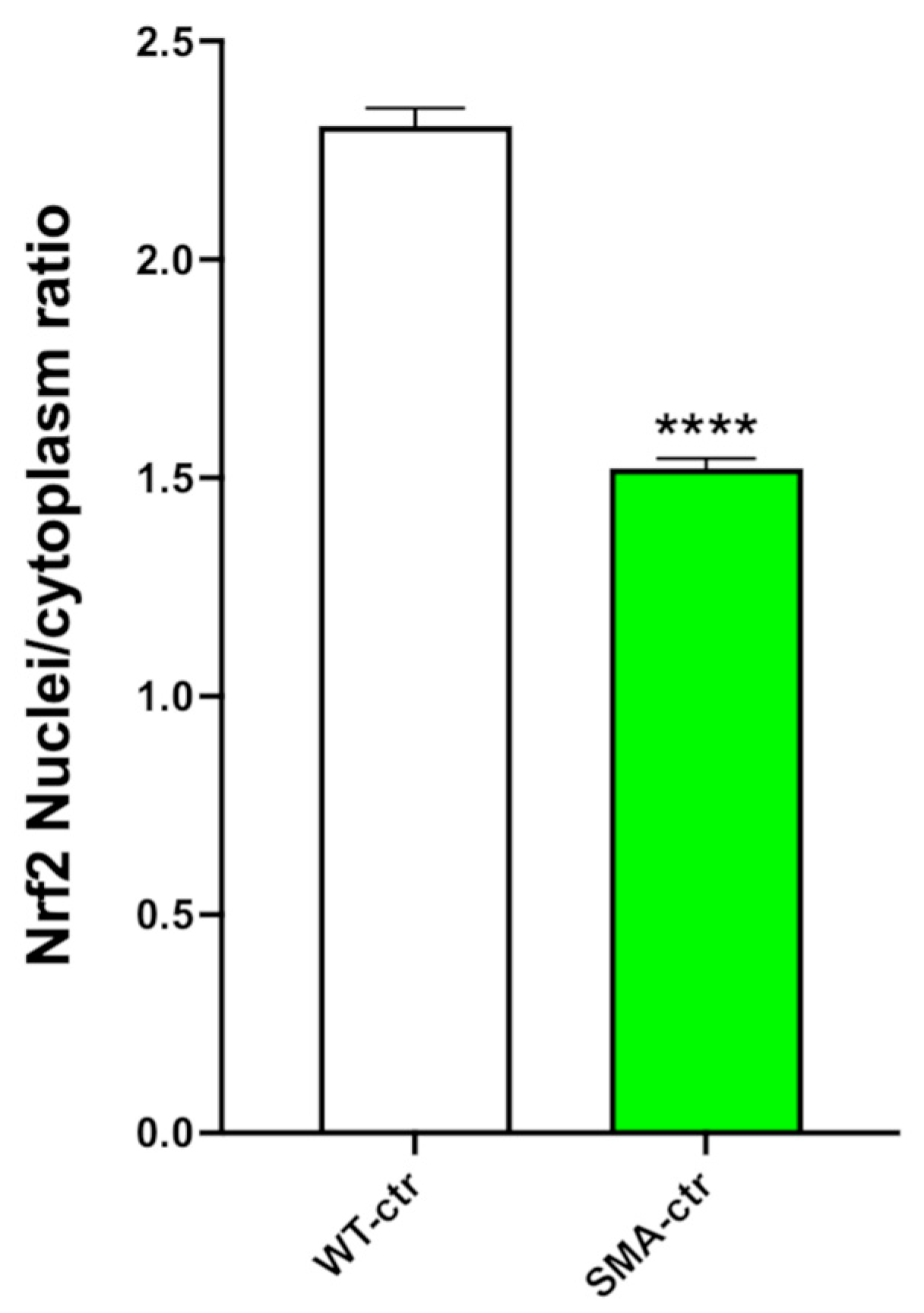
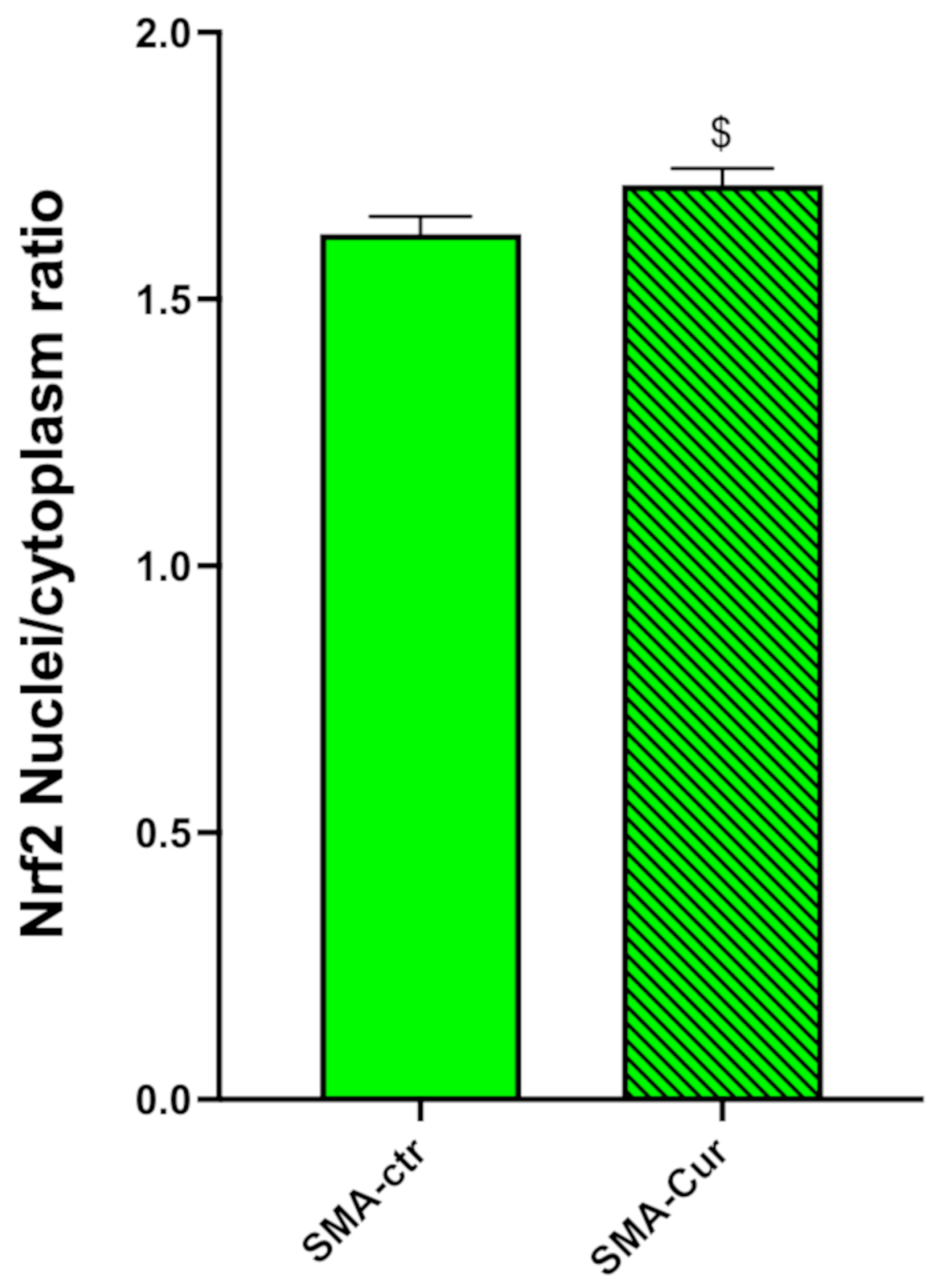
2.6. NRF2 Analysis
2.7. Curcumin Treatment of NSCs
2.8. Curcumin Increases Proliferation Capability, Doubling Time, and Self-Maintenance
2.9. Curcumin Does Not Alter the Expression of Stemness Markers
2.10. Curcumin Significantly Increases Metabolic Activity
2.11. Curcumin Significantly Increases the Expression Levels of SMN
2.12. Curcumin Significantly Increases the Nuclear/Cytoplasmic NRF2 Ratio
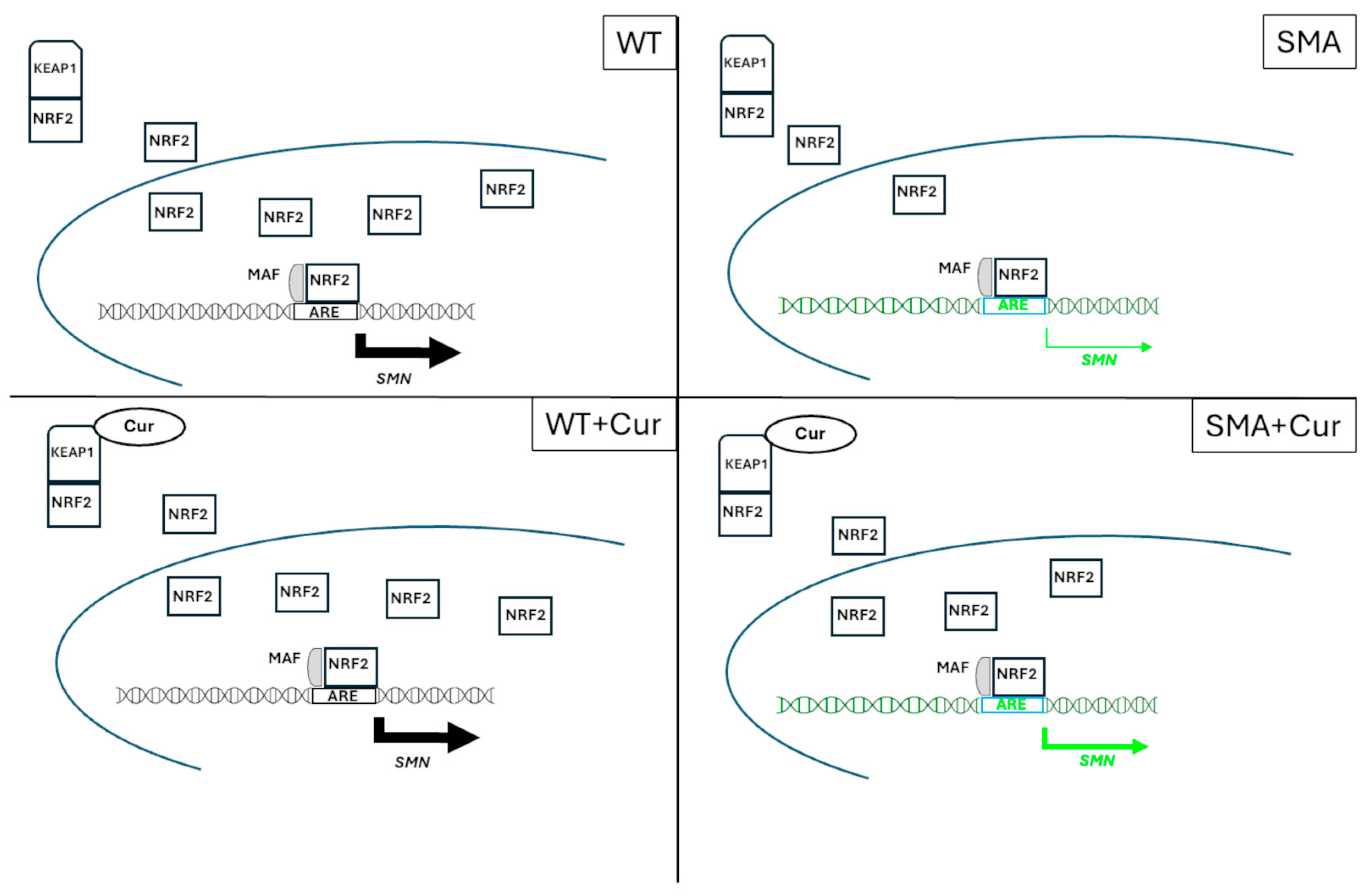
3. Discussion
4. Materials and Methods
4.1. Mouse Models
4.2. Isolation and Culture of NSCs
4.3. Growth Analysis
4.4. Clonal Analyses
4.5. MTT Assay
4.6. Undifferentiated Cell Immunofluorescence
4.7. NRF2 Staining
4.8. NRF2 Nuclear Staining
4.9. Protein Extraction, Preparation, and Quantification and Western Blot Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Werdnig, G. Two early infantile hereditary cases of progressive muscular atrophy simulating dystrophy, but on a neural basis. 1891. Arch. Neurol. 1971, 25, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Adami, R.; Scesa, G.; Bottai, D. Stem cell transplantation in neurological diseases: Improving effectiveness in animal models. Front. Cell Dev. Biol. 2014, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Bottai, D.; Adami, R. Spinal muscular atrophy: New findings for an old pathology. Brain Pathol. 2013, 23, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Talbot, K.; Tizzano, E.F. The clinical landscape for SMA in a new therapeutic era. Gene Ther. 2017, 24, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Bertini, E.; Iannaccone, S.T. Childhood spinal muscular atrophy: Controversies and challenges. Lancet Neurol. 2012, 11, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Finkel, R.S.; McDermott, M.P.; Kaufmann, P.; Darras, B.T.; Chung, W.K.; Sproule, D.M.; Kang, P.B.; Foley, A.R.; Yang, M.L.; Martens, W.B.; et al. Observational study of spinal muscular atrophy type I and implications for clinical trials. Neurology 2014, 83, 810–817. [Google Scholar] [CrossRef]
