Evaluation of a Novel Immunochromatographic Device for Detecting Porphyromonas gingivalis in Patients with Periodontal Disease
Abstract
1. Introduction
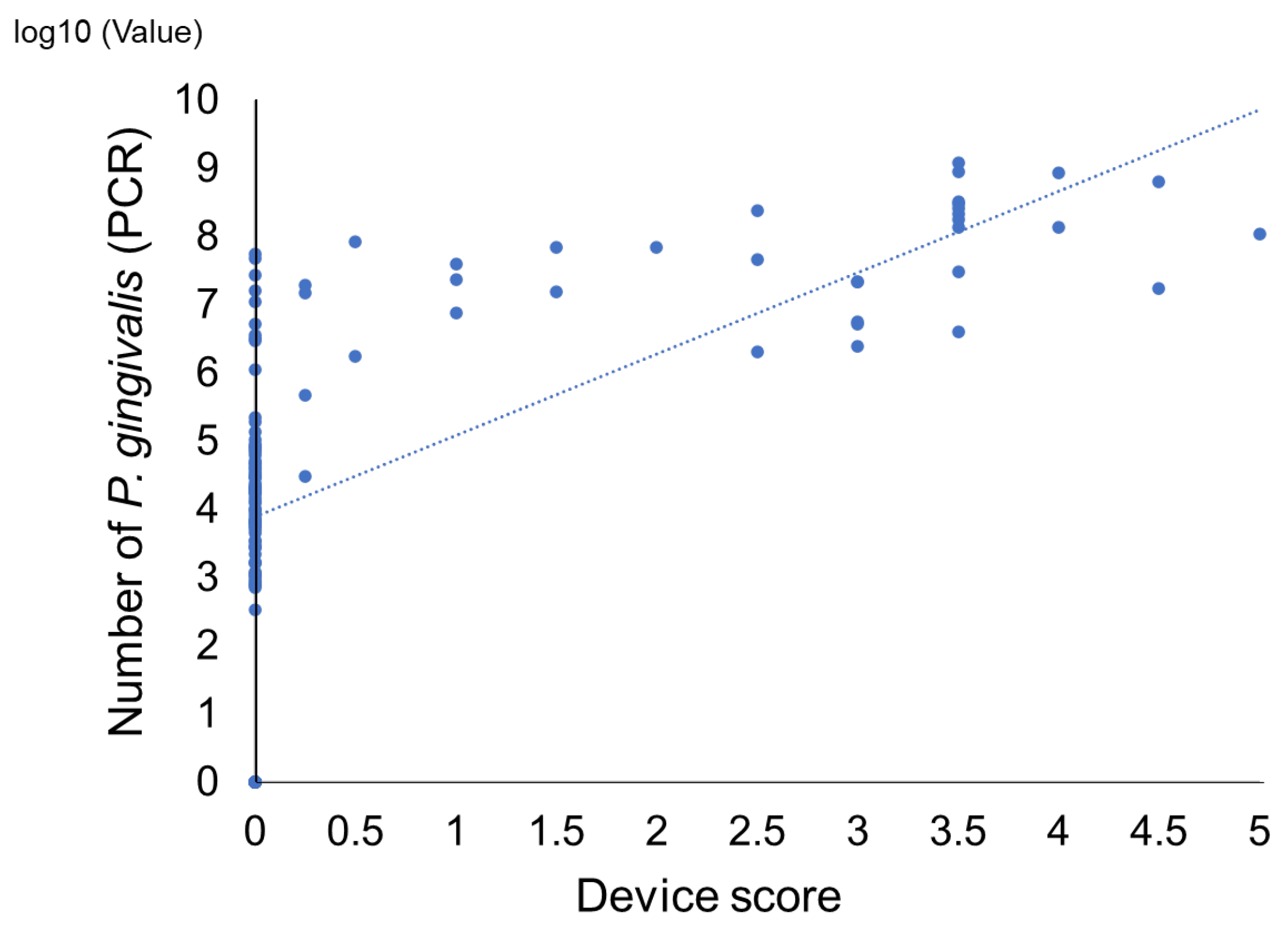
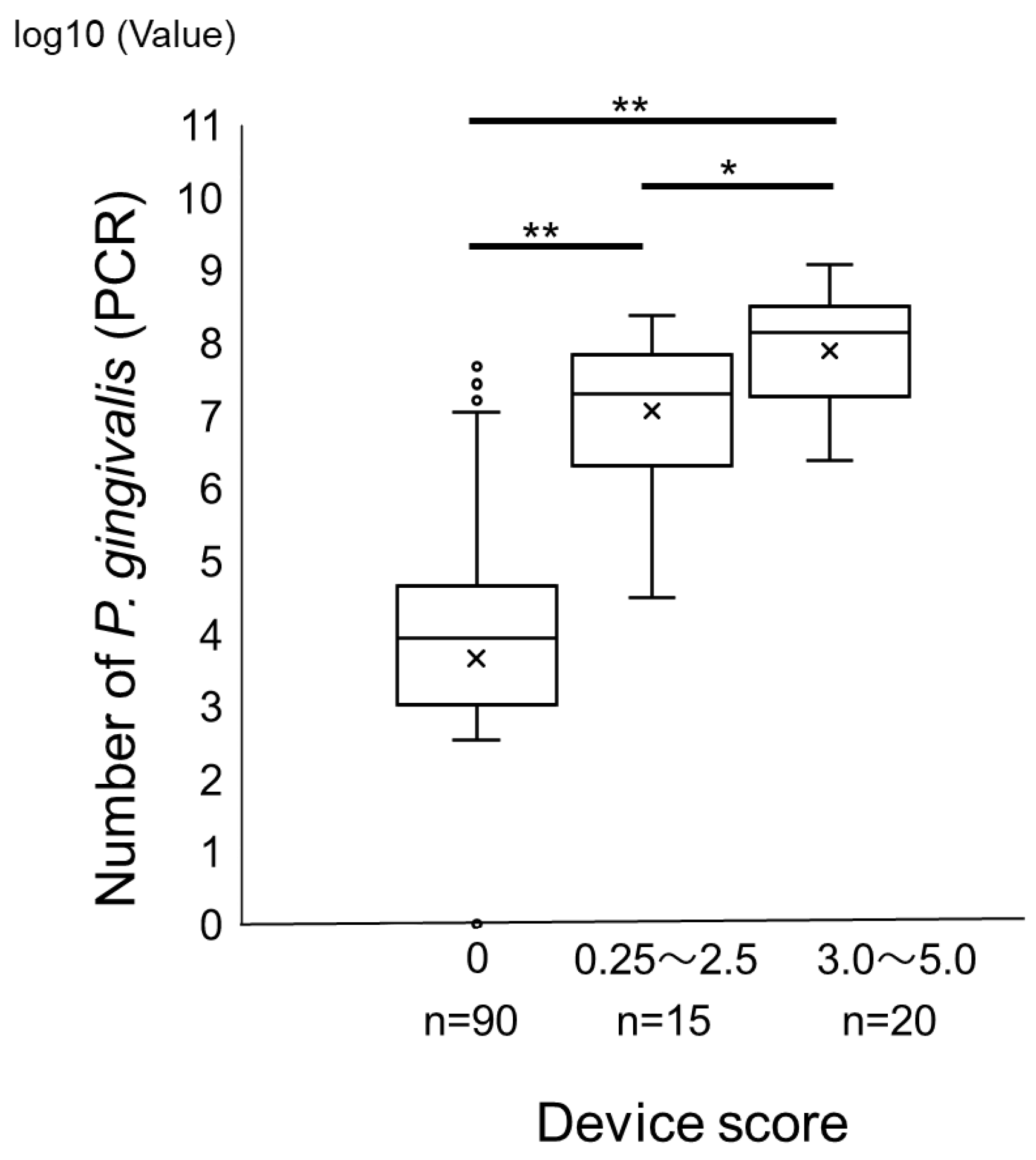
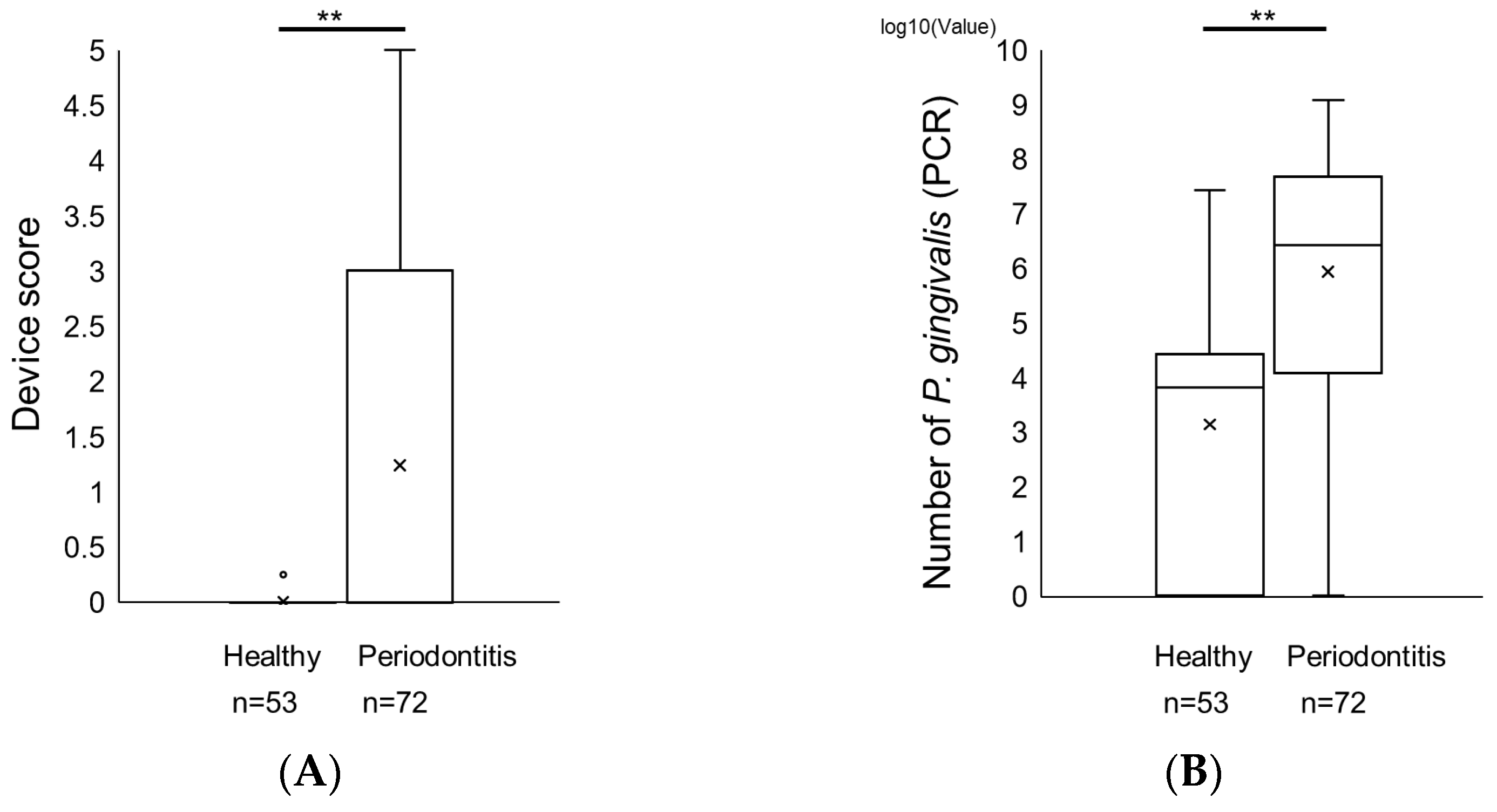
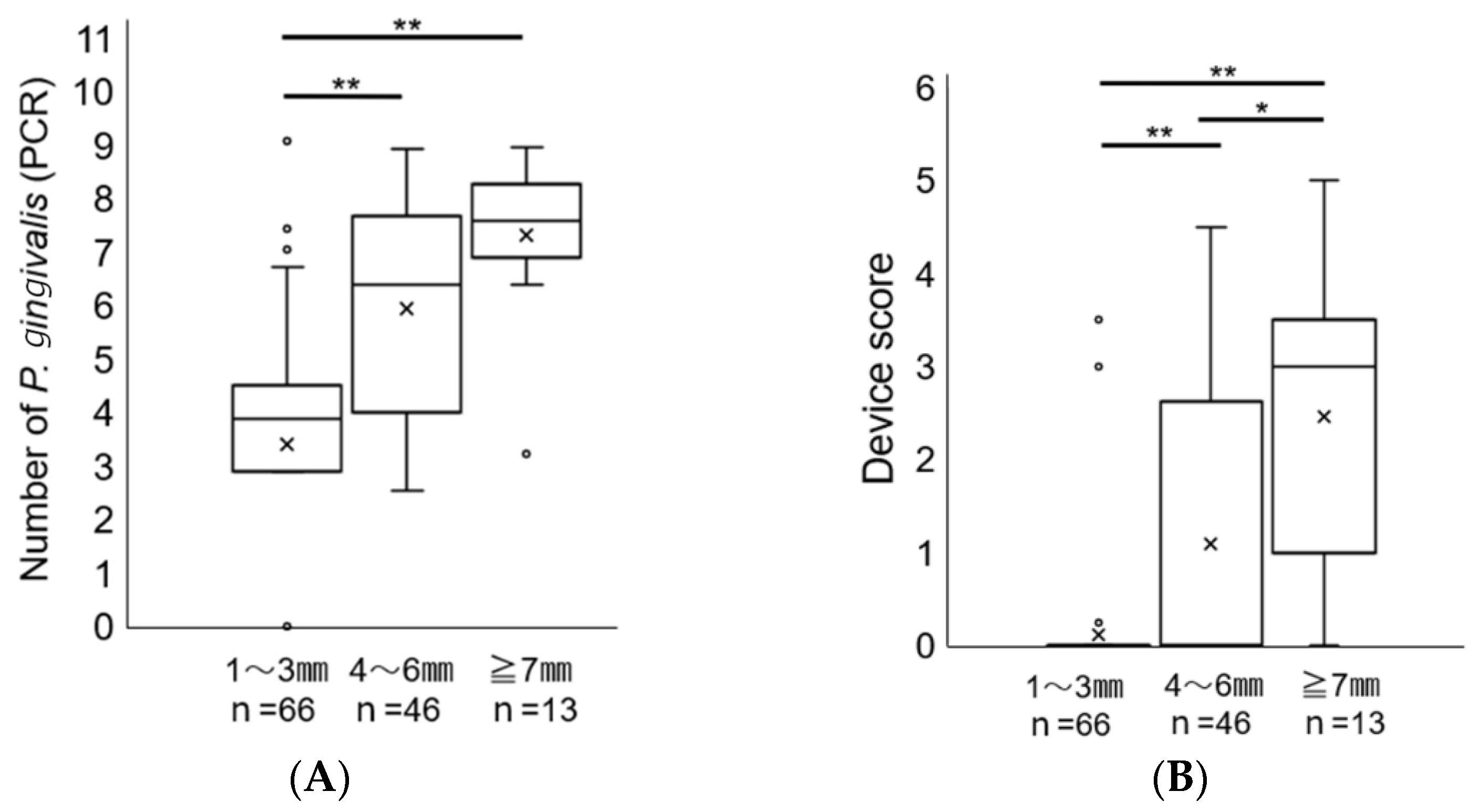
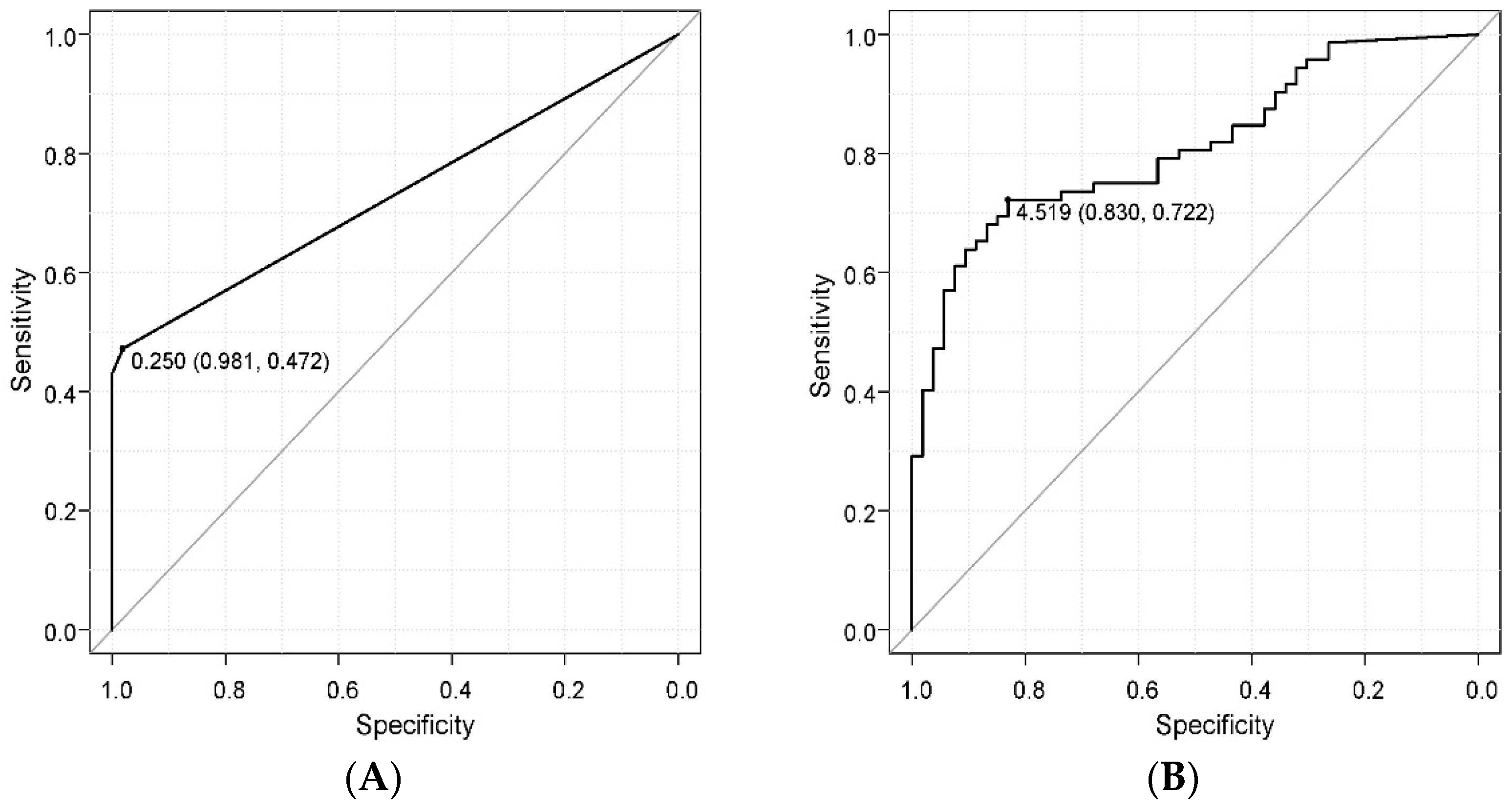
2. Results
Comparison of a Novel Immunochromatographic Device and Real-Time PCR Method in P. gingivalis Detection
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Subgingival Plaque Sampling
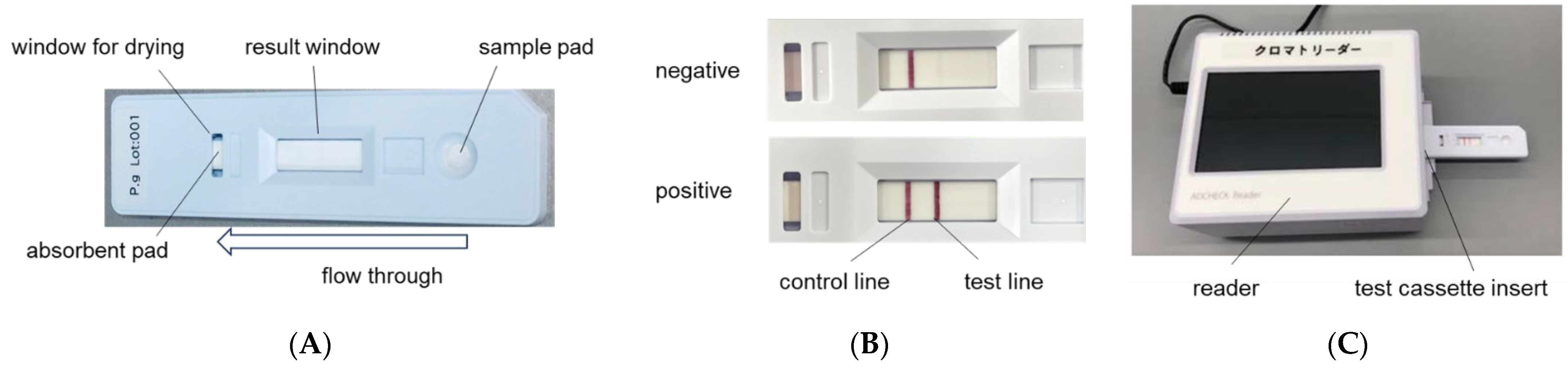
4.3. Immunochromatographic Device for P. gingivalis
4.4. Real-Time PCR Method
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Usui, M.; Onizuka, S.; Sato, T.; Kokabu, S.; Ariyoshi, W.; Nakashima, K. Mechanism of alveolar bone destruction in periodontitis—Periodontal bacteria and inflammation. Jpn. Dent. Sci. Rev. 2021, 57, 201–208. [Google Scholar] [CrossRef]
- Tsukasaki, M. Rankl and osteoimmunology in periodontitis. J. Bone Miner. Metab. 2021, 39, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Teles, R.P.; Haffajee, A.D.; Socransky, S.S. Microbiological goals of periodontal therapy. Periodontol. 2000 2006, 42, 180–218. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Sowa, M.G.; Iacopino, A.M.; Maev, R.G.; Hewko, M.D.; Man, A.; Liu, K.Z. An update on novel non-invasive approaches for periodontal diagnosis. J. Periodontol. 2010, 81, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Sixou, M. Diagnostic testing as a supportive measure of treatment strategy. Oral Dis. 2003, 9 (Suppl. S1), 54–62. [Google Scholar] [CrossRef] [PubMed]
- Shahi, S.; Zununi Vahed, S.; Fathi, N.; Sharifi, S. Polymerase chain reaction (PCR)-based methods: Promising molecular tools in dentistry. Int. J. Biol. Macromol. 2018, 117, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Eick, S.; Straube, A.; Guentsch, A.; Pfister, W.; Jentsch, H. Comparison of real-time polymerase chain reaction and DNA-strip technology in microbiological evaluation of periodontitis treatment. Diagn. Microbiol. Infect. Dis. 2011, 69, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Lopatin, D.E.; Giordano, J.; Alcoforado, G.; Hujoel, P. Comparison of the benzoyl-DL-arginine-naphthylamide (BANA) test, DNA probes, and immunological reagents for ability to detect anaerobic periodontal infections due to Porphyromonas gingivalis, Treponema denticola, and Bacteroides forsythus. J. Clin. Microbiol. 1992, 30, 427–433. [Google Scholar] [CrossRef]
