MiR-223-3p in Cancer Development and Cancer Drug Resistance: Same Coin, Different Faces
Abstract
1. Introduction
2. Physiological and Pathophysiological Roles of MiR-223-3p: Inflammation and Immune Cell Differentiation
3. Role of miR-223 in Specific Types of Cancers
3.1. MiR-223 in Colorectal Cancer (CRC)
3.1.1. MiR-223 as an Onco-Suppressor in CRC
3.1.2. MiR-223 as an Onco-miRNA in CRC
3.1.3. Role of miR-223 in Drug Resistance in CRC
3.1.4. MiR-223 as a Biomarker in CRC
3.2. MiR-223 in Lung Cancer
3.2.1. MiR-223 as an Onco-Suppressor in NSCLC
3.2.2. MiR-223 as an Onco-miRNA in NSCLC
3.2.3. Role of miR-223 in Drug Resistance of NSCLC Cells
3.2.4. MiR-223 as Biomarker of NSCLC
3.3. MiR-223 in Breast Cancer
3.3.1. MiR-223 Role as an Onco-Suppressor in Breast Cancer
3.3.2. MiR-223 as an Onco-miRNA in Breast Cancer
3.3.3. MiR-223 in Breast Cancer Drug and Radiotherapy Resistance
3.3.4. MiR-223 as Biomarker of Breast Cancer
3.4. MiR-223 in Ovarian Cancer
3.4.1. MiR-223 as an Onco-microRNA in Ovarian Cancer
3.4.2. MiR-223 in Ovarian Cancer Drug Resistance and Prognosis
3.4.3. MiR-223 as a Biomarker of Ovarian Cancer
3.5. MiR-223 in Prostate Cancer
3.5.1. MiR-223 as an Onco-Suppressor in Prostate Cancer
3.5.2. MiR-223 as an Onco-miRNA in Prostate Cancer
3.5.3. Role of miR-223 in Drug Resistance and Prognosis of Prostate Cancer
3.5.4. MiR-223 as Biomarker of Prostate Cancer
3.6. MiR-223 in Glioma and Glioblastoma
3.6.1. MiR-223 as an Onco-Suppressor in Glioma and GBM
3.6.2. MiR-223 as an Onco-miRNA in GBM
3.6.3. MiR-223 in Drug Resistance and Prognosis of Gliomas and GBM
3.6.4. MiR-223 as Biomarker of Gliomas and GBM
3.7. MiR-223 in Pancreatic Cancer
3.7.1. MiR-223 as an Onco-miRNA in PC
3.7.2. Role of miR-223 in Drug Resistance and Prognosis of PC
3.7.3. Role of miR-223 as a Biomarker in PC
3.8. MiR-223 in Hematological Malignancies
3.8.1. MiR-223 in Hematopoiesis
3.8.2. MiR-223 as an Onco-Suppressor in Hematological Malignancies
MiR-223 as an Onco-Suppressor in ALL and AML (Table 1)
Oncogenic Pathways Which Act by Lowering miR-223 Levels in ALL and AML
MiR-223 as an Onco-Suppressor in CLL and CML
3.8.3. Role of miR-223 as an Onco-miRNA in Hematological Malignancies
3.8.4. Role of miR-223 in Prognosis and Drug Resistance of Hematological Malignancies
3.8.5. Role of miR-223 as a Biomarker in Hematological Malignancies
| Organ | Cancer Type | Function | Direct Targets and Their Dysregulation in Cancer | Downstream Molecules Implicated | Signaling Pathway Affected | Cell Process Affected | Sponging Activity vs. miR-223 | Ref. |
|---|---|---|---|---|---|---|---|---|
| Intestine | Colorectal carcinoma | Oncosuppression | FOXO1 | cyt FOXO1 ↓; nuclear FOXO1 ↑ | cyclin D1/p21/p27 | Cell proliferation ↓; Cell proliferation and invasion ↑ | lncRNA ROR | [61,62] |
| Onco-miR | FOXO3 ↓; FBXW7 ↓; RhoB ↓; RASA1 ↓; SLC4A4 ↓; PRDM1 | BIM ↓ | FOXO3a/BIM signaling; FBXW7; PRDM1; Notch and Akt/mTOR pathways | Apoptosis ↓ and cell proliferation ↑; EMT ↑; tumor growth in vivo ↑ | lncRNA DRAIC; circLRCH3 | [69,70,71,72,73,74,75,76,77] | ||
| Chemoresistance to doxorubicin | miR-223 ↑; FBXW7 ↓ | N/A | MiR-223/FBXW7 axis | EMT ↑ | N/A | [78] | ||
| Lung | (1) Non-small-cell lung cancer (NSCLC) | Oncosuppression | NLRP3 ↑ | N/A | NLRP3-mediated inflammasome | Cell invasion and migration ↑; inflammation; and innate immunity | lncRNA SLCO4A1-AS1 | [96] |
| (i) Adenocarcinoma (ADC) | Oncosuppression | murine PARP1 ↑ | N/A | PARP1-miR-223 negative feedback loop | Tumor burden ↑; cell proliferation ↑; oxidative stress ↓; antioxidant enzymes, especially catalase ↑; apoptosis and autophagy of cancer cells ↓ | N/A | [99,100] | |
| (ii) Squamous cell carcinoma (SCC) | Oncosuppression | Mutant TP53 ↑; PTN ↑ | N/A | Mutant TP53 represses the transcription of miR-223 feedback loop | Cell proliferation and metastasis ↑; cell growth ↑; stemness ↑; and angiogenesis ↑ | lncRNA PITPNA-AS1 | [95,98] | |
| NSCLC | Onco-miR | RhoB ↓; EPB41L3 ↓; TGFBR3 ↓ | N/A | Cell viability ↑; migration ↑; proliferation ↑; and invasion ↑ | ADAMTS9-AS2 | [101,102,103] | ||
| Chemoresistance to cisplatin and doxorubicin | miR-223 ↑; FBXW7 ↓ (contradictory reports) | N/A | Apoptosis ↓ | N/A | [104,105] | |||
| Chemoresistance to erlotinib | miR-223 ↑; FBXW7 ↓ in HCC827/ER cells (contradictory reports) | N/A | Apoptosis ↓; colony formation potential ↑; EMT ↑ | N/A | [106] | |||
| Chemoresistance to cisplatin | miR-223 ↓; HNMT ↑ (contradictory reports) | N/A | HER2 signaling pathway | ↑CSCs maintenance and antioxidant properties | N/A | [107] | ||
| Chemoresistance to erlotinib | miR-223 ↓; IGF1R ↑ in PC9/ER cells and PC9/CD133+ cells (contradictory reports) | N/A | PI3K/Akt signaling pathway | Apoptosis ↓ | N/A | [108,109] | ||
| Breast | Breast Cancer | Oncosuppression | ECT2 ↑, PFN2 ↑; NLRP3 ↑; STIM1 ↑; HAX ↑; FABP7 ↑; SCARB1 ↑; HMGCS1 ↑ | IL-1β and IL-18, caspase-9, caspase-7, and caspase-3, ABCA1 | NLRP3 inflammasome | Tumor growth in vivo ↑; cell proliferation, invasion and migration ↑; glycolysis ↑; inflammation ↑; apoptosis ↓; EMT ↑; cholesterol biosynthesis ↑; cholesterol efflux ↓; cancer cell stemness ↑ | circABCB10; circZFR | [131,132,133,137,138,141,143] |
| Onco-miR | Mef2c ↓; FBXW7 ↓ | Notch signaling; Mef2c-β-catenin pathway | Cells invasiveness and metastasization ↑ | N/A | [52,150] | |||
| Sensitization to intraoperative RT | miR-223 ↑; EGF ↓ | N/A | EGF-EGFR pathway (autocrine/paracrine stimulation loop) | N/A | [154] | |||
| Resistance to RT | miR-223 ↓; PFN2 ↑ | N/A | Glycolytic pathway (Warburg effect) | Cell viability ↑; glucose consumption, lactic acid production, LDH-A activity, and ATP production ↑ | circABCB10 | [133] | ||
| Ovary | Ovarian cancer | Onco-miR | SOX11 ↓; LARP4 ↓; FBXW7 ↓ | N/A | circBNC2/miR-223/LARP4 and circBNC2/miR-223/ FBXW7 axes | Cell viability, cell cycle progression; migration, invasion and tumor growth ↑ | circBNC2/hsa_circ_0008732 | [13,159,161] |
| Chemoresistance to cisplatin | PTEN ↓ | PI3K/AKT | PTEN/PI3K/AKT pathway ↑ (in vitro and in vivo) | Apoptosis ↓; cell viability ↑ | N/A | [51] | ||
| Prostate | Prostate cancer | Oncosuppression | EYA3 ↑ | c-Myc ↑; CDK2/CDK4 ↑; p21 ↓; p27 ↓ | c-Myc signaling pathway ↑ | Cell proliferation ↑; cell migration and invasion ↑; tumor growth in vivo ↑ | circGNG4 | [168] |
| Onco-miR | SEPTIN6 ↓ | N/A | Cell proliferation ↑; apoptosis ↓; cell invasion ↑ | N/A | [169] | |||
| Chemosensitivity to docetaxel | miR-223 ↓; FOXO3 ↑ | N/A | Apoptosis (in vitro) ↑; tumor growth (in vivo) ↑ | N/A | [170] | |||
| Resistance to RT | miR-223 ↑; FOXO3 ↓ | Glut1 ↑; HK-2 ↑; LDH-A ↑ | Glycolysis ↑; apoptosis ↓ | N/A | [171] | |||
| Central Nervous System | Glioma and glioblastoma | Oncosuppression | NFIA ↑; MSI2 ↑; CTNND1 ↑; EGFR ↑; NLRP3 ↑ | p-21 ↑; IL-1β, MCP-1, IL-18, IL-8, and caspase1 ↑ | Wnt/β-catenin pathway ↑; PI3K/AKT pathway ↑ | Cell cycle progression ↑; chemoresistant CSCs differentiation ↓; cell proliferation and migration ↑; EMT ↑; inflammation ↑; apoptosis ↓ | lncSNHG29 and PITPNA-AS-1 | [186,187,188,189,190] |
| Onco-miR | PAX6 ↓ | MMP2, MMP9, and VEGFA ↑ | Cell viability and invasiveness ↑ | N/A | [192] | |||
| Sensitization to RT | miR-223 ↑; ATM ↓ | N/A | Tumor growth (in vitro and in vivo) ↓ | N/A | [195] | |||
| Chemoresistance to temozolomide | miR-223 ↑; PAX6 ↓ | N/A | PI3K/AKT pathway ↑ | N/A | [193,194] | |||
| Pancreas | PDAC | Onco-miR | PDS5B ↓; FBXW7 ↓; SLC4A4 ↓ | PTCH2 ↓; HNRNPK ↑; E-cadherin ↓; Vimentin ↑; MMP2, MMP9, and VEGFA ↑ | IL6/STAT3/CCND1 axis ↑; Sonic Hedgehog ↑ | Cell growth, migration and invasion ↑; angiogenesis ↑; apoptosis ↓; tumor growth (in vivo) ↑ | hsa_circ_001587 | [220,221,223,224,225,226] |
| Chemoresistance to cisplatin | miR-223 ↑; FOXO3 ↓ | N/A | Cell proliferation ↑; apoptosis ↓ | N/A | [229] | |||
| Chemoresistance to gemcitabine | miR-223 ↑; FBXW7 ↓ | E-cadherin ↓; N-cadherin, vimentin, Snail, Slug, ZEB1 and ZEB2 ↑ | Notch signaling pathway ↑ | Cell proliferation and migration ↑; apoptosis ↓ | N/A | [222] | ||
| Blood | (1) Myeloid cancer | |||||||
| (a) AML | Oncosuppression | FBXW7 ↑; PRMT4 ↑; E2F1 ↑ | N/A | Apoptosis ↓; myeloid differentiation of human stem/progenitor cells ↓ | N/A | [258,259,265] | ||
| Onco-miR | MOZ ↓ | N/A | monocyte differentiation ↓; cell proliferation ↑; stemness ↑ | N/A | [272] | |||
| Resistance to cisplatin | miR-223 ↑; MOZ ↓ | N/A | Apoptosis ↓ | N/A | [272] | |||
| (b) CML | Oncosuppression | MEF2C ↑; PTBP2 ↑; FLT3 ↑ | Bcl-xL ↑; MMP2 | PI3K/AKT pathway | Cell proliferation ↑; abnormal splicing ↑; cell viability ↑; apoptosis ↓; ROS ↑; tumor growth (in vivo) | N/A | [270,271] | |
| (2) Lymphoid cancer | ||||||||
| (a) ALL | Oncosuppression | STAT1 ↑ | BCL2 ↑ | Cell proliferation ↑ | N/A | [268] | ||
| Onco-miR | ARRB1 ↓; FBXW7 ↓ | TAL1 ↑; JAK2 ↑; HES1 ↑; HES2 ↑; PPARA ↓; DNM1 ↓; GRK4 ↓; MYC ↑; MYB ↑; NOTCH1 ↑; CCNE1 ↑ | Notch signaling pathway ↑ | Cell proliferation and invasion of T-ALL cells ↑; apoptosis ↓ | Circ_0000094 | [273,276,279] | ||
| Chemoresistance to γ-secretase inhibitor | miR-223 ↑; FBXW7 ↓ | N/A | Notch signaling pathway ↑ | Cell proliferation ↑ | N/A | [277] | ||
| Chemosensitivity to glucocorticoids | miR-223 ↑ | N/A | Apoptosis ↑ | N/A | [283] | |||
| (b) CLL | Oncosuppression | STAT3 ↑ | N/A | N/A | [269] |
| Organ/Tissue | Cancer Type | Change in miR-223 Levels in Cancer Patients | Biomarker Indication | Accuracy as Biomarker Values (Confidence Interval) 1 | Involved Downstream Molecules | References |
|---|---|---|---|---|---|---|
| Intestine | CRC 2 | ↑ serum/plasma | a Diagnosis; b prognosis (poor) | a AUC = 0.963, Se = 97.1%, Sp = 96.7% [79]; b AUC = 0.593, Se = 33.3%; Sp = 54.6% [80]; a AUC = 0.890 (0.833–0.933) [81]; a AUC = 0.838 (0.627–1.000) [82]; a AUC = 1, Se = 100%; Sp = 100% in 1st validation set; AUCs = 0.632 and 0.680 in 2nd and 3rd validation sets [85] | N/A | [80,81,82,83,86] |
| ↓ plasma of CRC patients after surgery | Follow-up | N/A | N/A | [81] | ||
| ↑ tumor | Prognosis: shorter overall survival; worse TNM staging; higher probability to develop metastases | HR = 1.374 (0.708–1.98) | N/A | [79] | ||
| ↑ stool and plasma | Diagnosis | AUC = 0.796 (0.734–0.858, stool), 0,707 (0.646–0.768, plasma) [83]; AUC = 0.939 (0.825–0.988, stool), Se = 76.5%, Sp = 96.4% [84] | N/A | [84,85] | ||
| Lung | NSCLC | ↑ NSCLC tissue | Prognosis (poor survival) | N/A | N/A | [101] |
| ↑ platelets and platelets-derived MVs of NSCLC patients | Diagnosis | N/A | N/A | [102] | ||
| ↓ miR-223 and ↑ HNMT expression in NSCLC tissue | Prognosis (poor) | N/A | N/A | [107] | ||