- Sugarman, E.A.; Nagan, N.; Zhu, H.; Akmaev, V.R.; Zhou, Z.; Rohlfs, E.M.; Flynn, K.; Hendrickson, B.C.; Scholl, T.; Sirko-Osadsa, D.A.; et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: Clinical laboratory analysis of >72,400 specimens. Eur. J. Hum. Genet. 2012, 20, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Adami, R.; Bottai, D. Spinal Muscular Atrophy Modeling and Treatment Advances by Induced Pluripotent Stem Cells Studies. Stem Cell Rev. Rep. 2019, 15, 795–813. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E. Spinal muscular atrophy: From rags to riches. Neuromuscul. Disord. 2021, 31, 998–1003. [Google Scholar] [CrossRef]
- Dubowitz, V. Chaos in classification of the spinal muscular atrophies of childhood. Neuromuscul. Disord. 1991, 1, 77–80. [Google Scholar] [CrossRef]
- Adami, R.; Bottai, D. NSC Physiological Features in Spinal Muscular Atrophy: SMN Deficiency Effects on Neurogenesis. Int. J. Mol. Sci. 2022, 23, 15209. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Dreyfuss, G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010, 24, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zheng, Y.; Guo, M.; Ares, I.; Martínez, M.; Lopez-Torres, B.; Martínez-Larrañaga, M.R.; Wang, X.; Anadón, A.; Martínez, M.A. Oxidative stress, the blood-brain barrier and neurodegenerative diseases: The critical beneficial role of dietary antioxidants. Acta Pharm. Sin. B 2023, 13, 3988–4024. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Seo, J.; Rahn, S.J.; Singh, R.N. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS ONE 2012, 7, e49595. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seo, J.; Singh, N.N.; Ottesen, E.W.; Sivanesan, S.; Shishimorova, M.; Singh, R.N. Oxidative Stress Triggers Body-Wide Skipping of Multiple Exons of the Spinal Muscular Atrophy Gene. PLoS ONE 2016, 11, e0154390. [Google Scholar] [CrossRef]
- Cui, Q.; Wang, W.; Namani, A.; Wang, H.; Hammad, A.; Huang, P.; Gao, Y.; Elshaer, M.; Wu, Y.; Wang, X.J.; et al. NRF2 has a splicing regulatory function involving the survival of motor neuron (SMN) in non-small cell lung cancer. Oncogene 2023, 42, 2751–2763. [Google Scholar] [CrossRef] [PubMed]
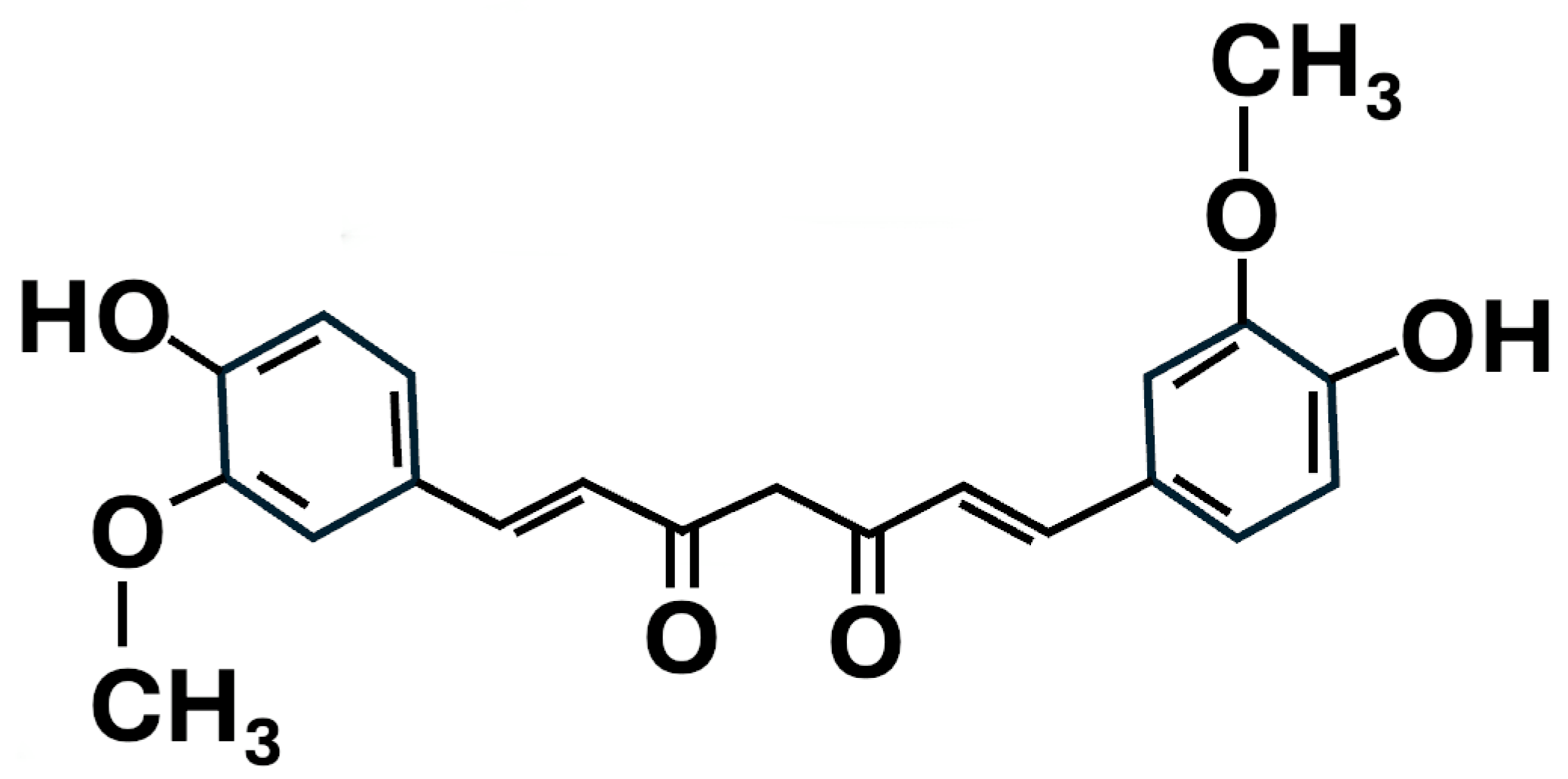