- Usui, M.; Iwasaki, M.; Ariyoshi, W.; Kobayashi, K.; Kasai, S.; Yamanaka, R.; Nakashima, K.; Nishihara, T. The ability of a novel trypsin-lie peptidase activity assay kit to detect red-complex species. Diagnostics 2022, 12, 2172. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. The bacterial etiology of destructive periodontal disease: Current concepts. J. Periodontol. 1992, 63, 322–331. [Google Scholar] [CrossRef]
- Nuñez-Belmar, J.; Morales-Olavarria, M.; Vicencio, E.; Vernal, R.; Cárdenas, J.P.; Cortez, C. Contribution of -Omics technologies in the study of Porphyromonas gingivalis during periodontitis pathogenesis: A minireview. Int. J. Mol. Sci. 2022, 24, 620. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. Dental biofilms: Difficult therapeutic targets. Periodontol. 2000 2002, 28, 12–55. [Google Scholar] [CrossRef]
- Lunar Silva, I.; Cascales, E. Molecular strategies underlying Porphyromonas gingivalis virulence. J. Mol. Biol. 2021, 433, 166836. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Lamont, R.J. Breaking bad: Manipulation of the host response by Porphyromonas gingivalis. Eur. J. Immunol. 2014, 44, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.C.; Ebersole, J.L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 2005, 38, 72–122. [Google Scholar] [CrossRef] [PubMed]
