Revisiting Epigenetics Fundamentals and Its Biomedical Implications
Abstract
1. Ancestry of Epigenetic Concept
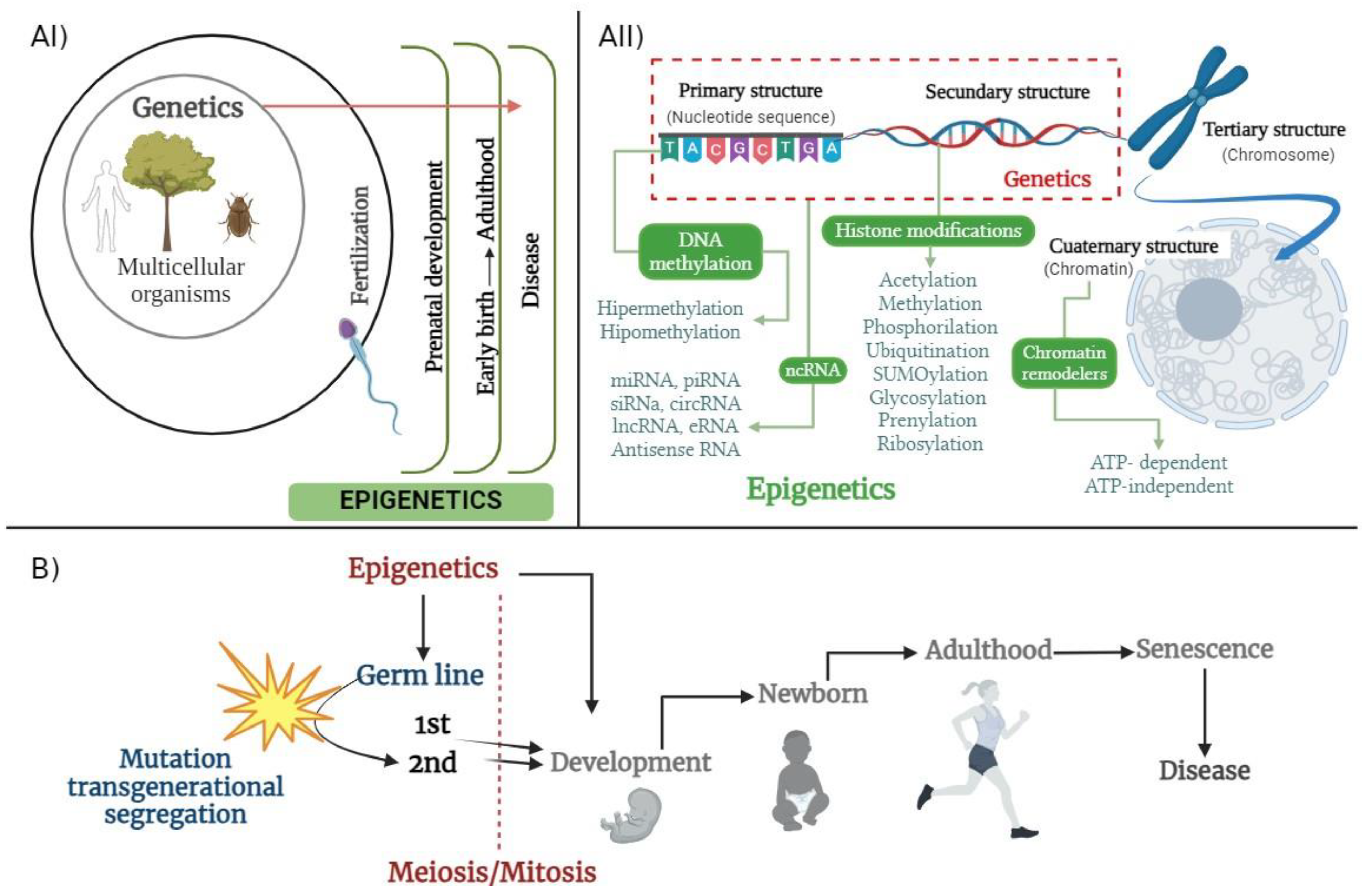
2. Contemporary Epigenetic Scopes
…“The work has quite correctly taken on a “Lamarckian” flavour and is quite frankly another milestone in the paradigm shift now underway in molecular and cell biology, which is now impacting on a clearly observable trait, regulation of coat colour”….[39]
- (A)
- As described earlier, Conrad Waddington was the first to describe the concept of epigenetics in the early 1940s, as “the branch of biology, which studies the causal interactions between genes and their products which bring the phenotype into being” [31].