| ↑ sputum/serum/plasma of NSCLC patients | a Screening/diagnosis; b prognosis | a within a 12-miR-panel AUC = 0.821 (0.792–0.850), distinguishes NSCLC from HC and COPD [110]; b higher risk of progression in ADC patients [110]; a AUC = 0.94 (0.91–0.96), Se = 87%, Sp = 86% [111]; a within a 3-miR-panel AUC = 0.951 (0.926–0.976), Se = 84.35, Sp = 90.83 [112]; a AUC = 0.809 (0.749–0.860), Se = 69.8%, Sp = 84.3% [113]; a AUC = 0.744 (0.668–0.811), Se = 76.9%, Sp = 80% [114]; a AUC = 0.808 (0.712–0.884), Se = 74.3%, Sp = 78.2% [115]; a within an 11-miR-panel, accuracies ranging from 0.756 to 0.963 in validation set depending on data-mining technique [116]; N/A [117]; a AUC = 0.828 (0.763–0.881) for ADC, Se = 76.8%, Sp = 84.4% [118]; N/A [120]; a AUC = 0.90 (0.81–0.99), Se = 82%, Sp = 95% [121]; a AUC = 0.693, Se = 82.1%, Sp = 52% [122]; | N/A | [111,112,113,114,115,116,117,118,119,121,122,123] | ||
| ↓ serum | Diagnosis of early-stage NSCLC | AUC = 0.79 (0.64–0.95) | [120] | |||
| SCC | ↓ plasma | Earlier disease progression in patients treated with Nivolumab | N/A | N/A | [124] | |
| Breast | Breast cancer | ↑ plasma-derived exosomes of IDC patients | Progression from DCIS to IDC | N/A | N/A | [144] |
| Ovaries | OC | ↑ relapsed ovarian serous adenocarcinoma tissue | Relapse of ovarian cancer | N/A | N/A | [164] |
| ↑ OC tissue | Presence of lymph node metastasis, histological tumor grade, and FIGO stage | N/A | SOX11 ↓; FBXW7 ↓ | [13,159] | ||
| ↑ in OC tissue compared to normal tissue; ↑ serum circulating exosomal miR-223 | Resistance to cisplatin-based therapy; increased probability of recurrence | N/A | N/A | [51] | ||
| ↑ in TAM-derived exosomes | Associated with shorter progression-free survival (PFS) | N/A | PTEN ↓; PI3K/AKT ↑ (in recipient OC cells) | [51] | ||
| Prostate | Prostate Cancer | ↑ urine and PCa cells | Associated with radioresistance | N/A | N/A | [171] |
| ↓ serum of PCa and chronic prostatitis patients vs. BPH patients | Diagnosis | AUC PCa vs. non-cancer = 0.817, Se = 81%, Sp = 71%; AUC PCa vs. BPH = 0.938, Se= 88%, Sp = 88%; AUC CP vs. BPH = 0.880, Se = 70%, Sp = 92% | N/A | [173] | ||
| CNS | GBM and Glioma | ↑ glioma and GBM tissue | Associated with longer survival | N/A | MAPK signaling | [197] |
| GBM mesenchymal subtype | ↑ GBM tissue | Associated with longer survival in the mesenchymal subtype | N/A | N/A | [198] | |
| GBM | ↑ GBM tissue | Associated with shorter survival | N/A | NFKBIZ; PSCD4; BCL3 SLC16A3; PLEKHQ1 LHFPL2; LSP1; URP2; CTSL1; ISG20; HMOX1; IL17RA; RBMX; FZD7; CCNB1IP1; LITAF | [199] | |
| LGG | ↑ LGG tissue | Prognosis (poor for high levels) | In ROC curves for survival prediction: 1-year survival: AUC = 0.638 3-year survival: AUC = 0.577 5-year survival: AUC = 0.578; better accuracy of a 5-miR signature including miR-223 | N/A | [200] | |
| ↑ serum of LGG and GBM patients | a Diagnosis; b follow-up | a All gliomas vs. HC: AUC = 0.7771 for miR-223 only, AUC = 0.998 within a 4-miR panel; b Positive correlation with post-operative MRI score in LGG | N/A | [202] | ||
| Diffuse LGG | ↑ whole blood of diffuse LGG (grade II) patients vs. HC | Diagnosis | AUC = 0.827 | N/A | [201] | |
| GBM | ↑ whole blood of GBM patients vs. HC | Diagnosis | AUC = 0.804 | N/A | [203] | |
| Pancreas | PC | ↑ whole blood of PC patients vs. HC or CPa | Diagnosis | Within 4-miR panel: AUC = 0.86 (0.82–0.90), Se = 85%, Sp = 64% in training PC vs. controls; AUC = 0.83 (0.76–0.90), Se = 85%, Sp = 45% in validation PC vs. controls + CPa; other data available | [234] | |
| PC and IPMN | ↑ tissue and plasma of PC vs. HC and ↑ malignant IPMN patients vs. benign IPMN patients; ↓ in post-operative samples | Diagnosis, prognosis | a with plasma, AUC = 0.834, Se = 62%, Sp = 94.1%; AUC = 0.789, Se = 82.4%, Sp = 62.9% in distinguishing malignant IPMN and PIDC; | N/A | [235] | |
| IPMN | ↑ IPMN tissue vs. normal pancreatic tissue (FFPE); ↑ in poor vs. good prognosis IPMN and IPMN vs. CPa | Diagnosis, prognosis | N/A | N/A | [236] | |
| PDAC | ↑ urine stage I (but not stages II-IV) PDAC patients vs. HC | Diagnosis | AUC = 0.795 (0.586–1.000), Se = 83.3%, Sp = 76.9% | N/A | [237] | |
| Blood | ALL | ↓ Bone marrow of relapsed pediatric ALL patients vs. complete remission patients | Lower relapse-free survival | N/A | E2F1 | [280] |
| ALL | ↓ in bone marrow and blood of T-ALL and B-ALL pediatric patients compared to HC | Diagnosis; follow-up | N/A | N/A | [281] | |
| B-ALL | ↓ plasma B-ALL vs. HC | Diagnosis | B-ALL vs. HC Se = 89%, Sp = 100% | N/A | [287] | |
| AML | ↓ serum; ↑ in post-operative samples; shorter OS and PFS with low levels | a Diagnosis; b poor prognosis | a AUC = 0.849, Se = 83.2%, Sp = 81.4% [284] b RR = 3.54 (1.47–5.79), univariate analysis [284]; | N/A | [284] | |
| ALL | ↓ bone marrow in ALL patients vs. HC | Diagnosis | N/A | N/A | [253] | |
| ALL/AML | ↑ bone marrow | Diagnosis; differential diagnosis (AML > ALL > HC) | N/A [253]; AUC miR-223 only (AML + ALL vs. HC) = 0.853; AUC 3-miR panel = 0.994, Se = 95%, Sp = 98.14% [254]; Diagnostic OR 2-miR panel (from metanalysis) = 546, 95% (73.38–4041.28) [285]; Accuracy (AML + ALL vs. HC) > 95, Se = 96%, Sp = 95% [286] | N/A | [254,255,285,286] | |
| CLL | ↓ miR-223/ ↑ HSP90AB1 (associated to polymorphism rs2307842) | Poor prognosis; shorter time to treatment | 17 months (5–28.9) without treatment vs. 104 months of cases without HSP90AB1 overexpression | HSP90AB1 | [293] | |
| B-CLL | high levels in B cells from peripheral blood | Higher overall survival, treatment-free survival, progression free survival | 137.2 months OS in low vs. not reached in high miR-223 (RR 4.9); 24.1 months TFS in low vs. 107.2 in high miR-223 (RR 2.7) [289] 40 months OS in low vs. not reached in high miR-223; 13 months PFS in low vs. not reached in high miR-223 [290] | N/A | [289,290] | |
| CML | ↓ in plasma | Increased disease risk | N/A | N/A | [270] |
4. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin Structure Analyses Identify miRNA Promoters. Genes Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Lin, L.; Li, T.; Yang, J.; Wei, Y. The Role of miRNA-223 in Cancer: Function, Diagnosis and Therapy. Gene 2017, 616, 1–7. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The Role and Mechanisms of Action of microRNAs in Cancer Drug Resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Annese, T.; Tamma, R.; De Giorgis, M.; Ribatti, D. microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 2020, 10, 581007. [Google Scholar] [CrossRef]
- ElKhouly, A.M.; Youness, R.A.; Gad, M.Z. MicroRNA-486-5p and microRNA-486-3p: Multifaceted Pleiotropic Mediators in Oncological and Non-Oncological Conditions. Non-Coding RNA Res. 2020, 5, 11–21. [Google Scholar] [CrossRef]
- Peterson, S.M.; Thompson, J.A.; Ufkin, M.L.; Sathyanarayana, P.; Liaw, L.; Congdon, C.B. Common Features of microRNA Target Prediction Tools. Front. Genet. 2014, 5, 23. [Google Scholar] [CrossRef]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in Plasma Exosome Is Stable under Different Storage Conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef]
- miRBase: The microRNA Database the Archive for microRNA Sequences and Annotations. Available online: https://mirbase.org/ (accessed on 1 September 2023).
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian mRNAs Are Conserved Targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Zeng, Z.-M.; Chen, Y.-Y.; Wen, X.-C.; Geng, X.-C.; Zhu, Y.-X.; Hao, L.-C.; Dong, Z.-S.; Yang, J.-F.; Wang, T.-T.; Zhang, R.-B.; et al. Whole-Transcriptome Sequencing with ceRNA Regulation Network Construction and Verification in Glioblastoma. Am. J. Transl. Res. 2023, 15, 4291–4313. [Google Scholar]
- Liu, T.; Yuan, L.; Zou, X. Circular RNA Circ-BNC2 (Hsa_circ_0008732) Inhibits the Progression of Ovarian Cancer through microRNA-223-3p/ FBXW7 Axis. J. Ovarian Res. 2022, 15, 95. [Google Scholar] [CrossRef]
- Nahand, J.S.; Jamshidi, S.; Hamblin, M.R.; Mahjoubin-Tehran, M.; Vosough, M.; Jamali, M.; Khatami, A.; Moghoofei, M.; Baghi, H.B.; Mirzaei, H. Circular RNAs: New Epigenetic Signatures in Viral Infections. Front. Microbiol. 2020, 11, 1853. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, Expression and Potential Functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef]
- Verduci, L.; Strano, S.; Yarden, Y.; Blandino, G. The CircRNA–MicroRNA Code: Emerging Implications for Cancer Diagnosis and Treatment. Mol. Oncol. 2019, 13, 669–680. [Google Scholar] [CrossRef]
- Merulla, A.E.; Stella, M.; Barbagallo, C.; Battaglia, R.; Caponnetto, A.; Broggi, G.; Altieri, R.; Certo, F.; Caltabiano, R.; Ragusa, M.; et al. circSMARCA5 Is an Upstream Regulator of the Expression of miR-126-3p, miR-515-5p, and Their mRNA Targets, Insulin-like Growth Factor Binding Protein 2 (IGFBP2) and NRAS Proto-Oncogene, GTPase (NRAS) in Glioblastoma. Int. J. Mol. Sci. 2022, 23, 13676. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Lei, M.; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and Functional Roles of Circular RNAs in Human Cancer. Mol. Cancer 2020, 19, 30. [Google Scholar] [CrossRef]
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. Epigenetic Alteration and microRNA Dysregulation in Cancer. Front. Genet. 2013, 4, 258. [Google Scholar] [CrossRef]
- Ferragut Cardoso, A.P.; Banerjee, M.; Nail, A.N.; Lykoudi, A.; States, J.C. miRNA Dysregulation Is an Emerging Modulator of Genomic Instability. Semin. Cancer Biol. 2021, 76, 120–131. [Google Scholar] [CrossRef]
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized High-Throughput microRNA Expression Profiling Provides Novel Biomarker Assessment of Clinical Prostate and Breast Cancer Biopsies. Mol. Cancer 2006, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hu, Y.; Tong, S.; Williams, B.R.G.; Smyth, G.K.; Gantier, M.P. The Use of miRNA Microarrays for the Analysis of Cancer Samples with Global miRNA Decrease. RNA 2013, 19, 876–888. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q.; Wang, S. MicroRNAs: A New Key in Lung Cancer. Cancer Chemother. Pharmacol. 2014, 74, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA Expression Profiles Classify Human Cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, F.; Tadayyon Tabrizi, A.; Hashemian, P.; Alijannejad, S.; Rahdar, H.A.; Ferns, G.A.; Hassanian, S.M.; Shahidsales, S.; Avan, A. Role of Regulatory miRNAs of the Wnt/ β-Catenin Signaling Pathway in Tumorigenesis of Breast Cancer. Gene 2020, 754, 144892. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The Role of MicroRNAs in Human Cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Dong, C.; Ji, C. MicroRNA and Drug Resistance. Cancer Gene Ther. 2010, 17, 523–531. [Google Scholar] [CrossRef]
- Gandham, S.K.; Rao, M.; Shah, A.; Trivedi, M.S.; Amiji, M.M. Combination microRNA-Based Cellular Reprogramming with Paclitaxel Enhances Therapeutic Efficacy in a Relapsed and Multidrug-Resistant Model of Epithelial Ovarian Cancer. Mol. Ther. Oncolytics 2022, 25, 57–68. [Google Scholar] [CrossRef]
- Shu, J.; Silva, B.V.R.E.; Gao, T.; Xu, Z.; Cui, J. Dynamic and Modularized MicroRNA Regulation and Its Implication in Human Cancers. Sci. Rep. 2017, 7, 13356. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- NCBI Gene. Available online: https://www.ncbi.nlm.nih.gov/gene/407008 (accessed on 10 July 2024).