- Mineoka, T.; Awano, S.; Rikimaru, T.; Kurata, H.; Yoshida, A.; Ansai, T.; Takehara, T. Site-specific development of periodontal disease is associated with increased levels of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia in subgingival plaque. J. Periodontol. 2008, 79, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Fujise, O.; Hamachi, T.; Inoue, K.; Miura, M.; Maeda, K. Microbiological markers for prediction and assessment of treatment outcome following non-surgical periodontal therapy. J. Periodontol. 2002, 73, 1253–1259. [Google Scholar] [CrossRef]
- Tomita, S.; Komiya-Ito, A.; Imamura, K.; Kita, D.; Ota, K.; Takayama, S.; Makino-Oi, A.; Kinumatsu, T.; Ota, M.; Saito, A. Prevalence of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia in Japanese patients with generalized chronic and aggressive periodontitis. Microb. Pathog. 2013, 61, 11–15. [Google Scholar] [CrossRef]
- Beck, J.D.; Papapanou, P.N.; Philips, K.H.; Offenbacher, S. Periodontal medicine: 100 years of progress. J. Dent. Res. 2019, 98, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Chavakis, T. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat. Rev. Immunol. 2021, 21, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Kapila, Y.L. Oral Health’s inextricable connection to systemic health: Special populations bring to bear multimodal relationships and factors connecting periodontal disease to systemic diseases and conditions. Periodontol. 2000 2021, 87, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Inoue, M.; Usui, M.; Ariyoshi, W.; Nakashima, K.; Nagai-Yoshioka, Y.; Nishihara, T. The association between trypsin-like protease activity in the oral cavity and kidney function in Japanese workers. J. Clin. Periodontol. 2024, 51, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Dominy, S.S.; Lynch, C.; Ermini, F.; Benedyk, M.; Marczyk, A.; Konradi, A.; Nguyen, M.; Haditsch, U.; Raha, D.; Griffin, C.; et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 2019, 5, eaau3333. [Google Scholar] [CrossRef]
- Wu, Z.; Ni, J.; Liu, Y.; Teeling, J.L.; Takayama, F.; Collcutt, A.; Ibbett, P.; Nakanishi, H. Cathepsin B plays a critical role in inducing Alzheimer’s disease-like phenotypes following chronic systemic exposure to lipopolysaccharide from Porphyromonas gingivalis in mice. Brain Behav. Immun. 