- (B)
- Robin Holliday stated in 1990 that there is temporal and spatial control of gene activity during the development of multicellular organisms. From this point of view, whether mitotically and/or meiotically, heritable diverse gene functions, without a specific origin, are not related to changes in the DNA sequence [37,38].
- (C)
- Bartolomei in 1991, found diverted expression (imprinting) although DNA sequence remains constant in mice [40].
- (D)
- Epigenetic modification occurs from ncRNAs [41].
- (E)
- Through its mechanism, generally, epigenetics is determined by modifications in gene expression independent from the DNA sequence: “the study of changes in gene function that are mitotically and/or meiotically heritable, and that do not entail a change in DNA sequence” [41].
- (F)
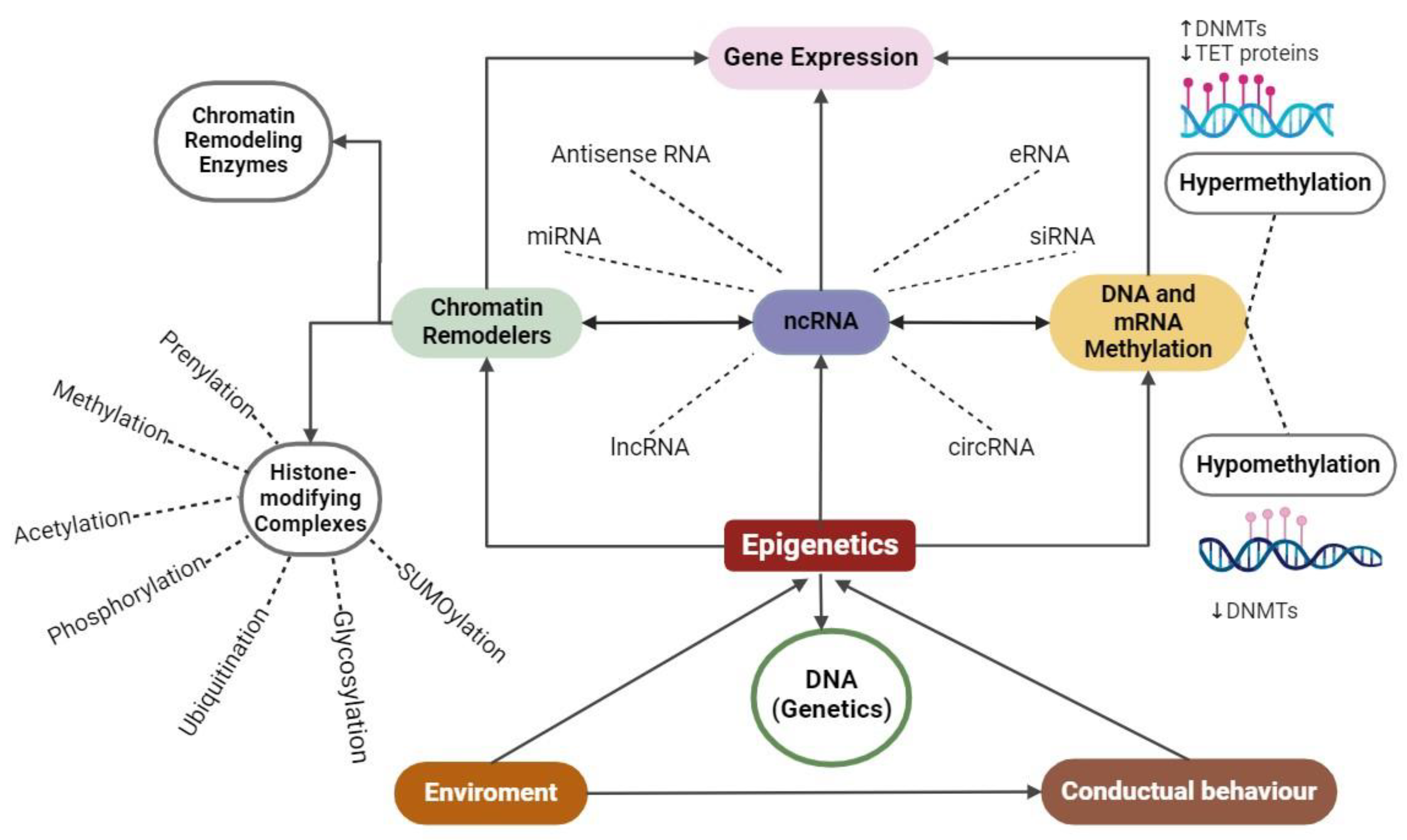
3. Epigenetic Biomolecular Mechanisms
3.1. Methylation
3.2. Histone Modifications
3.3. Interference RNA in Epigenetics
3.4. Antisense Transcripts
3.5. Special Cases: Piwi-Interacting RNAs (piRNAs)
3.6. Small Interference RNA
3.7. Circular RNA
3.8. eRNA
4. Epigenetics and Disease
4.1. Chronic Inflammatory Diseases
4.2. Infectious Diseases
4.3. Developmental Diseases
5. Epigenetics and Environment
6. Integrative Omics Studies
7. Conclusions and Discussion
Future Research Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johannsen, W. Elemente der Exakten Erblichkeitslehre; Cambridge University Press: Cambridge, UK, 1909. [Google Scholar] [CrossRef]
- Johannsen, W. The genotype conception of heredity. Am. Nat. 1911, 45, 129–159. [Google Scholar] [CrossRef]
- Johannsen, W. Some remarks about units in heredity. Hereditas 2010, 4, 133–141. [Google Scholar] [CrossRef]
- Sutton, W.S. The chromosomes in heredity. Biol. Bull. 1903, 4, 231–250. [Google Scholar] [CrossRef]
- Boveri, T.H. Ergebnisse über die Konstitution der Chromatischen Substanz des Zelkerns; Gustav Fischer Jena: Würzburg, Germany, 1904. [Google Scholar] [CrossRef]