- Adami, R.; Bottai, D. Curcumin and neurological diseases. Nutr. Neurosci. 2022, 25, 441–461. [Google Scholar] [CrossRef] [PubMed]
- Azzini, E.; Peña-Corona, S.I.; Hernández-Parra, H.; Chandran, D.; Saleena, L.A.K.; Sawikr, Y.; Peluso, I.; Dhumal, S.; Kumar, M.; Leyva-Gómez, G.; et al. Neuroprotective and anti-inflammatory effects of curcumin in Alzheimer’s disease: Targeting neuroinflammation strategies. Phytother. Res. 2024, 38, 3169–3189. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Pham, L.T.; Butchbach, M.E.; Zhang, H.L.; Monani, U.R.; Coovert, D.D.; Gavrilina, T.O.; Xing, L.; Bassell, G.J.; Burghes, A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005, 14, 845–857. [Google Scholar] [CrossRef]
- Fosang, A.J.; Colbran, R.J. Transparency Is the Key to Quality. J. Biol. Chem. 2015, 290, 29692–29694. [Google Scholar] [CrossRef]
- Shafey, D.; MacKenzie, A.E.; Kothary, R. Neurodevelopmental abnormalities in neurosphere-derived neural stem cells from SMN-depleted mice. J. Neurosci. Res. 2008, 86, 2839–2847. [Google Scholar] [CrossRef] [PubMed]
- Yeo, C.J.J.; Darras, B.T. Overturning the Paradigm of Spinal Muscular Atrophy as Just a Motor Neuron Disease. Pediatr. Neurol. 2020, 109, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Sumner, C.J.; Muntoni, F.; Darras, B.T.; Finkel, R.S. Spinal muscular atrophy. Nat. Rev. Dis. Primers 2022, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H. New and Developing Therapies in Spinal Muscular Atrophy: From Genotype to Phenotype to Treatment and Where Do We Stand? Int. J. Mol. Sci. 2020, 21, 3297. [Google Scholar] [CrossRef] [PubMed]
- James, R.; Chaytow, H.; Ledahawsky, L.M.; Gillingwater, T.H. Revisiting the role of mitochondria in spinal muscular atrophy. Cell. Mol. Life Sci. 2021, 78, 4785–4804. [Google Scholar] [CrossRef] [PubMed]
- Cadile, F.; Recchia, D.; Ansaldo, M.; Rossi, P.; Rastelli, G.; Boncompagni, S.; Brocca, L.; Pellegrino, M.A.; Canepari, M. Diaphragm Fatigue in SMNΔ7 Mice and Its Molecular Determinants: An Underestimated Issue. Int. J. Mol. Sci. 2023, 24, 14953. [Google Scholar] [CrossRef] [PubMed]