2017, 65, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Nonaka, S.; Wu, Z. Microglial cathepsin B and Porphyromonas gingivalis gingipains as potential therapeutic targets for sporadic Alzheimer’s disease. CNS Neurol. Disord. Drug Targets 2020, 19, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Gabarrini, G.; Grasso, S.; van Winkelhoff, A.J.; van Dijl, J.M. Gingimaps: Protein localization in the oral pathogen Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 2020, 84, e00032-19. [Google Scholar] [CrossRef]
- Ahmadi, P.; Mahmoudi, M.; Kheder, R.K.; Faraj, T.A.; Mollazadeh, S.; Abdulabbas, H.S.; Esmaeili, S.A. Impacts of Porphyromonas gingivalis periodontitis on rheumatoid arthritis autoimmunity. Int. Immunopharmacol. 2023, 118, 109936. [Google Scholar] [CrossRef]
- Li, Y.; Guo, R.; Oduro, P.K.; Sun, T.; Chen, H.; Yi, Y.; Zeng, W.; Wang, Q.; Leng, L.; Yang, L.; et al. The relationship between Porphyromonas gingivalis and rheumatoid arthritis: A meta-analysis. Front. Cell. Infect. Microbiol. 2022, 12, 956417. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89 (Suppl. S1), S159–S172. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Takeuchi, Y.; Umeda, M.; Ishikawa, I.; Benno, Y. Rapid Detection and Quantification of Five Periodontopathic Bacteria by Real-Time PCR. Microbiol. Immunol. 2001, 45, 39–44. [Google Scholar] [CrossRef] [PubMed]