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of Progenitor Cell Proliferation and Granulocyte Function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-H.; Zhang, L.; Wu, D.-S.; Zhang, Z.; Huang, F.-F.; Zhang, J.; Chen, X.-P.; Liang, D.-S.; Zeng, H.; Chen, F.-P. MiR-223 Regulates Human Embryonic Stem Cell Differentiation by Targeting the IGF-1R/Akt Signaling Pathway. PLoS ONE 2013, 8, e78769. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sugatani, T.; Hruska, K.A. MicroRNA-223 Is a Key Factor in Osteoclast Differentiation. J. Cell. Biochem. 2007, 101, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Berg, N.; Lee, J.W.; Le, T.-T.; Neudecker, V.; Jing, N.; Eltzschig, H. MicroRNA miR-223 as Regulator of Innate Immunity. J. Leukoc. Biol. 2018, 104, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Roffel, M.P.; Bracke, K.R.; Heijink, I.H.; Maes, T. miR-223: A Key Regulator in the Innate Immune Response in Asthma and COPD. Front. Med. 2020, 7, 196. [Google Scholar] [CrossRef]
- Fazi, F.; Rosa, A.; Fatica, A.; Gelmetti, V.; De Marchis, M.L.; Nervi, C.; Bozzoni, I. A Minicircuitry Comprised of MicroRNA-223 and Transcription Factors NFI-A and C/EBPα Regulates Human Granulopoiesis. Cell 2005, 123, 819–831. [Google Scholar] [CrossRef]
- Vian, L.; Di Carlo, M.; Pelosi, E.; Fazi, F.; Santoro, S.; Cerio, A.M.; Boe, A.; Rotilio, V.; Billi, M.; Racanicchi, S.; et al. Transcriptional Fine-Tuning of microRNA-223 Levels Directs Lineage Choice of Human Hematopoietic Progenitors. Cell Death Differ. 2014, 21, 290–301. [Google Scholar] [CrossRef]
- Bauernfeind, F.; Rieger, A.; Schildberg, F.A.; Knolle, P.A.; Schmid-Burgk, J.L.; Hornung, V. NLRP3 Inflammasome Activity Is Negatively Controlled by miR-223. J. Immunol. 2012, 189, 4175–4181. [Google Scholar] [CrossRef]
- Feng, Z.; Qi, S.; Zhang, Y.; Qi, Z.; Yan, L.; Zhou, J.; He, F.; Li, Q.; Yang, Y.; Chen, Q.; et al. Ly6G+ Neutrophil-Derived miR-223 Inhibits the NLRP3 Inflammasome in Mitochondrial DAMP-Induced Acute Lung Injury. Cell Death Dis. 2017, 8, e3170. [Google Scholar] [CrossRef]
- Liao, T.-L.; Chen, Y.-M.; Tang, K.-T.; Chen, P.-K.; Liu, H.-J.; Chen, D.-Y. MicroRNA-223 Inhibits Neutrophil Extracellular Traps Formation through Regulating Calcium Influx and Small Extracellular Vesicles Transmission. Sci. Rep. 2021, 11, 15676. [Google Scholar] [CrossRef]
- Ye, D.; Yao, J.; Du, W.; Chen, C.; Yang, Y.; Yan, K.; Li, J.; Xu, Y.; Zang, S.; Zhang, Y.; et al. Neutrophil Extracellular Traps Mediate Acute Liver Failure in Regulation of miR-223/Neutrophil Elastase Signaling in Mice. Cell. Mol. Gastroenterol. Hepatol. 2022, 14, 587–607. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, J.; Zhou, W.; Hsu, A.Y.; Deng, Q. miRNA-223 at the Crossroads of Inflammation and Cancer. Cancer Lett. 2019, 451, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, H.; Liu, Y.; Song, Y.; Lai, L.; Han, Q.; Cao, X.; Wang, Q. Inducible MicroRNA-223 Down-Regulation Promotes TLR-Triggered IL-6 and IL-1β Production in Macrophages by Targeting STAT3. PLoS ONE 2012, 7, e42971. [Google Scholar] [CrossRef]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.-S.; Liu, Z. MicroRNAs Modulate the Noncanonical Transcription Factor NF-κB Pathway by Regulating Expression of the Kinase IKKα during Macrophage Differentiation. Nat. Immunol. 2010, 11, 799–805. [Google Scholar] [CrossRef]
- Wang, J.; Bai, X.; Song, Q.; Fan, F.; Hu, Z.; Cheng, G.; Zhang, Y. miR-223 Inhibits Lipid Deposition and Inflammation by Suppressing Toll-Like Receptor 4 Signaling in Macrophages. Int. J. Mol. Sci. 2015, 16, 24965–24982. [Google Scholar] [CrossRef]
- Ying, W.; Tseng, A.; Chang, R.C.-A.; Morin, A.; Brehm, T.; Triff, K.; Nair, V.; Zhuang, G.; Song, H.; Kanameni, S.; et al. MicroRNA-223 Is a Crucial Mediator of PPARγ-Regulated Alternative Macrophage Activation. J. Clin. Investig. 2015, 125, 4149–4159. [Google Scholar] [CrossRef]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-Box Protein 1 Is Required to Sort microRNAs into Exosomes in Cells and in a Cell-Free Reaction. eLife 2016, 5, e19276. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, H.; Yin, X.; Yang, M.; Wei, H.; Chen, Q.; Feng, F.; Liu, Y.; Xu, W.; Li, Y. Macrophages Derived Exosomes Deliver miR-223 to Epithelial Ovarian Cancer Cells to Elicit a Chemoresistant Phenotype. J. Exp. Clin. Cancer Res. 2019, 38, 81. [Google Scholar] [CrossRef]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.-D.; Song, E. Microvesicles Secreted by Macrophages Shuttle Invasion-Potentiating microRNAs into Breast Cancer Cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA. Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Patterns and Trends in Colorectal Cancer Incidence and Mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.H.; Ellis, I.; Allison, K.; Brogi, E.; Fox, S.B.; Lakhani, S.; Lazar, A.J.; Morris, E.A.; Sahin, A.; Salgado, R.; et al. The 2019 World Health Organization Classification of Tumours of the Breast. Histopathology 2020, 77, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A Genetic Model for Colorectal Tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef] [PubMed]
- van Engeland, M.; Derks, S.; Smits, K.M.; Meijer, G.A.; Herman, J.G. Colorectal Cancer Epigenetics: Complex Simplicity. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.F.; Ibrahim, A.E.K.; Arends, M.J. Molecular Pathological Classification of Colorectal Cancer. Virchows Arch. Int. J. Pathol. 2016, 469, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Okayama, H.; Harris, C.C. The Role of microRNAs in Colorectal Cancer. Cancer J. Sudbury Mass 2012, 18, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, H.; Jia, C.Y.; Cheng, W.; Yu, M.; Peng, M.; Zhu, Y.; Zhao, Q.; Dong, Y.W.; Shao, K.; et al. MicroRNA-223 Regulates FOXO1 Expression and Cell Proliferation. FEBS Lett. 2012, 586, 1038–1043. [Google Scholar] [CrossRef]
- Ma, Y.-L.; Wang, C.-Y.; Guan, Y.-J.; Gao, F.-M. Long Noncoding RNA ROR Promotes Proliferation and Invasion of Colorectal Cancer by Inhibiting Tumor Suppressor Gene NF2 through Interacting with miR-223-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2401–2411. [Google Scholar] [CrossRef]
- Chen, J.; Vitetta, L. Is miR-223 Upregulation in Inflammatory Bowel Diseases a Protective Response? Front. Biosci. Elite Ed. 2023, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Josse, C.; Bouznad, N.; Geurts, P.; Irrthum, A.; Huynh-Thu, V.A.; Servais, L.; Hego, A.; Delvenne, P.; Bours, V.; Oury, C. Identification of a microRNA Landscape Targeting the PI3K/Akt Signaling Pathway in Inflammation-Induced Colorectal Carcinogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G229–G243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Luo, X.; Li, H.; Yue, X.; Deng, L.; Cui, Y.; Lu, Y. MicroRNA-223 Functions as an Oncogene in Human Colorectal Cancer Cells. Oncol. Rep. 2014, 32, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Earle, J.S.L.; Luthra, R.; Romans, A.; Abraham, R.; Ensor, J.; Yao, H.; Hamilton, S.R. Association of microRNA Expression with Microsatellite Instability Status in Colorectal Adenocarcinoma. J. Mol. Diagn. JMD 2010, 12, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Tang, W.; Du, P.; Wang, G.; Chen, W.; Li, J.; Zhu, Y.; Gao, J.; Cui, L. Identifying microRNA-mRNA Regulatory Network in Colorectal Cancer by a Combination of Expression Profile and Bioinformatics Analysis. BMC Syst. Biol. 2012, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Chai, B.; Guo, Y.; Cui, X.; Liu, J.; Suo, Y.; Dou, Z.; Li, N. MiR-223-3p Promotes the Proliferation, Invasion and Migration of Colon Cancer Cells by Negative Regulating PRDM1. Am. J. Transl. Res. 2019, 11, 4516–4523. [Google Scholar] [PubMed]
- Cheng, Z.; Cao, Y.; Ni, Q.; Qin, J. miR-223 Promotes Proliferation of Colon Cancer Cells by Down-Regulating Bcl-2-Like Protein 11 (BIM) Expression. J. Biomater. Tissue Eng. 2019, 9, 1424–1428. [Google Scholar] [CrossRef]
- Ju, H.; Tan, J.-Y.; Cao, B.; Song, M.-Q.; Tian, Z.-B. Effects of miR-223 on Colorectal Cancer Cell Proliferation and Apoptosis through Regulating FoxO3a/BIM. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3771–3778. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ma, T.; Duan, J.; Liu, X.; Liu, L. MicroRNA-223-induced Inhibition of the FBXW7 Gene Affects the Proliferation and Apoptosis of Colorectal Cancer Cells via the Notch and Akt/mTOR Pathways. Mol. Med. Rep. 2020, 23, 154. [Google Scholar] [CrossRef]
- Yeh, C.-H.; Bellon, M.; Nicot, C. FBXW7: A Critical Tumor Suppressor of Human Cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- Wei, L.-J.; Li, J.-A.; Bai, D.-M.; Song, Y. miR-223-RhoB Signaling Pathway Regulates the Proliferation and Apoptosis of Colon Adenocarcinoma. Chem.-Biol. Interact. 2018, 289, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wang, C.; Long, S.; Ma, Y.; Guo, Y.; Huang, Z.; Chen, X.; Zhang, C.; Chen, J.; Zhang, J. C/EBP-β-Activated microRNA-223 Promotes Tumour Growth through Targeting RASA1 in Human Colorectal Cancer. Br. J. Cancer 2015, 112, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Ranjbaran, J.; Safarpour, H.; Nomiri, S.; Tavakoli, T.; Rezaei, Z.; Salmani, F.; Larki, P.; Chamani, E. Experimental Validation of in Silico Analysis Estimated the Reverse Effect of Upregulated Hsa-miR-106a-5p and Hsa-miR-223-3p on SLC4A4 Gene Expression in Iranian Patients with Colorectal Adenocarcinoma by RT-qPCR. Cancer Med. 2023, 12, 7005–7018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, Y.; Wang, P.; Jiang, L.; Xiao, X. LncRNA DRAIC Promotes Apoptosis and Inhibits Proliferation of Colorectal Cancer via Regulating MiR-223. Minerva Med. 2021; online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, D.; Tao, K.; Wang, G. Circular RNA circLRCH3 Inhibits Proliferation, Migration, and Invasion of Colorectal Cancer Cells Through miRNA-223/LPP Axis. OncoTargets Ther. 2022, 15, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhao, Z.; Song, J.; Luo, B.; Huang, L. MiR-223 Promotes the Doxorubicin Resistance of Colorectal Cancer Cells via Regulating Epithelial-Mesenchymal Transition by Targeting FBXW7. Acta Biochim. Biophys. Sin. 2018, 50, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, Y.; Du, L.; Dong, Z.; Wang, L.; Zhang, X.; Zhou, X.; Zheng, G.; Qu, A.; Wang, C. Overexpression of miR-223 Correlates with Tumor Metastasis and Poor Prognosis in Patients with Colorectal Cancer. Med. Oncol. 2014, 31, 256. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, H.A.; El Amin, H.A.; Ahmed, E.S.M.; Kenawy, A.G.; El-Ebidi, A.M.; ElNakeeb, I.; Kholef, E.F.M.; Elsewify, W.A.E. Role of MicroRNA-223 and MicroRNA-182 as Novel Biomarkers in Early Detection of Colorectal Cancer. Int. J. Gen. Med. 2022, 15, 3281–3291. [Google Scholar] [CrossRef]
- Pesta, M.; Kucera, R.; Topolcan, O.; Karlikova, M.; Houfkova, K.; Polivka, J.; Macanova, T.; Machova, I.; Slouka, D.; Kulda, V. Plasma microRNA Levels Combined with CEA and CA19-9 in the Follow-Up of Colorectal Cancer Patients. Cancers 2019, 11, 864. [Google Scholar] [CrossRef]