- Viscomi, C.; Bottani, E.; Civiletto, G.; Cerutti, R.; Moggio, M.; Fagiolari, G.; Schon, E.A.; Lamperti, C.; Zeviani, M. In vivo correction of COX deficiency by activation of the AMPK/PGC-1α axis. Cell Metab. 2011, 14, 80–90. [Google Scholar] [CrossRef]
- Zheng, B.; Liao, Z.; Locascio, J.J.; Lesniak, K.A.; Roderick, S.S.; Watt, M.L.; Eklund, A.C.; Zhang-James, Y.; Kim, P.D.; Hauser, M.A.; et al. PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci. Transl. Med. 2010, 2, 52ra73. [Google Scholar] [CrossRef] [PubMed]
- Montesano, A.; Luzi, L.; Senesi, P.; Mazzocchi, N.; Terruzzi, I. Resveratrol promotes myogenesis and hypertrophy in murine myoblasts. J. Transl. Med. 2013, 11, 310. [Google Scholar] [CrossRef]
- Brandalise, F.; Cesaroni, V.; Gregori, A.; Repetti, M.; Romano, C.; Orrù, G.; Botta, L.; Girometta, C.; Guglielminetti, M.L.; Savino, E.; et al. Dietary Supplementation of Hericium erinaceus Increases Mossy Fiber-CA3 Hippocampal Neurotransmission and Recognition Memory in Wild-Type Mice. Evid. Based Complement. Alternat. Med. 2017, 2017, 3864340. [Google Scholar] [CrossRef]
- Luchetti, A.; Ciafrè, S.A.; Murdocca, M.; Malgieri, A.; Masotti, A.; Sanchez, M.; Farace, M.G.; Novelli, G.; Sangiuolo, F. A Perturbed MicroRNA Expression Pattern Characterizes Embryonic Neural Stem Cells Derived from a Severe Mouse Model of Spinal Muscular Atrophy (SMA). Int. J. Mol. Sci. 2015, 16, 18312–18327. [Google Scholar] [CrossRef] [PubMed]
- Attari, F.; Zahmatkesh, M.; Aligholi, H.; Mehr, S.E.; Sharifzadeh, M.; Gorji, A.; Mokhtari, T.; Khaksarian, M.; Hassanzadeh, G. Curcumin as a double-edged sword for stem cells: Dose, time and cell type-specific responses to curcumin. DARU J. Pharm. Sci. 2015, 23, 33. [Google Scholar] [CrossRef] [PubMed]
- Son, S.; Kim, K.T.; Cho, D.C.; Kim, H.J.; Sung, J.K.; Bae, J.S. Curcumin Stimulates Proliferation of Spinal Cord Neural Progenitor Cells via a Mitogen-Activated Protein Kinase Signaling Pathway. J. Korean Neurosurg. Soc. 2014, 56, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Son, T.G.; Park, H.R.; Park, M.; Kim, M.S.; Kim, H.S.; Chung, H.Y.; Mattson, M.P.; Lee, J. Curcumin stimulates proliferation of embryonic neural progenitor cells and neurogenesis in the adult hippocampus. J. Biol. Chem. 2008, 283, 14497–14505. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.X.; Liu, J.; Wang, C.M.; Zhou, J.P.; He, Z.Z.; Lin, H. Low-dose curcumin stimulates proliferation of rat embryonic neural stem cells through glucocorticoid receptor and STAT3. CNS Neurosci. Ther. 2018, 24, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Dhawan, G.; Kapoor, R.; Mattson, M.P.; Rattan, S.I. Curcumin and hormesis with particular emphasis on neural cells. Food Chem. Toxicol. 2019, 129, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Araki, S.; Arai, N.; Kumada, S.; Itoh, M.; Tamagawa, K.; Oda, M.; Morimatsu, Y. Oxidative stress and disturbed glutamate transport in spinal muscular atrophy. Brain Dev. 2002, 24, 770–775. [Google Scholar] [CrossRef]
- Araki, S.; Hayashi, M.; Tamagawa, K.; Saito, M.; Kato, S.; Komori, T.; Sakakihara, Y.; Mizutani, T.; Oda, M. Neuropathological analysis in spinal muscular atrophy type II. Acta Neuropathol. 2003, 106, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Zilio, E.; Piano, V.; Wirth, B. Mitochondrial Dysfunction in Spinal Muscular Atrophy. Int. J. Mol. Sci. 2022, 23, 10878. [Google Scholar] [CrossRef]
- Monani, U.R.; McPherson, J.D.; Burghes, A.H. Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT). Biochim. Biophys. Acta 1999, 1445, 330–336. [Google Scholar] [CrossRef]
- Boda, B.; Mas, C.; Giudicelli, C.; Nepote, V.; Guimiot, F.; Levacher, B.; Zvara, A.; Santha, M.; LeGall, I.; Simonneau, M. Survival motor neuron SMN1 and SMN2 gene promoters: Identical sequences and differential expression in neurons and non-neuronal cells. Eur. J. Hum. Genet. 2004, 12, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.W.; Chun, K.S.; Kim, D.H.; Kim, S.J.; Kim, S.H.; Cho, N.C.; Na, H.K.; Surh, Y.J. Curcumin induces stabilization of Nrf2 protein through Keap1 cysteine modification. Biochem. Pharmacol. 2020, 173, 113820. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, M.; Venkidasamy, B.; Subramanian, U.; Samynathan, R.; Ali Shariati, M.; Rebezov, M.; Girish, S.; Thangavel, S.; Dhanapal, A.R.; Fedoseeva, N.; et al. Bioactive Compounds in Oxidative Stress-Mediated Diseases: Targeting the NRF2/ARE Signaling Pathway and Epigenetic Regulation. Antioxidants 2021, 10, 1859. [Google Scholar] [CrossRef]