| Comparison of the Novel Immunochromatographic Device and Real-Time PCR (Real-Time PCR) in Subgingival Plaque Samples | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| real-time PCR | sensitivity | specificity | PPV | NPV | accuracy | ||||
| POS | NEG | TOTAL | 77% | 98% | 94% | 89% | 90% | ||
| TEST DEVICE | POS | 33 | 2 | 35 | |||||
| NEG | 10 | 80 | 90 | ||||||
| TOTAL | 43 | 82 | 88 | ||||||
| X2 (p value) | p < 0.01 | ||||||||
| Parameters | Device Score (Test Device) | Number of P. gingivalis (Real-Time PCR) |
|---|---|---|
| PPD (mm) | 0.612 | 0.594 |
| CAL (mm) | 0.605 | 0.606 |
| BOP | 0.403 | 0.365 |
| Healthy Subjects | Patients with Periodontitis | Difference | |
|---|---|---|---|
| Number Male Female | n = 53 23 30 | n = 72 36 36 | |
| Age (average) | 47.51 ± 21.10 | 69.81 ± 11.82 | p < 0.01 |
| PPD (mm) | 2.32 ± 0.51 | 4.96 ± 1.54 | p < 0.01 |
| CAL (mm) | 0.74 ± 1.07 | 5.60 ± 1.97 | p < 0.01 |
| BOP | 0.06 ± 0.23 | 0.40 ± 0.49 | p < 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamanaka, R.; Usui, M.; Kobayashi, K.; Onizuka, S.; Kasai, S.; Sano, K.; Hironaka, S.; Yamasaki, R.; Yoshii, S.; Sato, T.; et al. Evaluation of a Novel Immunochromatographic Device for Detecting Porphyromonas gingivalis in Patients with Periodontal Disease. Int. J. Mol. Sci. 2024, 25, 8187. https://doi.org/10.3390/ijms25158187