- Zheng, G.; Du, L.; Yang, X.; Zhang, X.; Wang, L.; Yang, Y.; Li, J.; Wang, C. Serum microRNA Panel as Biomarkers for Early Diagnosis of Colorectal Adenocarcinoma. Br. J. Cancer 2014, 111, 1985–1992. [Google Scholar] [CrossRef]
- Zekri, A.-R.N.; Youssef, A.S.E.-D.; Lotfy, M.M.; Gabr, R.; Ahmed, O.S.; Nassar, A.; Hussein, N.; Omran, D.; Medhat, E.; Eid, S.; et al. Circulating Serum miRNAs as Diagnostic Markers for Colorectal Cancer. PLoS ONE 2016, 11, e0154130. [Google Scholar] [CrossRef]
- Chang, P.-Y.; Chen, C.-C.; Chang, Y.-S.; Tsai, W.-S.; You, J.-F.; Lin, G.-P.; Chen, T.-W.; Chen, J.-S.; Chan, E.-C. MicroRNA-223 and microRNA-92a in Stool and Plasma Samples Act as Complementary Biomarkers to Increase Colorectal Cancer Detection. Oncotarget 2016, 7, 10663–10675. [Google Scholar] [CrossRef]
- Phua, L.C.; Chue, X.P.; Koh, P.K.; Cheah, P.Y.; Chan, E.C.Y.; Ho, H.K. Global Fecal microRNA Profiling in the Identification of Biomarkers for Colorectal Cancer Screening among Asians. Oncol. Rep. 2014, 32, 97–104. [Google Scholar] [CrossRef]
- Hishida, A.; Yamada, H.; Ando, Y.; Okugawa, Y.; Shiozawa, M.; Miyagi, Y.; Daigo, Y.; Toiyama, Y.; Shirai, Y.; Tanaka, K.; et al. Investigation of miRNA Expression Profiles Using Cohort Samples Reveals Potential Early Detectability of Colorectal Cancers by Serum miR-26a-5p before Clinical Diagnosis. Oncol. Lett. 2022, 23, 87. [Google Scholar] [CrossRef]
- Francisci, S.; Minicozzi, P.; Pierannunzio, D.; Ardanaz, E.; Eberle, A.; Grimsrud, T.K.; Knijn, A.; Pastorino, U.; Salmerón, D.; Trama, A.; et al. Survival Patterns in Lung and Pleural Cancer in Europe 1999–2007: Results from the EUROCARE-5 Study. Eur. J. Cancer 2015, 51, 2242–2253. [Google Scholar] [CrossRef]
- Weeden, C.E.; Gayevskiy, V.; Marceaux, C.; Batey, D.; Tan, T.; Yokote, K.; Ribera, N.T.; Clatch, A.; Christo, S.; Teh, C.E.; et al. Early Immune Pressure Initiated by Tissue-Resident Memory T Cells Sculpts Tumor Evolution in Non-Small Cell Lung Cancer. Cancer Cell 2023, 41, 837–852.e6. [Google Scholar] [CrossRef]
- Pavel, A.B.; Campbell, J.D.; Liu, G.; Elashoff, D.; Dubinett, S.; Smith, K.; Whitney, D.; Lenburg, M.E.; Spira, A.; AEGIS Study Team. Alterations in Bronchial Airway miRNA Expression for Lung Cancer Detection. Cancer Prev. Res. 2017, 10, 651–659. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational Landscape and Significance across 12 Major Cancer Types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef]
- Schembri, F.; Sridhar, S.; Perdomo, C.; Gustafson, A.M.; Zhang, X.; Ergun, A.; Lu, J.; Liu, G.; Zhang, X.; Bowers, J.; et al. MicroRNAs as Modulators of Smoking-Induced Gene Expression Changes in Human Airway Epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 2319–2324. [Google Scholar] [CrossRef]
- Izzotti, A.; Calin, G.A.; Arrigo, P.; Steele, V.E.; Croce, C.M.; De Flora, S. Downregulation of microRNA Expression in the Lungs of Rats Exposed to Cigarette Smoke. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2009, 23, 806–812. [Google Scholar] [CrossRef]
- Kong, X.; Gao, M.; Liu, Y.; Zhang, P.; Li, M.; Ma, P.; Shang, P.; Wang, W.; Liu, H.; Zhang, Q.; et al. GSDMD-miR-223-NLRP3 Axis Involved in B(a)P-Induced Inflammatory Injury of Alveolar Epithelial Cells. Ecotoxicol. Environ. Saf. 2022, 232, 113286. [Google Scholar] [CrossRef]
- Yu, N.; Yong, S.; Kim, H.K.; Choi, Y.-L.; Jung, Y.; Kim, D.; Seo, J.; Lee, Y.E.; Baek, D.; Lee, J.; et al. Identification of Tumor Suppressor miRNAs by Integrative miRNA and mRNA Sequencing of Matched Tumor-Normal Samples in Lung Adenocarcinoma. Mol. Oncol. 2019, 13, 1356–1368. [Google Scholar] [CrossRef]
- Luo, P.; Wang, Q.; Ye, Y.; Zhang, J.; Lu, D.; Cheng, L.; Zhou, H.; Xie, M.; Wang, B. MiR-223-3p Functions as a Tumor Suppressor in Lung Squamous Cell Carcinoma by miR-223-3p-Mutant P53 Regulatory Feedback Loop. J. Exp. Clin. Cancer Res. 2019, 38, 74. [Google Scholar] [CrossRef]
- Zhu, S.; Kong, X.; Song, M.; Chi, M.; Liu, Y.; Zhang, P.; Zhang, Q.; Shang, P.; Feng, F. MiR-223-3p Attenuates the Migration and Invasion of NSCLC Cells by Regulating NLRP3. Front. Oncol. 2022, 12, 985962. [Google Scholar] [CrossRef]
- Li, Q.; Jiang, B.; Qi, Y.; Zhang, H.; Ma, H. Long Non-Coding RNA SLCO4A1-AS1 Drives the Progression of Non-Small-Cell Lung Cancer by Modulating miR-223-3p/IKKα/NF-κB Signaling. Cancer Biol. Ther. 2020, 21, 806–814. [Google Scholar] [CrossRef]
- Peng, B.-H.; Ji, Y.-F.; Qiu, X.-J. LncRNA PITPNA-AS1/miR-223-3p/PTN Axis Regulates Malignant Progression and Stemness in Lung Squamous Cell Carcinoma. J. Clin. Lab. Anal. 2022, 36, e24506. [Google Scholar] [CrossRef]
- Mateu-Jiménez, M.; Cucarull-Martínez, B.; Yelamos, J.; Barreiro, E. Reduced Tumor Burden through Increased Oxidative Stress in Lung Adenocarcinoma Cells of PARP-1 and PARP-2 Knockout Mice. Biochimie 2016, 121, 278–286. [Google Scholar] [CrossRef]
- Meloche, J.; Le Guen, M.; Potus, F.; Vinck, J.; Ranchoux, B.; Johnson, I.; Antigny, F.; Tremblay, E.; Breuils-Bonnet, S.; Perros, F.; et al. miR-223 Reverses Experimental Pulmonary Arterial Hypertension. Am. J. Physiol. Cell Physiol. 2015, 309, C363–C372. [Google Scholar] [CrossRef]
- Li, S.; Feng, Y.; Huang, Y.; Liu, Y.; Wang, Y.; Liang, Y.; Zeng, H.; Qu, H.; Wei, L. MiR-223-3p Regulates Cell Viability, Migration, Invasion, and Apoptosis of Non-Small Cell Lung Cancer Cells by Targeting RHOB. Open Life Sci. 2020, 15, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Yan, X.; Pan, Y.; Wang, Y.; Wang, N.; Li, L.; Liu, Y.; Chen, X.; Zhang, C.-Y.; Gu, H.; et al. MicroRNA-223 Delivered by Platelet-Derived Microvesicles Promotes Lung Cancer Cell Invasion via Targeting Tumor Suppressor EPB41L3. Mol. Cancer 2015, 14, 58. [Google Scholar] [CrossRef]
- Liu, C.; Yang, Z.; Deng, Z.; Zhou, Y.; Gong, Q.; Zhao, R.; Chen, T. Upregulated lncRNA ADAMTS9-AS2 Suppresses Progression of Lung Cancer through Inhibition of miR-223-3p and Promotion of TGFBR3. IUBMB Life 2018, 70, 536–546. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Zhang, S.; Zheng, X.; Xie, S.; Mao, J.; Cai, Y.; Lu, X.; Hu, L.; Shen, J.; et al. MiR-223 Regulates Autophagy Associated with Cisplatin Resistance by Targeting FBXW7 in Human Non-Small Cell Lung Cancer. Cancer Cell Int. 2020, 20, 258. [Google Scholar] [CrossRef]
- Li, R.; Wu, S.; Chen, X.; Xu, H.; Teng, P.; Li, W. miR-223/FBW7 Axis Regulates Doxorubicin Sensitivity through Epithelial Mesenchymal Transition in Non-Small Cell Lung Cancer. Am. J. Transl. Res. 2016, 8, 2512–2524. [Google Scholar] [PubMed]
- Zhang, H.; Chen, F.; He, Y.; Yi, L.; Ge, C.; Shi, X.; Tang, C.; Wang, D.; Wu, Y.; Nian, W. Sensitivity of Non-Small Cell Lung Cancer to Erlotinib Is Regulated by the Notch/miR-223/FBXW7 Pathway. Biosci. Rep. 2017, 37, BSR20160478. [Google Scholar] [CrossRef] [PubMed]
- Kuo, K.-T.; Lin, C.-H.; Wang, C.-H.; Pikatan, N.W.; Yadav, V.K.; Fong, I.-H.; Yeh, C.-T.; Lee, W.-H.; Huang, W.-C. HNMT Upregulation Induces Cancer Stem Cell Formation and Confers Protection against Oxidative Stress through Interaction with HER2 in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2022, 23, 1663. [Google Scholar] [CrossRef]
- Han, J.; Zhao, F.; Zhang, J.; Zhu, H.; Ma, H.; Li, X.; Peng, L.; Sun, J.; Chen, Z. miR-223 Reverses the Resistance of EGFR-TKIs through IGF1R/PI3K/Akt Signaling Pathway. Int. J. Oncol. 2016, 48, 1855–1867. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.-Y.; Han, J.; Chen, X.-W.; Wang, J.; Wang, X.-D.; Sun, J.-G.; Chen, Z.-T. miR-223 Enhances the Sensitivity of Non-Small Cell Lung Cancer Cells to Erlotinib by Targeting the Insulin-like Growth Factor-1 Receptor. Int. J. Mol. Med. 2016, 38, 183–191. [Google Scholar] [CrossRef]
- Mussbacher, M.; Pirabe, A.; Brunnthaler, L.; Schrottmaier, W.C.; Assinger, A. Horizontal MicroRNA Transfer by Platelets–Evidence and Implications. Front. Physiol. 2021, 12, 678362. [Google Scholar] [CrossRef] [PubMed]
- Sanfiorenzo, C.; Ilie, M.I.; Belaid, A.; Barlési, F.; Mouroux, J.; Marquette, C.-H.; Brest, P.; Hofman, P. Two Panels of Plasma microRNAs as Non-Invasive Biomarkers for Prediction of Recurrence in Resectable NSCLC. PLoS ONE 2013, 8, e54596. [Google Scholar] [CrossRef]
- Geng, Q.; Fan, T.; Zhang, B.; Wang, W.; Xu, Y.; Hu, H. Five microRNAs in Plasma as Novel Biomarkers for Screening of Early-Stage Non-Small Cell Lung Cancer. Respir. Res. 2014, 15, 149. [Google Scholar] [CrossRef]
- Lv, S.; Xue, J.; Wu, C.; Wang, L.; Wu, J.; Xu, S.; Liang, X.; Lou, J. Identification of A Panel of Serum microRNAs as Biomarkers for Early Detection of Lung Adenocarcinoma. J. Cancer 2017, 8, 48–56. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, F.; Shen, T.; Luo, Q.; Ding, Z.; Qian, L.; Huang, J. Plasma miR-145, miR-20a, miR-21 and miR-223 as Novel Biomarkers for Screening Early-Stage Non-Small Cell Lung Cancer. Oncol. Lett. 2017, 13, 669–676. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, K.; Zhou, Y.; Hu, Z.; Chen, S.; Huang, Y. Application of Serum microRNA-9-5p, 21–25p, and 223-3p Combined with Tumor Markers in the Diagnosis of Non-Small-Cell Lung Cancer in Yunnan in Southwestern China. OncoTargets Ther. 2018, 11, 587–597. [Google Scholar] [CrossRef] [PubMed]
- D’Antona, P.; Cattoni, M.; Dominioni, L.; Poli, A.; Moretti, F.; Cinquetti, R.; Gini, E.; Daffrè, E.; Noonan, D.M.; Imperatori, A.; et al. Serum miR-223: A Validated Biomarker for Detection of Early-Stage Non-Small Cell Lung Cancer. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2019, 28, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ding, M.; Duan, X.; Feng, X.; Wang, P.; Jiang, Q.; Cheng, Z.; Zhang, W.; Yu, S.; Yao, W.; et al. Diagnostic Value of Plasma MicroRNAs for Lung Cancer Using Support Vector Machine Model. J. Cancer 2019, 10, 5090–5098. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yu, L.; Lin, X.; Zheng, Q.; Zhang, S.; Chen, D.; Pan, X.; Huang, Y. Combination of Serum miRNAs with Serum Exosomal miRNAs in Early Diagnosis for Non-Small-Cell Lung Cancer. Cancer Manag. Res. 2020, 12, 485–495. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C.; Peng, X.; Liu, K.; Zhao, L.; Chen, X.; Yu, H.; Lai, Y. A Combination of Four Serum miRNAs for Screening of Lung Adenocarcinoma. Hum. Cell 2020, 33, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Bedard, E.L.R.; Abraham, A.G.; Joy, A.A.; Ghosh, S.; Wang, X.; Lim, A.; Shao, D.; Loebenberg, R.; Roa, W.H. A Novel Composite Biomarker Panel For Detection Of Early Stage Non-Small Cell Lung Cancer. Clin. Investig. Med. Med. Clin. Exp. 2021, 44, E15–E24. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jin, Y.; Feng, Y. Evaluation of Plasma Extracellular Vesicle MicroRNA Signatures for Lung Adenocarcinoma and Granuloma With Monte-Carlo Feature Selection Method. Front. Genet. 2019, 10, 367. [Google Scholar] [CrossRef]
- Bagheri, A.; Khorram Khorshid, H.R.; Mowla, S.J.; Mohebbi, H.A.; Mohammadian, A.; Yaseri, M.; Solaymani-Dodaran, M.; Sherafatian, M.; Tavallaie, M. Altered miR-223 Expression in Sputum for Diagnosis of Non-Small Cell Lung Cancer. Avicenna J. Med. Biotechnol. 2017, 9, 189–195. [Google Scholar]