- Khor, T.O.; Huang, Y.; Wu, T.Y.; Shu, L.; Lee, J.; Kong, A.N. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem. Pharmacol. 2011, 82, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Furlan, V.; Bren, U. Insight into Inhibitory Mechanism of PDE4D by Dietary Polyphenols Using Molecular Dynamics Simulations and Free Energy Calculations. Biomolecules 2021, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Tsouri, S.; Tselo, E.; Premetis, G.E.; Furlan, V.; Pantiora, P.D.; Mavroidi, B.; Matiadis, D.; Pelecanou, M.; Papageorgiou, A.C.; Bren, U.; et al. A Monocarbonyl Curcuminoid Derivative Inhibits the Activity of Human Glutathione Transferase A4-4 and Chemosensitizes Glioblastoma Cells to Temozolomide. Pharmaceuticals 2024, 17, 365. [Google Scholar] [CrossRef] [PubMed]
- Corenblum, M.J.; Ray, S.; Remley, Q.W.; Long, M.; Harder, B.; Zhang, D.D.; Barnes, C.A.; Madhavan, L. Reduced Nrf2 expression mediates the decline in neural stem cell function during a critical middle-age period. Aging Cell 2016, 15, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Anandhan, A.; Kirwan, K.R.; Corenblum, M.J.; Madhavan, L. Enhanced NRF2 expression mitigates the decline in neural stem cell function during aging. Aging Cell 2021, 20, e13385. [Google Scholar] [CrossRef] [PubMed]
- Starvaggi Cucuzza, L.; Motta, M.; Miretti, S.; Accornero, P.; Baratta, M. Curcuminoid-phospholipid complex induces apoptosis in mammary epithelial cells by STAT-3 signaling. Exp. Mol. Med. 2008, 40, 647–657. [Google Scholar] [CrossRef]
- Bottai, D.; Spreafico, M.; Pistocchi, A.; Fazio, G.; Adami, R.; Grazioli, P.; Canu, A.; Bragato, C.; Rigamonti, S.; Parodi, C.; et al. Modeling Cornelia de Lange syndrome in vitro and in vivo reveals a role for cohesin complex in neuronal survival and differentiation. Hum. Mol. Genet. 2019, 28, 64–73. [Google Scholar] [CrossRef]