Yamanaka R, Usui M, Kobayashi K, Onizuka S, Kasai S, Sano K, Hironaka S, Yamasaki R, Yoshii S, Sato T, et al. Evaluation of a Novel Immunochromatographic Device for Detecting Porphyromonas gingivalis in Patients with Periodontal Disease. International Journal of Molecular Sciences. 2024; 25(15):8187. https://doi.org/10.3390/ijms25158187
Chicago/Turabian StyleYamanaka, Rieko, Michihiko Usui, Kaoru Kobayashi, Satoru Onizuka, Shingo Kasai, Kotaro Sano, Shou Hironaka, Ryota Yamasaki, Shinji Yoshii, Tsuyoshi Sato, and et al. 2024. "Evaluation of a Novel Immunochromatographic Device for Detecting Porphyromonas gingivalis in Patients with Periodontal Disease" International Journal of Molecular Sciences 25, no. 15: 8187. https://doi.org/10.3390/ijms25158187
APA StyleYamanaka, R., Usui, M., Kobayashi, K., Onizuka, S., Kasai, S., Sano, K., Hironaka, S., Yamasaki, R., Yoshii, S., Sato, T., Fujii, W., Iwasaki, M., Ariyoshi, W., Nakashima, K., & Nishihara, T. (2024). Evaluation of a Novel Immunochromatographic Device for Detecting Porphyromonas gingivalis in Patients with Periodontal Disease. International Journal of Molecular Sciences, 25(15), 8187. https://doi.org/10.3390/ijms25158187