- Aiso, T.; Ohtsuka, K.; Ueda, M.; Karita, S.; Yokoyama, T.; Takata, S.; Matsuki, N.; Kondo, H.; Takizawa, H.; Okada, A.A.; et al. Serum Levels of Candidate microRNA Diagnostic Markers Differ among the Stages of Non-Small-Cell Lung Cancer. Oncol. Lett. 2018, 16, 6643–6651. [Google Scholar] [CrossRef] [PubMed]
- Monastirioti, A.; Papadaki, C.; Kalapanida, D.; Rounis, K.; Michaelidou, K.; Papadaki, M.A.; Mavroudis, D.; Agelaki, S. Plasma-Based microRNA Expression Analysis in Advanced Stage NSCLC Patients Treated with Nivolumab. Cancers 2022, 14, 4739. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K. Heterogeneity in Breast Cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Primer 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Łukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanisławek, A. Breast Cancer—Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef]
- Harbeck, N.; Gnant, M. Breast Cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, L.; Huang, B.; Wang, Y.; Ji, L.; Wu, J.; Di, G.; Liu, G.; Yu, K.; Shao, Z.; et al. The Prognostic and Predictive Potential of Ki-67 in Triple-Negative Breast Cancer. Sci. Rep. 2020, 10, 225. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Zang, Y.; Wang, F. NLRP3 Inflammasome Inactivation Driven by miR-223-3p Reduces Tumor Growth and Increases Anticancer Immunity in Breast Cancer. Mol. Med. Rep. 2019, 19, 2180–2188. [Google Scholar] [CrossRef]
- Wang, X.; Tong, Z.; Liu, H. MiR-223-3p Targeting Epithelial Cell Transforming Sequence 2 Oncogene Inhibits the Activity, Apoptosis, Invasion and Migration of MDA-MB-468 Breast Cancer Cells. OncoTargets Ther. 2019, 12, 7675–7684. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhong, R.; Deng, C.; Zhou, Z. Circle RNA circABCB10 Modulates PFN2 to Promote Breast Cancer Progression, as Well as Aggravate Radioresistance through Facilitating Glycolytic Metabolism Via miR-223-3p. Cancer Biother. Radiopharm. 2021, 36, 477–490. [Google Scholar] [CrossRef]
- Terme, M.; Ullrich, E.; Aymeric, L.; Meinhardt, K.; Desbois, M.; Delahaye, N.; Viaud, S.; Ryffel, B.; Yagita, H.; Kaplanski, G.; et al. IL-18 Induces PD-1–Dependent Immunosuppression in Cancer. Cancer Res. 2011, 71, 5393–5399. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Zhao, G.; Liu, L.; Liu, F.; Gong, W.; Liu, X.; Yang, L.; Wang, J.; Hou, Y. Pre-Treatment with IL-1β Enhances the Efficacy of MSC Transplantation in DSS-Induced Colitis. Cell. Mol. Immunol. 2012, 9, 473–481. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, Z.; Ma, N.; Wang, B.; Liu, J.; Zhang, L.; Gu, L. MicroRNA-223 Targeting STIM1 Inhibits the Biological Behavior of Breast Cancer. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 45, 856–866. [Google Scholar] [CrossRef]
- Sun, X.; Li, Y.; Zheng, M.; Zuo, W.; Zheng, W. MicroRNA-223 Increases the Sensitivity of Triple-Negative Breast Cancer Stem Cells to TRAIL-Induced Apoptosis by Targeting HAX-1. PLoS ONE 2016, 11, e0162754. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Yang, H.; Fang, Q.; Quan, H.; Lu, H.; Wang, X. Circ_ZFR Affects FABP7 Expression to Regulate Breast Cancer Progression by Acting as a Sponge for miR-223-3p. Thorac. Cancer 2022, 13, 1369–1380. [Google Scholar] [CrossRef]
- Tang, X.Y.; Umemura, S.; Tsukamoto, H.; Kumaki, N.; Tokuda, Y.; Osamura, R.Y. Overexpression of Fatty Acid Binding Protein-7 Correlates with Basal-like Subtype of Breast Cancer. Pathol.-Res. Pract. 2010, 206, 98–101. [Google Scholar] [CrossRef]
- Cordero, A.; Kanojia, D.; Miska, J.; Panek, W.K.; Xiao, A.; Han, Y.; Bonamici, N.; Zhou, W.; Xiao, T.; Wu, M.; et al. FABP7 Is a Key Metabolic Regulator in HER2+ Breast Cancer Brain Metastasis. Oncogene 2019, 38, 6445–6460. [Google Scholar] [CrossRef]
- Vickers, K.C.; Landstreet, S.R.; Levin, M.G.; Shoucri, B.M.; Toth, C.L.; Taylor, R.C.; Palmisano, B.T.; Tabet, F.; Cui, H.L.; Rye, K.-A.; et al. MicroRNA-223 Coordinates Cholesterol Homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 14518–14523. [Google Scholar] [CrossRef]
- Palma, G.B.H.; Kaur, M. miRNA-128 and miRNA-223 Regulate Cholesterol-mediated Drug Resistance in Breast Cancer. IUBMB Life 2023, 75, 743–764. [Google Scholar] [CrossRef]
- Baek, A.E.; Yu, Y.-R.A.; He, S.; Wardell, S.E.; Chang, C.-Y.; Kwon, S.; Pillai, R.V.; McDowell, H.B.; Thompson, J.W.; Dubois, L.G.; et al. The Cholesterol Metabolite 27 Hydroxycholesterol Facilitates Breast Cancer Metastasis through Its Actions on Immune Cells. Nat. Commun. 2017, 8, 864. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Iinuma, H.; Umemoto, Y.; Yanagisawa, T.; Matsumoto, A.; Jinno, H. Exosome-encapsulated microRNA-223-3p as a Minimally Invasive Biomarker for the Early Detection of Invasive Breast Cancer. Oncol. Lett. 2018, 15, 9584–9592. [Google Scholar] [CrossRef]
- Du, T.; Wang, D.; Wan, X.; Xu, J.; Xiao, Q.; Liu, B. Regulatory Effect of microRNA-223-3p on Breast Cancer Cell Processes via the Hippo/Yap Signaling Pathway. Oncol. Lett. 2021, 22, 516. [Google Scholar] [CrossRef]
- Wang, R.; Yang, Z.; Liang, L.; Feng, X.; Che, B.; Zhang, X.; Zheng, Q.; Yan, X.; Han, H. Notch Activation Suppresses Endothelial Cell Migration and Sprouting via miR-223-3p Targeting Fbxw7. Vitr. Cell. Dev. Biol.-Anim. 2022, 58, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Reedijk, M. Notch Signaling and Breast Cancer. In Notch Signaling in Embryology and Cancer; Reichrath, J., Reichrath, S., Eds.; Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2012; Volume 727, pp. 241–257. ISBN 978-1-4614-0898-7. [Google Scholar]
- Fryer, C.J.; White, J.B.; Jones, K.A. Mastermind Recruits CycC:CDK8 to Phosphorylate the Notch ICD and Coordinate Activation with Turnover. Mol. Cell 2004, 16, 509–520. [Google Scholar] [CrossRef]
- Tetzlaff, M.T.; Yu, W.; Li, M.; Zhang, P.; Finegold, M.; Mahon, K.; Harper, J.W.; Schwartz, R.J.; Elledge, S.J. Defective Cardiovascular Development and Elevated Cyclin E and Notch Proteins in Mice Lacking the Fbw7 F-Box Protein. Proc. Natl. Acad. Sci. USA 2004, 101, 3338–3345. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, S.; Wang, Y.; Zhang, X.; Liu, X.; Li, J.; Li, P.; Du, L.; Wang, C. miR-223-3p Targets FBXW7 to Promote Epithelial-mesenchymal Transition and Metastasis in Breast Cancer. Thorac. Cancer 2022, 13, 474–482. [Google Scholar] [CrossRef]
- Henriques Palma, G.B.; Kaur, M. Cholesterol Depletion Modulates Drug Resistance Pathways to Sensitize Resistant Breast Cancer Cells to Tamoxifen. Anticancer Res. 2022, 42, 565–579. [Google Scholar] [CrossRef]
- Torres-Roca, J.F.; Fulp, W.J.; Caudell, J.J.; Servant, N.; Bollet, M.A.; Van De Vijver, M.; Naghavi, A.O.; Harris, E.E.; Eschrich, S.A. Integration of a Radiosensitivity Molecular Signature into the Assessment of Local Recurrence Risk in Breast Cancer. Int. J. Radiat. Oncol. 2015, 93, 631–638. [Google Scholar] [CrossRef]
- Hall, J.S.; Iype, R.; Senra, J.; Taylor, J.; Armenoult, L.; Oguejiofor, K.; Li, Y.; Stratford, I.; Stern, P.L.; O’Connor, M.J.; et al. Investigation of Radiosensitivity Gene Signatures in Cancer Cell Lines. PLoS ONE 2014, 9, e86329. [Google Scholar] [CrossRef]
- Fabris, L.; Berton, S.; Citron, F.; D’Andrea, S.; Segatto, I.; Nicoloso, M.S.; Massarut, S.; Armenia, J.; Zafarana, G.; Rossi, S.; et al. Radiotherapy-Induced miR-223 Prevents Relapse of Breast Cancer by Targeting the EGF Pathway. Oncogene 2016, 35, 4914–4926. [Google Scholar] [CrossRef]
- Chase, D.; Perhanidis, J.; Gupta, D.; Kalilani, L.; Golembesky, A.; González-Martín, A. Association of Multiple High-Risk Factors on Observed Outcomes in Real-World Patients with Advanced Ovarian Cancer Treated with First-Line Therapy. JCO Clin. Cancer Inform. 2023, 7, e2200189. [Google Scholar] [CrossRef]
- Lopacinska-Jørgensen, J.; Oliveira, D.V.N.P.; Wayne Novotny, G.; Høgdall, C.K.; Høgdall, E.V. Integrated microRNA and mRNA Signatures Associated with Overall Survival in Epithelial Ovarian Cancer. PLoS ONE 2021, 16, e0255142. [Google Scholar] [CrossRef] [PubMed]
- Felder, M.; Kapur, A.; Gonzalez-Bosquet, J.; Horibata, S.; Heintz, J.; Albrecht, R.; Fass, L.; Kaur, J.; Hu, K.; Shojaei, H.; et al. MUC16 (CA125): Tumor Biomarker to Cancer Therapy, a Work in Progress. Mol. Cancer 2014, 13, 129. [Google Scholar] [CrossRef]
- Rahimian, N.; Razavi, Z.S.; Aslanbeigi, F.; Mirkhabbaz, A.M.; Piroozmand, H.; Shahrzad, M.K.; Hamblin, M.R.; Mirzaei, H. Non-Coding RNAs Related to Angiogenesis in Gynecological Cancer. Gynecol. Oncol. 2021, 161, 896–912. [Google Scholar] [CrossRef]
- Fang, G.; Liu, J.; Wang, Q.; Huang, X.; Yang, R.; Pang, Y.; Yang, M. MicroRNA-223-3p Regulates Ovarian Cancer Cell Proliferation and Invasion by Targeting SOX11 Expression. Int. J. Mol. Sci. 2017, 18, 1208. [Google Scholar] [CrossRef]
- Psilopatis, I.; Schaefer, J.I.; Arsenakis, D.; Bolovis, D.; Levidou, G. SOX11 and Epithelial-Mesenchymal Transition in Metastatic Serous Ovarian Cancer. Biomedicines 2023, 11, 2540. [Google Scholar] [CrossRef]
- Lu, M.; Gong, B.; Wang, Y.; Li, J. CircBNC2 Affects Epithelial Ovarian Cancer Progression through the miR-223-3p/LARP4 Axis. Anticancer Drugs 2023, 34, 384–394. [Google Scholar] [CrossRef]
- Al Hinai, M.; Malgundkar, S.; Gupta, I.; Lakhtakia, R.; Al Kalbani, M.; Burney, I.; Al Moundhri, M.; Okamoto, A.; Tamimi, Y. Epigenetic Status of FBXW7 Gene and Its Role in Ovarian Cancer Pathogenesis. Asian Pac. J. Cancer Prev. 2023, 24, 1583–1590. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, X.-X.; Li, M.-C.; Cao, C.-H.; Wan, D.-Y.; Xi, B.-X.; Tan, J.-H.; Wang, J.; Yang, Z.-Y.; Feng, X.-X.; et al. C/EBPβ Enhances Platinum Resistance of Ovarian Cancer Cells by Reprogramming H3K79 Methylation. Nat. Commun. 2018, 9, 1739. [Google Scholar] [CrossRef]
- Laios, A.; O’Toole, S.; Flavin, R.; Martin, C.; Kelly, L.; Ring, M.; Finn, S.P.; Barrett, C.; Loda, M.; Gleeson, N.; et al. Potential Role of miR-9 and miR-223 in Recurrent Ovarian Cancer. Mol. Cancer 2008, 7, 35. [Google Scholar] [CrossRef]
- Freedland, S.J.; Humphreys, E.B.; Mangold, L.A.; Eisenberger, M.; Dorey, F.J.; Walsh, P.C.; Partin, A.W. Risk of Prostate Cancer–Specific Mortality Following Biochemical Recurrence after Radical Prostatectomy. JAMA 2005, 294, 433. [Google Scholar] [CrossRef]
- Porkka, K.P.; Pfeiffer, M.J.; Waltering, K.K.; Vessella, R.L.; Tammela, T.L.J.; Visakorpi, T. MicroRNA Expression Profiling in Prostate Cancer. Cancer Res. 2007, 67, 6130–6135. [Google Scholar] [CrossRef]
- Rana, S.; Valbuena, G.N.; Curry, E.; Bevan, C.L.; Keun, H.C. MicroRNAs as Biomarkers for Prostate Cancer Prognosis: A Systematic Review and a Systematic Reanalysis of Public Data. Br. J. Cancer 2022, 126, 502–513. [Google Scholar] [CrossRef]
- Xu, S.; Lian, Z.; Zhang, S.; Xu, Y.; Zhang, H. CircGNG4 Promotes the Progression of Prostate Cancer by Sponging miR-223 to Enhance EYA3/c-Myc Expression. Front. Cell Dev. Biol. 2021, 9, 684125. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, J.; Yi, L.; Wang, Y.; Dong, Z.; Liu, Z.; Ou-yang, S.; Wu, H.; Zhong, Z.; Yin, Z.; et al. MiR-223-3p Targeting SEPT6 Promotes the Biological Behavior of Prostate Cancer. Sci. Rep. 2014, 4, 7546. [Google Scholar] [CrossRef]
- Feng, Q.; He, P.; Wang, Y. MicroRNA-223-3p Regulates Cell Chemo-Sensitivity by Targeting FOXO3 in Prostatic Cancer. Gene 2018, 658, 152–158. [Google Scholar] [CrossRef]