- Grazioli, P.; Parodi, C.; Mariani, M.; Bottai, D.; Di Fede, E.; Zulueta, A.; Avagliano, L.; Cereda, A.; Tenconi, R.; Wierzba, J.; et al. Lithium as a possible therapeutic strategy for Cornelia de Lange syndrome. Cell Death Discov. 2021, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Adami, R.; Pagano, J.; Colombo, M.; Platonova, N.; Recchia, D.; Chiaramonte, R.; Bottinelli, R.; Canepari, M.; Bottai, D. Reduction of Movement in Neurological Diseases: Effects on Neural Stem Cells Characteristics. Front. Neurosci. 2018, 12, 336. [Google Scholar] [CrossRef] [PubMed]
- Vescovi, A.L.; Galli, R.; Gritti, A. Clonal analyses and cryopreservation of neural stem cell cultures. In Neural Stem Cells: Methods and Protocols; Methods in Molecular Biology™; Humana Press: Totowa, NJ, USA, 2002; Volume 198, pp. 115–123. [Google Scholar] [CrossRef]
- Bottai, D.; Cigognini, D.; Nicora, E.; Moro, M.; Grimoldi, M.G.; Adami, R.; Abrignani, S.; Marconi, A.M.; Di Giulio, A.M.; Gorio, A. Third trimester amniotic fluid cells with the capacity to develop neural phenotypes and with heterogeneity among sub-populations. Restor. Neurol. Neurosci. 2012, 30, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Petit, A.C.; Legué, E.; Nicolas, J.F. Methods in clonal analysis and applications. Reprod. Nutr. Dev. 2005, 45, 321–339. [Google Scholar] [CrossRef] [PubMed]
- Gritti, A.; Galli, R.; Vescovi, A.L. Clonal analyses and cryopreservation of neural stem cell cultures. In Neural Stem Cells; Methods in Molecular Biology™; Humana Press: Totowa, NJ, USA, 2008; Volume 438, pp. 173–184. [Google Scholar] [CrossRef]
- Roda, E.; De Luca, F.; Priori, E.C.; Ratto, D.; Pinelli, S.; Corradini, E.; Mozzoni, P.; Poli, D.; Mazzini, G.; Bottone, M.G.; et al. The Designer Drug αPHP Affected Cell Proliferation and Triggered Deathly Mechanisms in Murine Neural Stem/Progenitor Cells. Biology 2023, 12, 1225. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Bottai, D.; Scesa, G.; Cigognini, D.; Adami, R.; Nicora, E.; Abrignani, S.; Di Giulio, A.M.; Gorio, A. Third trimester NG2-positive amniotic fluid cells are effective in improving repair in spinal cord injury. Exp. Neurol. 2014, 254, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, J.W.; McLaury, A.R.; Limberg, A.K.; Vargas-Hernandez, J.S.; Bayram, B.; Owen, A.R.; Berry, D.J.; Sanchez-Sotelo, J.; Morrey, M.E.; van Wijnen, A.J.; et al. Total Protein Staining is Superior to Classical or Tissue-Specific Protein Staining for Standardization of Protein Biomarkers in Heterogeneous Tissue Samples. Gene Rep. 2020, 19, 100641. [Google Scholar] [CrossRef]