- Zhou, K.; Wei, Y.; Li, X.; Yang, X. MiR-223-3p Targets FOXO3a to Inhibit Radiosensitivity in Prostate Cancer by Activating Glycolysis. Life Sci. 2021, 282, 119798. [Google Scholar] [CrossRef]
- Chang, X.; Liu, X.; Wang, H.; Yang, X.; Gu, Y. Glycolysis in the Progression of Pancreatic Cancer. Am. J. Cancer Res. 2022, 12, 861–872. [Google Scholar]
- Dülgeroğlu, Y.; Eroğlu, O. Serum Levels of miR-223-3p and miR-223-5p in Prostate Diseases. MicroRNA 2021, 9, 303–309. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro-Oncology 2019, 21, v1–v100. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Broggi, G.; Salvatorelli, L.; Barbagallo, D.; Certo, F.; Altieri, R.; Tirrò, E.; Massimino, M.; Vigneri, P.; Guadagno, E.; Maugeri, G.; et al. Diagnostic Utility of the Immunohistochemical Expression of Serine and Arginine Rich Splicing Factor 1 (SRSF1) in the Differential Diagnosis of Adult Gliomas. Cancers 2021, 13, 2086. [Google Scholar] [CrossRef]
- Barbagallo, D.; Caponnetto, A.; Barbagallo, C.; Battaglia, R.; Mirabella, F.; Brex, D.; Stella, M.; Broggi, G.; Altieri, R.; Certo, F.; et al. The GAUGAA Motif Is Responsible for the Binding between circSMARCA5 and SRSF1 and Related Downstream Effects on Glioblastoma Multiforme Cell Migration and Angiogenic Potential. Int. J. Mol. Sci. 2021, 22, 1678. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Caponnetto, A.; Brex, D.; Mirabella, F.; Barbagallo, C.; Lauretta, G.; Morrone, A.; Certo, F.; Broggi, G.; Caltabiano, R.; et al. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme through the Binding of SRSF1. Cancers 2019, 11, 194. [Google Scholar] [CrossRef]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.M.; Ragusa, M.; et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Condorelli, A.; Ragusa, M.; Salito, L.; Sammito, M.; Banelli, B.; Caltabiano, R.; Barbagallo, G.; Zappalà, A.; Battaglia, R.; et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 Axis Is Involved in Glioblastoma Multiforme. Oncotarget 2016, 7, 4746–4759. [Google Scholar] [CrossRef]
- Mellinghoff, I.K.; van den Bent, M.J.; Blumenthal, D.T.; Touat, M.; Peters, K.B.; Clarke, J.; Mendez, J.; Yust-Katz, S.; Welsh, L.; Mason, W.P.; et al. Vorasidenib in IDH1- or IDH2-Mutant Low-Grade Glioma. N. Engl. J. Med. 2023, 389, 589–601. [Google Scholar] [CrossRef]
- Altieri, R.; Barbagallo, D.; Certo, F.; Broggi, G.; Ragusa, M.; Di Pietro, C.; Caltabiano, R.; Magro, G.; Peschillo, S.; Purrello, M.; et al. Peritumoral Microenvironment in High-Grade Gliomas: From FLAIRectomy to Microglia-Glioma Cross-Talk. Brain Sci. 2021, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-M.; Huang, W.; Park, R.; Park, P.J.; Johnson, M.D. A Developmental Taxonomy of Glioblastoma Defined and Maintained by MicroRNAs. Cancer Res. 2011, 71, 3387–3399. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, S.M.; Laug, D.; Brawley, V.S.; Zhang, Z.; Corder, A.; Yin, Z.; Wong, S.T.C.; Li, X.-N.; Foster, A.E.; Ahmed, N.; et al. The miR-223/Nuclear Factor I-A Axis Regulates Glial Precursor Proliferation and Tumorigenesis in the CNS. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 13560–13568. [Google Scholar] [CrossRef]
- Evers, L.; Schäfer, A.; Pini, R.; Zhao, K.; Stei, S.; Nimsky, C.; Bartsch, J.W. Identification of Dysregulated microRNAs in Glioblastoma Stem-like Cells. Brain Sci. 2023, 13, 350. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Li, Z.; Jiang, Y.; Jiang, Z.; Tang, L. SNHG29 Regulates miR-223-3p/CTNND1 Axis to Promote Glioblastoma Progression via Wnt/β-Catenin Signaling Pathway. Cancer Cell Int. 2019, 19, 345. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.; Tu, S.; Fu, W.; Wang, J.; Bai, Z. LncRNA PITPNA-AS1 Stimulates Cell Proliferation and Suppresses Cell Apoptosis in Glioblastoma via Targeting miR-223-3p/EGFR Axis and Activating PI3K/AKT Signaling Pathway. Cell Cycle Georget. Tex 2021, 20, 1988–1998. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Shen, L.; Nie, X.; Lu, B.; Pan, X.; Su, Z.; Yan, A.; Yan, R.; Zhou, Y.; Li, L.; et al. MiR-223-3p Overexpression Inhibits Cell Proliferation and Migration by Regulating Inflammation-Associated Cytokines in Glioblastomas. Pathol. Res. Pract. 2018, 214, 1330–1339. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, A.; Liu, S.; Li, R.; Wang, X.; Yan, W.; Li, H.; You, Y. Genome-Wide Identification of Epithelial-Mesenchymal Transition-Associated microRNAs Reveals Novel Targets for Glioblastoma Therapy. Oncol. Lett. 2018, 15, 7625–7630. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.-S.; Luo, Q.-Z.; Han, Y.; Li, X.-B.; Cao, L.-J.; Wu, L.-X. microRNA-223 Promotes the Growth and Invasion of Glioblastoma Cells by Targeting Tumor Suppressor PAX6. Oncol. Rep. 2013, 30, 2263–2269. [Google Scholar] [CrossRef]
- Huang, B.-S.; Luo, Q.-Z.; Han, Y.; Huang, D.; Tang, Q.-P.; Wu, L.-X. MiR-223/PAX6 Axis Regulates Glioblastoma Stem Cell Proliferation and the Chemo Resistance to TMZ via Regulating PI3K/Akt Pathway. J. Cell. Biochem. 2017, 118, 3452–3461. [Google Scholar] [CrossRef]
- Cheng, Q.; Ma, X.; Cao, H.; Chen, Z.; Wan, X.; Chen, R.; Peng, R.; Huang, J.; Jiang, B. Role of miR-223/Paired Box 6 Signaling in Temozolomide Chemoresistance in Glioblastoma Multiforme Cells. Mol. Med. Rep. 2017, 15, 597–604. [Google Scholar] [CrossRef][Green Version]
- Liang, L.; Zhu, J.; Zaorsky, N.G.; Deng, Y.; Wu, X.; Liu, Y.; Liu, F.; Cai, G.; Gu, W.; Shen, L.; et al. MicroRNA-223 Enhances Radiation Sensitivity of U87MG Cells in Vitro and in Vivo by Targeting Ataxia Telangiectasia Mutated. Int. J. Radiat. Oncol. Biol. Phys. 2014, 88, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Mekala, J.R.; Kurappalli, R.K.; Ramalingam, P.; Moparthi, N.R. N-Acetyl l-Aspartate and Triacetin Modulate Tumor Suppressor MicroRNA and Class I and II HDAC Gene Expression Induce Apoptosis in Glioblastoma Cancer Cells In Vitro. Life Sci. 2021, 286, 120024. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, C.; Lu, W.; Chen, R.; Yu, M. Establishment of a Prognostic-Related microRNAs Risk Model for Glioma by Bioinformatics Analysis. Ann. Transl. Med. 2021, 9, 1022. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Gao, K.; Luo, H.; Wang, X.; Shi, Y.; Dong, Q.; Luan, W.; You, Y. Identification of Intrinsic Subtype-Specific Prognostic microRNAs in Primary Glioblastoma. J. Exp. Clin. Cancer Res. CR 2014, 33, 9. [Google Scholar] [CrossRef]
- Huang, Y.-T.; Hsu, T.; Kelsey, K.T.; Lin, C.-L. Integrative Analysis of Micro-RNA, Gene Expression, and Survival of Glioblastoma Multiforme. Genet. Epidemiol. 2015, 39, 134–143. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, C.; Qu, T.; Feng, Y. A Novel-Defined Necroptosis-Related miRNA Signature for Forecasting the Prognosis of Low-Grade Glioma. BioMed Res. Int. 2022, 2022, 9957604. [Google Scholar] [CrossRef] [PubMed]
- Gozé, C.; Reynes, C.; Forestier, L.; Sabatier, R.; Duffau, H. Pilot Study of Whole Blood MicroRNAs as Potential Tools for Diffuse Low-Grade Gliomas Detection. Cell. Mol. Neurobiol. 2018, 38, 715–725. [Google Scholar] [CrossRef]
- Morokoff, A.; Jones, J.; Nguyen, H.; Ma, C.; Lasocki, A.; Gaillard, F.; Bennett, I.; Luwor, R.; Stylli, S.; Paradiso, L.; et al. Serum microRNA Is a Biomarker for Post-Operative Monitoring in Glioma. J. Neuro-Oncol. 2020, 149, 391–400. [Google Scholar] [CrossRef]
- Roth, P.; Wischhusen, J.; Happold, C.; Chandran, P.A.; Hofer, S.; Eisele, G.; Weller, M.; Keller, A. A Specific miRNA Signature in the Peripheral Blood of Glioblastoma Patients. J. Neurochem. 2011, 118, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Genovese, G.; Ergun, A.; Shukla, S.A.; Campos, B.; Hanna, J.; Ghosh, P.; Quayle, S.N.; Rai, K.; Colla, S.; Ying, H.; et al. microRNA Regulatory Network Inference Identifies miR-34a as a Novel Regulator of TGF-β Signaling in Glioblastoma. Cancer Discov. 2012, 2, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Ilic, M.; Ilic, I. Epidemiology of Pancreatic Cancer. World J. Gastroenterol. 2016, 22, 9694–9705. [Google Scholar] [CrossRef] [PubMed]
- Quiñonero, F.; Mesas, C.; Doello, K.; Cabeza, L.; Perazzoli, G.; Jimenez-Luna, C.; Rama, A.R.; Melguizo, C.; Prados, J. The Challenge of Drug Resistance in Pancreatic Ductal Adenocarcinoma: A Current Overview. Cancer Biol. Med. 2019, 16, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.; Jansen, G.; Giovannetti, E. Drug Resistance in Pancreatic Cancer: Impact of Altered Energy Metabolism. Crit. Rev. Oncol. Hematol. 2017, 114, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Brehm, C.U.; Gress, T.M.; Buchholz, M.; Alashkar Alhamwe, B.; Von Strandmann, E.; Slater, E.P.; Bartsch, J.W.; Bauer, C.; Lauth, M. The Immune Microenvironment in Pancreatic Cancer. Int. J. Mol. Sci. 2020, 21, 7307. [Google Scholar] [CrossRef] [PubMed]
- Ceyhan, G.O.; Friess, H. Pancreatic Fibrosis and Standard Diagnostics. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Goral, V. Pancreatic Cancer: Pathogenesis and Diagnosis. Asian Pac. J. Cancer Prev. APJCP 2015, 16, 5619–5624. [Google Scholar] [CrossRef]
- Liot, S.; El Kholti, N.; Balas, J.; Genestier, L.; Verrier, B.; Valcourt, U.; Lambert, E. Development of Thymic Tumor in [LSL:KrasG12D; Pdx1-CRE] Mice, an Adverse Effect Associated with Accelerated Pancreatic Carcinogenesis. Sci. Rep. 2021, 11, 15075. [Google Scholar] [CrossRef]
- Deer, E.L.; González-Hernández, J.; Coursen, J.D.; Shea, J.E.; Ngatia, J.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. Phenotype and Genotype of Pancreatic Cancer Cell Lines. Pancreas 2010, 39, 425–435. [Google Scholar] [CrossRef]
- Miquel, M.; Zhang, S.; Pilarsky, C. Pre-Clinical Models of Metastasis in Pancreatic Cancer. Front. Cell Dev. Biol. 2021, 9, 748631. [Google Scholar] [CrossRef]
- Daoud, A.Z.; Mulholland, E.J.; Cole, G.; McCarthy, H.O. MicroRNAs in Pancreatic Cancer: Biomarkers, Prognostic, and Therapeutic Modulators. BMC Cancer 2019, 19, 1130. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Hogendorf, P. The Role of microRNA in Pancreatic Cancer. Biomedicines 2021, 9, 1322. [Google Scholar] [CrossRef]
- Fathi, M.; Ghafouri-Fard, S.; Abak, A.; Taheri, M. Emerging Roles of miRNAs in the Development of Pancreatic Cancer. Biomed. Pharmacother. Biomed. Pharmacother. 2021, 141, 111914. [Google Scholar] [CrossRef]
- Yonemori, K.; Kurahara, H.; Maemura, K.; Natsugoe, S. MicroRNA in Pancreatic Cancer. J. Hum. Genet. 2017, 62, 33–40. [Google Scholar] [CrossRef]
- Rawat, M.; Kadian, K.; Gupta, Y.; Kumar, A.; Chain, P.S.G.; Kovbasnjuk, O.; Kumar, S.; Parasher, G. MicroRNA in Pancreatic Cancer: From Biology to Therapeutic Potential. Genes 2019, 10, 752. [Google Scholar] [CrossRef] [PubMed]
- Rachagani, S.; Macha, M.A.; Menning, M.S.; Dey, P.; Pai, P.; Smith, L.M.; Mo, Y.-Y.; Batra, S.K. Changes in microRNA (miRNA) Expression during Pancreatic Cancer Development and Progression in a Genetically Engineered KrasG12D;Pdx1-Cre Mouse (KC) Model. Oncotarget 2015, 6, 40295–40309. [Google Scholar] [CrossRef]
- Ma, J.; Cao, T.; Cui, Y.; Zhang, F.; Shi, Y.; Xia, J.; Wang, Z.P. miR-223 Regulates Cell Proliferation and Invasion via Targeting PDS5B in Pancreatic Cancer Cells. Mol. Ther. Nucleic Acids 2019, 14, 583–592. [Google Scholar] [CrossRef]
- He, D.; Huang, C.; Zhou, Q.; Liu, D.; Xiong, L.; Xiang, H.; Ma, G.; Zhang, Z. HnRNPK/miR-223/FBXW7 Feedback Cascade Promotes Pancreatic Cancer Cell Growth and Invasion. Oncotarget 2017, 8, 20165–20178. [Google Scholar] [CrossRef]
- Ma, J.; Zeng, F.; Ma, C.; Pang, H.; Fang, B.; Lian, C.; Yin, B.; Zhang, X.; Wang, Z.; Xia, J. Synergistic Reversal Effect of Epithelial-to-Mesenchymal Transition by miR-223 Inhibitor and Genistein in Gemcitabine-Resistant Pancreatic Cancer Cells. Am. J. Cancer Res. 2016, 6, 1384–1395. [Google Scholar]
- Zhang, N.; Coutinho, L.E.; Pati, D. PDS5A and PDS5B in Cohesin Function and Human Disease. Int. J. Mol. Sci. 2021, 22, 5868. [Google Scholar] [CrossRef]
- Denes, V.; Pilichowska, M.; Makarovskiy, A.; Carpinito, G.; Geck, P. Loss of a Cohesin-Linked Suppressor APRIN (Pds5b) Disrupts Stem Cell Programs in Embryonal Carcinoma: An Emerging Cohesin Role in Tumor Suppression. Oncogene 2010, 29, 3446–3452. [Google Scholar] [CrossRef]
- Ma, J.; Cui, Y.; Cao, T.; Xu, H.; Shi, Y.; Xia, J.; Tao, Y.; Wang, Z.P. PDS5B Regulates Cell Proliferation and Motility via Upregulation of Ptch2 in Pancreatic Cancer Cells. Cancer Lett. 2019, 460, 65–74. [Google Scholar] [CrossRef]
- Zhang, X.; Tan, P.; Zhuang, Y.; Du, L. hsa_circRNA_001587 Upregulates SLC4A4 Expression to Inhibit Migration, Invasion, and Angiogenesis of Pancreatic Cancer Cells via Binding to microRNA-223. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G703–G717. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Q.; Zhai, G.; Ke, S.; Yu, X.; Guo, J. SLC4A4 Promotes Prostate Cancer Progression in Vivo and in Vitro via AKT-Mediated Signalling Pathway. Cancer Cell Int. 2022, 22, 127. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, X.; Wang, T.; Xing, J. MiR-223-3p Promotes Cell Proliferation and Metastasis by Downregulating SLC4A4 in Clear Cell Renal Cell Carcinoma. Aging 2019, 11, 615–633. [Google Scholar] [CrossRef]
- Huang, R.; Song, X.; Wang, C.-M. MiR-223 Regulates CDDP Resistance in Pancreatic Cancer via Targeting FoxO3a. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7892–7898. [Google Scholar] [CrossRef]
- Liu, Y.; Ao, X.; Ding, W.; Ponnusamy, M.; Wu, W.; Hao, X.; Yu, W.; Wang, Y.; Li, P.; Wang, J. Critical Role of FOXO3a in Carcinogenesis. Mol. Cancer 2018, 17, 104. [Google Scholar] [CrossRef]
- Feng, S.; Jiang, Z.-J.; Yu, D.; Li, J.; Liu, G.; Sun, J.-J. FOXO3a Expression and Its Diagnostic Value in Pancreatic Ductal Adenocarcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 5422–5429. [Google Scholar] [PubMed]
- Ma, J.; Fang, B.; Zeng, F.; Ma, C.; Pang, H.; Cheng, L.; Shi, Y.; Wang, H.; Yin, B.; Xia, J.; et al. Down-Regulation of miR-223 Reverses Epithelial-Mesenchymal Transition in Gemcitabine-Resistant Pancreatic Cancer Cells. Oncotarget 2015, 6, 1740–1749. [Google Scholar] [CrossRef]
- Ma, J.; Cheng, L.; Liu, H.; Zhang, J.; Shi, Y.; Zeng, F.; Miele, L.; Sarkar, F.H.; Xia, J.; Wang, Z. Genistein Down-Regulates miR-223 Expression in Pancreatic Cancer Cells. Curr. Drug Targets 2013, 14, 1150–1156. [Google Scholar] [CrossRef]
- Schultz, N.A.; Dehlendorff, C.; Jensen, B.V.; Bjerregaard, J.K.; Nielsen, K.R.; Bojesen, S.E.; Calatayud, D.; Nielsen, S.E.; Yilmaz, M.; Holländer, N.H.; et al. MicroRNA Biomarkers in Whole Blood for Detection of Pancreatic Cancer. JAMA 2014, 311, 392–404. [Google Scholar] [CrossRef]
- Komatsu, S.; Ichikawa, D.; Miyamae, M.; Kawaguchi, T.; Morimura, R.; Hirajima, S.; Okajima, W.; Ohashi, T.; Imamura, T.; Konishi, H.; et al. Malignant Potential in Pancreatic Neoplasm; New Insights Provided by Circulating miR-223 in Plasma. Expert Opin. Biol. Ther. 2015, 15, 773–785. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, J.; Sun, C.; Wang, L.; Jin, G.; Xin, L.; Jin, Z.; Wang, D.; Li, Z. MicroRNA Expression Levels as Diagnostic Biomarkers for Intraductal Papillary Mucinous Neoplasm. Oncotarget 2017, 8, 58765–58770. [Google Scholar] [CrossRef][Green Version]
- Debernardi, S.; Massat, N.J.; Radon, T.P.; Sangaralingam, A.; Banissi, A.; Ennis, D.P.; Dowe, T.; Chelala, C.; Pereira, S.P.; Kocher, H.M.; et al. Noninvasive Urinary miRNA Biomarkers for Early Detection of Pancreatic Adenocarcinoma. Am. J. Cancer Res. 2015, 5, 3455–3466. [Google Scholar]
- Hellberg, T.; Mohr, R.; Geisler, L.; Knorr, J.; Wree, A.; Demir, M.; Benz, F.; Lambrecht, J.; Loosen, S.H.; Tacke, F.; et al. Serum Levels of miR-223 but Not miR-21 Are Decreased in Patients with Neuroendocrine Tumors. PLoS ONE 2020, 15, e0244504. [Google Scholar] [CrossRef] [PubMed]
- Geisler, L.; Mohr, R.; Lambrecht, J.; Knorr, J.; Jann, H.; Loosen, S.H.; Özdirik, B.; Luedde, T.; Hammerich, L.; Tacke, F.; et al. The Role of miRNA in the Pathophysiology of Neuroendocrine Tumors. Int. J. Mol. Sci. 2021, 22, 8569. [Google Scholar] [CrossRef]
- Li, Y.; Deng, S.; Peng, J.; Wang, X.; Essandoh, K.; Mu, X.; Peng, T.; Meng, Z.-X.; Fan, G.-C. MicroRNA-223 Is Essential for Maintaining Functional β-Cell Mass during Diabetes through Inhibiting Both FOXO1 and SOX6 Pathways. J. Biol. Chem. 2019, 294, 10438–10448. [Google Scholar] [CrossRef]
- Méndez-Ferrer, S.; Bonnet, D.; Steensma, D.P.; Hasserjian, R.P.; Ghobrial, I.M.; Gribben, J.G.; Andreeff, M.; Krause, D.S. Bone Marrow Niches in Haematological Malignancies. Nat. Rev. Cancer 2020, 20, 285–298. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.-M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating Morphologic, Clinical, and Genomic Data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; de Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A Report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.d.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Chen, C.-Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Gentner, B.; Pochert, N.; Rouhi, A.; Boccalatte, F.; Plati, T.; Berg, T.; Sun, S.M.; Mah, S.M.; Mirkovic-Hösle, M.; Ruschmann, J.; et al. MicroRNA-223 Dose Levels Fine Tune Proliferation and Differentiation in Human Cord Blood Progenitors and Acute Myeloid Leukemia. Exp. Hematol. 2015, 43, 858–868.e7. [Google Scholar] [CrossRef] [PubMed]
- Fazi, F.; Racanicchi, S.; Zardo, G.; Starnes, L.M.; Mancini, M.; Travaglini, L.; Diverio, D.; Ammatuna, E.; Cimino, G.; Lo-Coco, F.; et al. Epigenetic Silencing of the Myelopoiesis Regulator microRNA-223 by the AML1/ETO Oncoprotein. Cancer Cell 2007, 12, 457–466. [Google Scholar] [CrossRef]
- Visone, R.; Rassenti, L.Z.; Veronese, A.; Taccioli, C.; Costinean, S.; Aguda, B.D.; Volinia, S.; Ferracin, M.; Palatini, J.; Balatti, V.; et al. Karyotype-Specific microRNA Signature in Chronic Lymphocytic Leukemia. Blood 2009, 114, 3872–3879. [Google Scholar] [CrossRef]
- Zardo, G.; Ciolfi, A.; Vian, L.; Starnes, L.M.; Billi, M.; Racanicchi, S.; Maresca, C.; Fazi, F.; Travaglini, L.; Noguera, N.; et al. Polycombs and microRNA-223 Regulate Human Granulopoiesis by Transcriptional Control of Target Gene Expression. Blood 2012, 119, 4034–4046. [Google Scholar] [CrossRef]
- Huo, X.-F.; Yu, J.; Peng, H.; Du, Z.-W.; Liu, X.-L.; Ma, Y.-N.; Zhang, X.; Zhang, Y.; Zhao, H.-L.; Zhang, J.-W. Differential Expression Changes in K562 Cells during the Hemin-Induced Erythroid Differentiation and the Phorbol Myristate Acetate (PMA)-Induced Megakaryocytic Differentiation. Mol. Cell. Biochem. 2006, 292, 155–167. [Google Scholar] [CrossRef]
- Yuan, J.-Y.; Wang, F.; Yu, J.; Yang, G.-H.; Liu, X.-L.; Zhang, J.-W. MicroRNA-223 Reversibly Regulates Erythroid and Megakaryocytic Differentiation of K562 Cells. J. Cell. Mol. Med. 2009, 13, 4551–4559. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, J.-H.; Zheng, Y.-S.; Zhang, P.; Chen, X.; Wu, J.; Xu, L.; Luo, X.-Q.; Ke, Z.-Y.; Zhou, H.; et al. Genome-Wide Analysis of Small RNA and Novel MicroRNA Discovery in Human Acute Lymphoblastic Leukemia Based on Extensive Sequencing Approach. PLoS ONE 2009, 4, e6849. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; He, C.; Wang, D.; Yuan, X.; Chen, J.; Jin, J. MicroRNAs Expression Signatures Are Associated with Lineage and Survival in Acute Leukemias. Blood Cells Mol. Dis. 2010, 44, 191–197. [Google Scholar] [CrossRef]
- Zhu, Y.-D.; Wang, L.; Sun, C.; Fan, L.; Zhu, D.-X.; Fang, C.; Wang, Y.-H.; Zou, Z.-J.; Zhang, S.-J.; Li, J.-Y.; et al. Distinctive microRNA Signature Is Associated with the Diagnosis and Prognosis of Acute Leukemia. Med. Oncol. 2012, 29, 2323–2331. [Google Scholar] [CrossRef]
- Chiaretti, S.; Messina, M.; Tavolaro, S.; Zardo, G.; Elia, L.; Vitale, A.; Fatica, A.; Gorello, P.; Piciocchi, A.; Scappucci, G.; et al. Gene Expression Profiling Identifies a Subset of Adult T-Cell Acute Lymphoblastic Leukemia with Myeloid-like Gene Features and over-Expression of miR-223. Haematologica 2010, 95, 1114–1121. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.; Chen, J.; Li, J.; He, L.; Wu, P.; Wang, M.; Tong, N.; Zhang, Z.; Fang, Y. Association of the 3′UTR FOXO3a Polymorphism Rs4946936 with an Increased Risk of Childhood Acute Lymphoblastic Leukemia in a Chinese Population. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2014, 34, 325–332. [Google Scholar] [CrossRef]
- Xiao, Y.; Su, C.; Deng, T. miR-223 Decreases Cell Proliferation and Enhances Cell Apoptosis in Acute Myeloid Leukemia via Targeting FBXW7. Oncol. Lett. 2016, 12, 3531–3536. [Google Scholar] [CrossRef]
- Pulikkan, J.A.; Dengler, V.; Peramangalam, P.S.; Peer Zada, A.A.; Müller-Tidow, C.; Bohlander, S.K.; Tenen, D.G.; Behre, G. Cell-Cycle Regulator E2F1 and microRNA-223 Comprise an Autoregulatory Negative Feedback Loop in Acute Myeloid Leukemia. Blood 2010, 115, 1768–1778. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, S.; Tyybäkinoja, A.; Borze, I.; Räty, R.; Saarinen-Pihkala, U.M.; Usvasalo, A.; Elonen, E.; Knuutila, S. Integrated Analysis of Gene Copy Number, Copy Neutral LOH, and microRNA Profiles in Adult Acute Lymphoblastic Leukemia. Cytogenet. Genome Res. 2012, 136, 246–255. [Google Scholar] [CrossRef]
- Kim, K.-T.; Carroll, A.P.; Mashkani, B.; Cairns, M.J.; Small, D.; Scott, R.J. MicroRNA-16 Is down-Regulated in Mutated FLT3 Expressing Murine Myeloid FDC-P1 Cells and Interacts with Pim-1. PLoS ONE 2012, 7, e44546. [Google Scholar] [CrossRef]
- Eyholzer, M.; Schmid, S.; Schardt, J.A.; Haefliger, S.; Mueller, B.U.; Pabst, T. Complexity of miR-223 Regulation by CEBPA in Human AML. Leuk. Res. 2010, 34, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Pichiorri, F.; Palumbo, T.; Visentini, M.; Aqeilan, R.; Cimmino, A.; Wang, H.; Sun, H.; Volinia, S.; Alder, H.; et al. MicroRNA Gene Expression during Retinoic Acid-Induced Differentiation of Human Acute Promyelocytic Leukemia. Oncogene 2007, 26, 4148–4157. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Rossetti, S.; Datta, A.; Eng, K.; Beghini, A.; Sacchi, N. miR-17 Deregulates a Core RUNX1-miRNA Mechanism of CBF Acute Myeloid Leukemia. Mol. Cancer 2015, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Perna, F.; Wang, L.; Voza, F.; Figueroa, M.E.; Tempst, P.; Erdjument-Bromage, H.; Gao, R.; Chen, S.; Paietta, E.; et al. PRMT4 Blocks Myeloid Differentiation by Assembling a Methyl-RUNX1-Dependent Repressor Complex. Cell Rep. 2013, 5, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Pan, Z.; Zhang, H.; Wu, M.; Gui, Z.; Yuan, Q.; Chen, J.; Peng, J.; Liu, Z.; Tan, Q.; et al. PARP-1 Modulates the Expression of miR-223 through Histone Acetylation to Involve in the Hydroquinone-Induced Carcinogenesis of TK6 Cells. J. Biochem. Mol. Toxicol. 2022, 36, e23142. [Google Scholar] [CrossRef] [PubMed]