- Maloy, A.; Alexander, S.; Andreas, A.; Nyunoya, T.; Chandra, D. Stain-Free total-protein normalization enhances the reproducibility of Western blot data. Anal. Biochem. 2022, 654, 114840. [Google Scholar] [CrossRef]
- Muntoni, F.; Bertini, E.; Comi, G.; Kirschner, J.; Lusakowska, A.; Mercuri, E.; Scoto, M.; van der Pol, W.L.; Vuillerot, C.; Burdeska, A.; et al. Long-term follow-up of patients with type 2 and non-ambulant type 3 spinal muscular atrophy (SMA) treated with olesoxime in the OLEOS trial. Neuromuscul. Disord. 2020, 30, 959–969. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adami, R.; Pezzotta, M.; Cadile, F.; Cuniolo, B.; Rovati, G.; Canepari, M.; Bottai, D. Physiological Features of the Neural Stem Cells Obtained from an Animal Model of Spinal Muscular Atrophy and Their Response to Antioxidant Curcumin. Int. J. Mol. Sci. 2024, 25, 8364. https://doi.org/10.3390/ijms25158364
Adami R, Pezzotta M, Cadile F, Cuniolo B, Rovati G, Canepari M, Bottai D. Physiological Features of the Neural Stem Cells Obtained from an Animal Model of Spinal Muscular Atrophy and Their Response to Antioxidant Curcumin. International Journal of Molecular Sciences. 2024; 25(15):8364. https://doi.org/10.3390/ijms25158364
Chicago/Turabian StyleAdami, Raffaella, Matteo Pezzotta, Francesca Cadile, Beatrice Cuniolo, Gianenrico Rovati, Monica Canepari, and Daniele Bottai. 2024. "Physiological Features of the Neural Stem Cells Obtained from an Animal Model of Spinal Muscular Atrophy and Their Response to Antioxidant Curcumin" International Journal of Molecular Sciences 25, no. 15: 8364. https://doi.org/10.3390/ijms25158364
APA StyleAdami, R., Pezzotta, M., Cadile, F., Cuniolo, B., Rovati, G., Canepari, M., & Bottai, D. (2024). Physiological Features of the Neural Stem Cells Obtained from an Animal Model of Spinal Muscular Atrophy and Their Response to Antioxidant Curcumin. International Journal of Molecular Sciences, 25(15), 8364. https://doi.org/10.3390/ijms25158364