- Bellon, M.; Lepelletier, Y.; Hermine, O.; Nicot, C. Deregulation of microRNA Involved in Hematopoiesis and the Immune Response in HTLV-I Adult T-Cell Leukemia. Blood 2009, 113, 4914–4917. [Google Scholar] [CrossRef] [PubMed]
- Moles, R.; Bellon, M.; Nicot, C. STAT1: A Novel Target of miR-150 and miR-223 Is Involved in the Proliferation of HTLV-I–Transformed and ATL Cells. Neoplasia 2015, 17, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Davari, N.; Ahmadpour, F.; Kiani, A.A.; Azadpour, M.; Asadi, Z.T. Evaluation of microRNA-223 and microRNA-125a Expression Association with STAT3 and Bcl2 Genes in Blood Leukocytes of CLL Patients: A Case–Control Study. BMC Res. Notes 2021, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Agatheeswaran, S.; Singh, S.; Biswas, S.; Biswas, G.; Chandra Pattnayak, N.; Chakraborty, S. BCR-ABL Mediated Repression of miR-223 Results in the Activation of MEF2C and PTBP2 in Chronic Myeloid Leukemia. Leukemia 2013, 27, 1578–1580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yu, H.; Li, J.; Fan, J.; Chen, J. 2-Methoxyestradiol Combined with Ascorbic Acid Facilitates the Apoptosis of Chronic Myeloid Leukemia Cells via the microRNA-223/Fms-like Tyrosine Kinase 3/Phosphatidylinositol-3 Kinase/Protein Kinase B Axis. Bioengineered 2022, 13, 3470–3485. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J.; Qian, L.; Miao, Y.; Song, W.; Liu, H.; Li, R. MOZ Forms an Autoregulatory Feedback Loop with miR-223 in AML and Monocyte/Macrophage Development. iScience 2019, 11, 189–204. [Google Scholar] [CrossRef]
- Shu, Y.; Wang, Y.; Lv, W.-Q.; Peng, D.-Y.; Li, J.; Zhang, H.; Jiang, G.-J.; Yang, B.-J.; Liu, S.; Zhang, J.; et al. ARRB1-Promoted NOTCH1 Degradation Is Suppressed by OncomiR miR-223 in T-Cell Acute Lymphoblastic Leukemia. Cancer Res. 2020, 80, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Zhou, X.; Qi, X.; Liu, S.; Li, K.; Tan, J.; Liu, Z.; Yu, J.; Zhang, P.; Zou, L. β-Arrestin1 Promotes the Self-Renewal of the Leukemia-Initiating Cell-Enriched Subpopulation in B-Lineage Acute Lymphoblastic Leukemia Related to DNMT1 Activity. Cancer Lett. 2015, 357, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, H.; Qin, R.; Shu, Y.; Liu, Z.; Zhang, P.; Duan, C.; Hong, D.; Yu, J.; Zou, L. The Cellular Senescence of Leukemia-Initiating Cells from Acute Lymphoblastic Leukemia Is Postponed by β-Arrestin1 Binding with P300-Sp1 to Regulate hTERT Transcription. Cell Death Dis. 2017, 8, e2756. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Sun, J.; Huang, J.; Yao, F.; Chen, X.; Zhu, B.; Zhao, D. Circular RNA circRNA_0000094 Sponges microRNA-223-3p and up-Regulate F-Box and WD Repeat Domain Containing 7 to Restrain T Cell Acute Lymphoblastic Leukemia Progression. Hum. Cell 2021, 34, 977–989, Erratum in Hum. Cell 2021, 34, 1584. [Google Scholar] [CrossRef]
- Kumar, V.; Palermo, R.; Talora, C.; Campese, A.F.; Checquolo, S.; Bellavia, D.; Tottone, L.; Testa, G.; Miele, E.; Indraccolo, S.; et al. Notch and NF-kB Signaling Pathways Regulate miR-223/FBXW7 Axis in T-Cell Acute Lymphoblastic Leukemia. Leukemia 2014, 28, 2324–2335. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Glass, M.; Wahnschaffe, L.; Otte, M.; Mayer, P.; Franitza, M.; Altmüller, J.; Hallek, M.; Hüttelmaier, S.; Schrader, A.; et al. Micro-RNA Networks in T-Cell Prolymphocytic Leukemia Reflect T-Cell Activation and Shape DNA Damage Response and Survival Pathways. Haematologica 2022, 107, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.R.; Sanda, T.; Lawton, L.N.; Li, X.; Kreslavsky, T.; Novina, C.D.; Brand, M.; Gutierrez, A.; Kelliher, M.A.; Jamieson, C.H.M.; et al. The TAL1 Complex Targets the FBXW7 Tumor Suppressor by Activating miR-223 in Human T Cell Acute Lymphoblastic Leukemia. J. Exp. Med. 2013, 210, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Han, B.-W.; Feng, D.-D.; Li, Z.-G.; Luo, X.-Q.; Zhang, H.; Li, X.-J.; Zhang, X.-J.; Zheng, L.-L.; Zeng, C.-W.; Lin, K.-Y.; et al. A Set of miRNAs That Involve in the Pathways of Drug Resistance and Leukemic Stem-Cell Differentiation Is Associated with the Risk of Relapse and Glucocorticoid Response in Childhood ALL. Hum. Mol. Genet. 2011, 20, 4903–4915. [Google Scholar] [CrossRef] [PubMed]
- Nemes, K.; Csóka, M.; Nagy, N.; Márk, Á.; Váradi, Z.; Dankó, T.; Kovács, G.; Kopper, L.; Sebestyén, A. Expression of Certain Leukemia/Lymphoma Related microRNAs and Its Correlation with Prognosis in Childhood Acute Lymphoblastic Leukemia. Pathol. Oncol. Res. POR 2015, 21, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Gusscott, S.; Kuchenbauer, F.; Humphries, R.K.; Weng, A.P. Notch-Mediated Repression of miR-223 Contributes to IGF1R Regulation in T-ALL. Leuk. Res. 2012, 36, 905–911. [Google Scholar] [CrossRef]
- Rainer, J.; Ploner, C.; Jesacher, S.; Ploner, A.; Eduardoff, M.; Mansha, M.; Wasim, M.; Panzer-Grümayer, R.; Trajanoski, Z.; Niederegger, H.; et al. Glucocorticoid-Regulated microRNAs and Mirtrons in Acute Lymphoblastic Leukemia. Leukemia 2009, 23, 746–752. [Google Scholar] [CrossRef]
- Yu, G.; Yin, Z.; He, H.; Zheng, Z.; Chai, Y.; Xuan, L.; Lin, R.; Wang, Q.; Li, J.; Xu, D. Low Serum miR-223 Expression Predicts Poor Outcome in Patients with Acute Myeloid Leukemia. J. Clin. Lab. Anal. 2020, 34, e23096. [Google Scholar] [CrossRef] [PubMed]
- Rashed, W.M.; Hammad, A.M.; Saad, A.M.; Shohdy, K.S. MicroRNA as a Diagnostic Biomarker in Childhood Acute Lymphoblastic Leukemia; Systematic Review, Meta-Analysis and Recommendations. Crit. Rev. Oncol. Hematol. 2019, 136, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Lu, J.; Sun, M.; Li, Z.; Zhang, H.; Neilly, M.B.; Wang, Y.; Qian, Z.; Jin, J.; Zhang, Y.; et al. MicroRNA Expression Signatures Accurately Discriminate Acute Lymphoblastic Leukemia from Acute Myeloid Leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 19971–19976. [Google Scholar] [CrossRef] [PubMed]
- Luna-Aguirre, C.M.; De La Luz Martinez-Fierro, M.; Mar-Aguilar, F.; Garza-Veloz, I.; Treviño-Alvarado, V.; Rojas-Martinez, A.; Jaime-Perez, J.C.; Malagon-Santiago, G.I.; Gutierrez-Aguirre, C.H.; Gonzalez-Llano, O.; et al. Circulating microRNA Expression Profile in B-Cell Acute Lymphoblastic Leukemia. Cancer Biomark. 2015, 15, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lin, J.; Huang, W.; Cao, Y.; Liu, Y.; Wang, T.; Zhong, W.; Wang, D.; Mao, R.; Chen, X. The Effect of Circulating miR-223 on Surveillance of Different Cancers: A Meta-Analysis. OncoTargets Ther. 2017, 10, 3193–3201. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stamatopoulos, B.; Meuleman, N.; Haibe-Kains, B.; Saussoy, P.; Van Den Neste, E.; Michaux, L.; Heimann, P.; Martiat, P.; Bron, D.; Lagneaux, L. microRNA-29c and microRNA-223 down-Regulation Has in Vivo Significance in Chronic Lymphocytic Leukemia and Improves Disease Risk Stratification. Blood 2009, 113, 5237–5245. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Yi, S.; Yu, Z.; Li, Z.; Wang, Y.; Zou, D.; Qi, J.; Zhao, Y.; Qiu, L. MicroRNA-223 Expression Is Uniformly down-Regulated in B Cell Lymphoproliferative Disorders and Is Associated with Poor Survival in Patients with Chronic Lymphocytic Leukemia. Leuk. Lymphoma 2012, 53, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, T.J.; Davis, Z.; Gardiner, A.; Oscier, D.G.; Stevenson, F.K. Unmutated Ig V(H) Genes Are Associated with a More Aggressive Form of Chronic Lymphocytic Leukemia. Blood 1999, 94, 1848–1854. [Google Scholar] [CrossRef]
- Prizment, A.E.; Linabery, A.M.; Lutsey, P.L.; Selvin, E.; Nelson, H.H.; Folsom, A.R.; Church, T.R.; Drake, C.G.; Platz, E.A.; Joshu, C. Circulating Beta-2 Microglobulin and Risk of Cancer: The Atherosclerosis Risk in Communities Study (ARIC). Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2016, 25, 657–664. [Google Scholar] [CrossRef]
- Rodríguez-Vicente, A.E.; Quwaider, D.; Benito, R.; Misiewicz-Krzeminska, I.; Hernández-Sánchez, M.; de Coca, A.G.; Fisac, R.; Alonso, J.-M.; Zato, C.; de Paz, J.F.; et al. MicroRNA-223 Is a Novel Negative Regulator of HSP90B1 in CLL. BMC Cancer 2015, 15, 238. [Google Scholar] [CrossRef]
- Liu, T.-Y.; Chen, S.-U.; Kuo, S.-H.; Cheng, A.-L.; Lin, C.-W. E2A-Positive Gastric MALT Lymphoma Has Weaker Plasmacytoid Infiltrates and Stronger Expression of the Memory B-Cell-Associated miR-223: Possible Correlation with Stage and Treatment Response. Mod. Pathol. 2010, 23, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.; Dalla-Favera, R. Germinal Centres: Role in B-Cell Physiology and Malignancy. Nat. Rev. Immunol. 2008, 8, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Hernández, S.S.; Jakobsen, M.R.; Bak, R.O. Plasmacytoid Dendritic Cells as a Novel Cell-Based Cancer Immunotherapy. Int. J. Mol. Sci. 2022, 23, 11397. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Newman, S.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Kalinski, P.; Endo, I.; Takabe, K. Plasmacytoid Dendritic Cell (pDC) Infiltration Correlate with Tumor Infiltrating Lymphocytes, Cancer Immunity, and Better Survival in Triple Negative Breast Cancer (TNBC) More Strongly than Conventional Dendritic Cell (cDC). Cancers 2020, 12, 3342. [Google Scholar] [CrossRef] [PubMed]
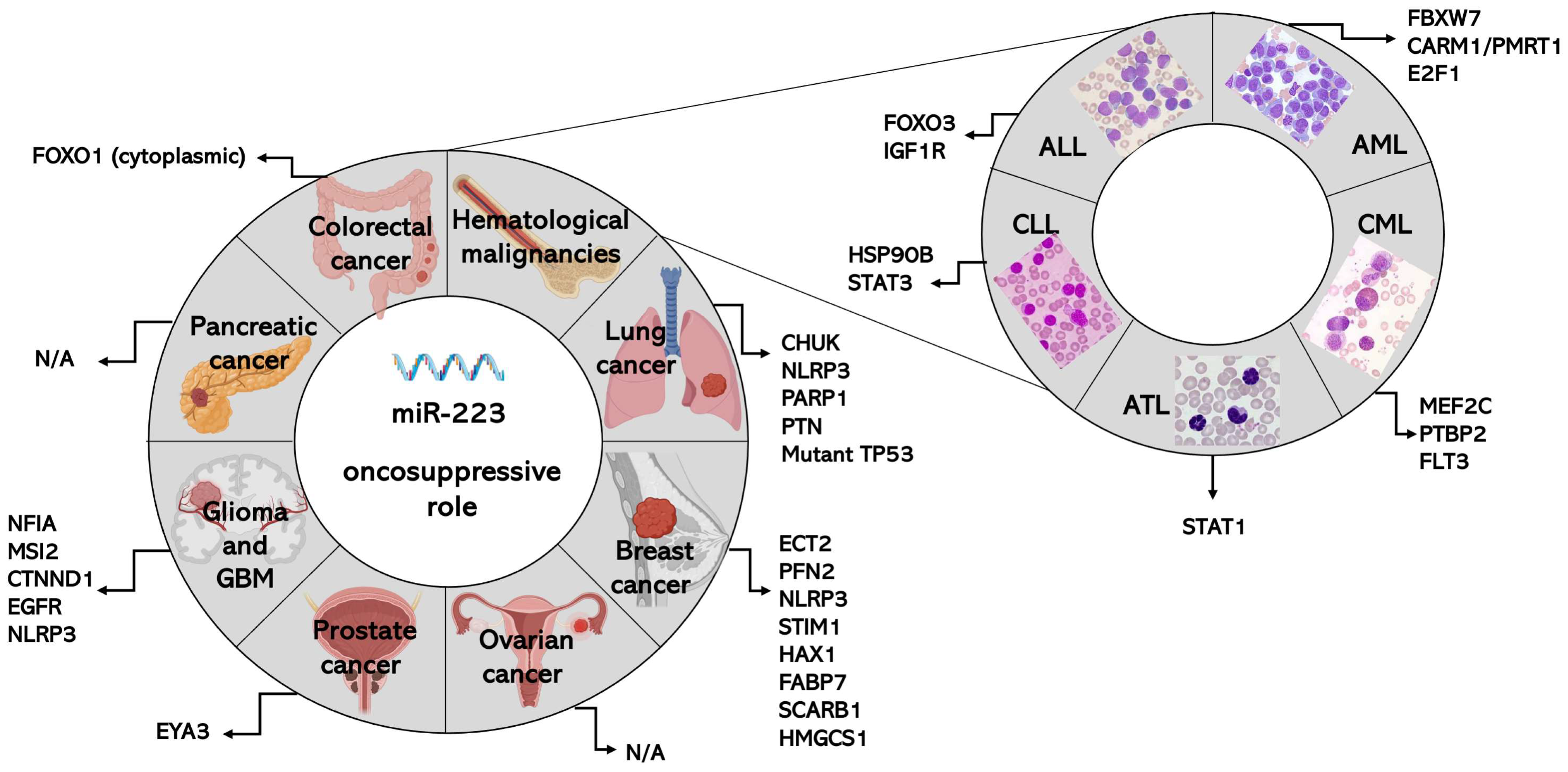
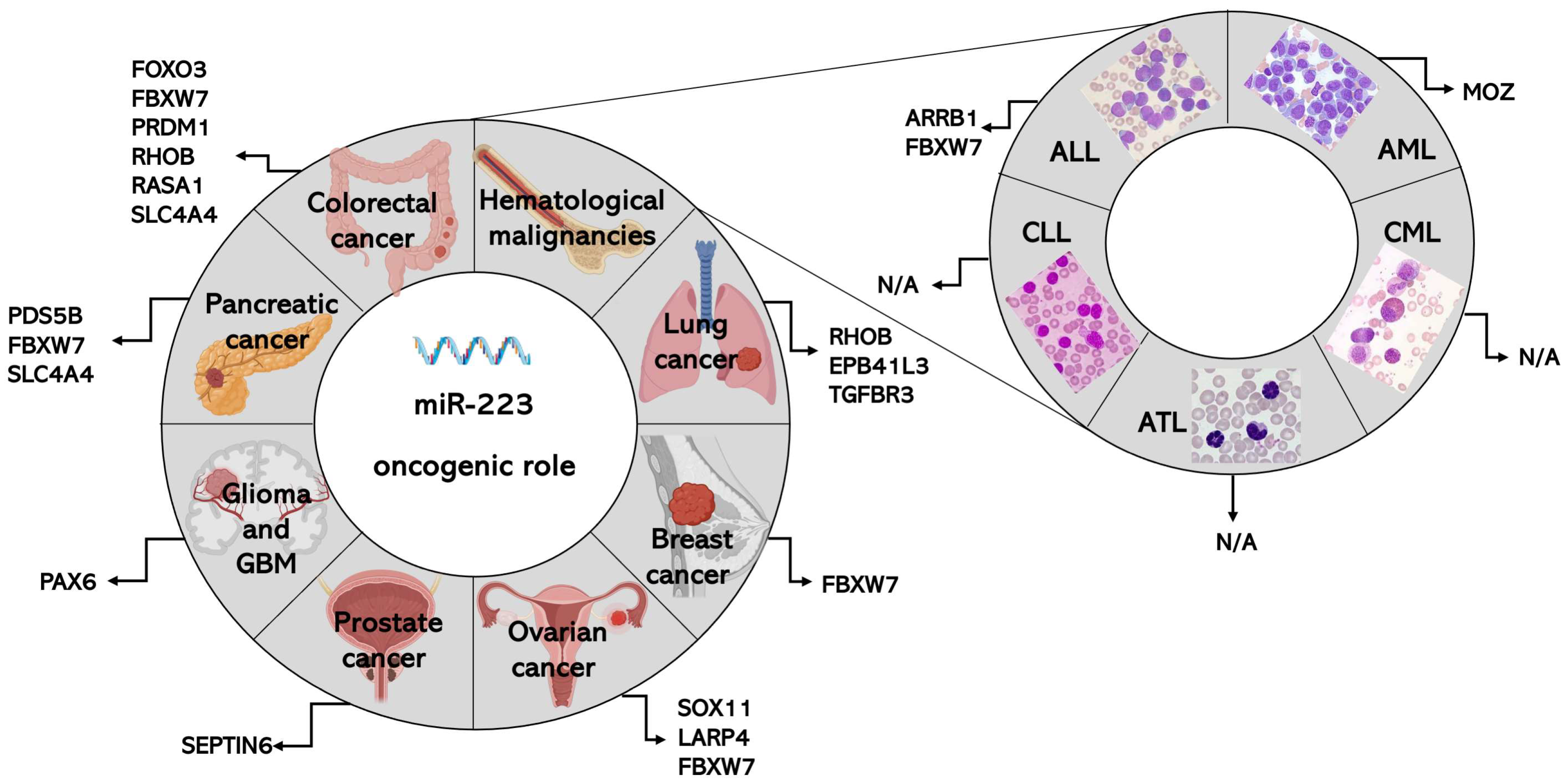
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbagallo, D.; Ponti, D.; Bassani, B.; Bruno, A.; Pulze, L.; Akkihal, S.A.; George-William, J.N.; Gundamaraju, R.; Campomenosi, P. MiR-223-3p in Cancer Development and Cancer Drug Resistance: Same Coin, Different Faces. Int. J. Mol. Sci. 2024, 25, 8191. https://doi.org/10.3390/ijms25158191
Barbagallo D, Ponti D, Bassani B, Bruno A, Pulze L, Akkihal SA, George-William JN, Gundamaraju R, Campomenosi P. MiR-223-3p in Cancer Development and Cancer Drug Resistance: Same Coin, Different Faces. International Journal of Molecular Sciences. 2024; 25(15):8191. https://doi.org/10.3390/ijms25158191
Chicago/Turabian StyleBarbagallo, Davide, Donatella Ponti, Barbara Bassani, Antonino Bruno, Laura Pulze, Shreya A. Akkihal, Jonahunnatha N. George-William, Rohit Gundamaraju, and Paola Campomenosi. 2024. "MiR-223-3p in Cancer Development and Cancer Drug Resistance: Same Coin, Different Faces" International Journal of Molecular Sciences 25, no. 15: 8191. https://doi.org/10.3390/ijms25158191
APA StyleBarbagallo, D., Ponti, D., Bassani, B., Bruno, A., Pulze, L., Akkihal, S. A., George-William, J. N., Gundamaraju, R., & Campomenosi, P. (2024). MiR-223-3p in Cancer Development and Cancer Drug Resistance: Same Coin, Different Faces. International Journal of Molecular Sciences, 25(15), 8191. https://doi.org/10.3390/ijms25158191