- Bjornsson, H.T. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015, 25, 1473–1481. [Google Scholar] [CrossRef]
- Burbano, H.A. Epigenetics and genetic determinism. História Ciências Saúde—Manguinhos 2006, 13, 851–863. [Google Scholar] [CrossRef][Green Version]
- Van Speybroeck, L.; De Waele, D.; Van de Vijver, G. Theories in Early Embryology. Ann. N. Y. Acad. Sci. 2002, 981, 7–49. [Google Scholar] [CrossRef] [PubMed]
- Carver, R.B.; Castéra, J.; Gericke, N.; Evangelista, N.A.M.; El-Hani, C.N. Young adults’ belief in genetic determinism, and knowledge and attitudes towards modern genetics and genomics: The PUGGS Questionnaire. PLoS ONE 2017, 12, e0169808. [Google Scholar] [CrossRef] [PubMed]
- Aristotle. Generation of Animals; Barnes, J., Ed.; Princeton Univ.: Princeton, NJ, USA, 1995; Volume 1, pp. 1111–1218. [Google Scholar]
- Aristotle. On the Generation of Animals; Platt, A., Translator; University of Adelaide: Adelaide, Australia, 2007; Available online: http://ebooks.adelaide.edu.au/a/aristotle/generation/complete.html (accessed on 22 April 2024).
- Pasipoularides, A. Some Notable Pioneers. In Heart’s Vortex: Intracardiac Blood Flow Phenomena; People’s Medical Publishing House: Shelton, CT, USA, 2010; pp. 115–164. [Google Scholar]
- Focher, F. The logic of life in Maupertuis: From Newtonian attraction to psychobiological determinism. DOAJ 2020, 12, 1–14. [Google Scholar] [CrossRef]
- Glass, H.B. Maupertuis, a Forgotten Genius. Sci. Am. 1955, 193, 100–111. [Google Scholar] [CrossRef]
- Buffon, G.L.L.; Daubenton, D.M.; Liard Philibert, G.; CéPèDe, L.; De SèVe, J.; De SèVe, J.E.; Buvé, E.; Panckoucke, C.J.; Plassan, P. Histoire Naturelle, Générale et Particulière; À Paris De l’Imprimerie royale MDCCXLIX: Paris, France, 1756; p. 6. Available online: https://www.biodiversitylibrary.org/item/128362 (accessed on 22 February 2024).
- Needham, J. A History of Embriology; Abelard-Schuman: New York, NY, USA, 1959. [Google Scholar]
- Aulie, R.P. Caspar Friedrich Wolff and his “Theoria Generationis”, 1759. J. Hist. Med. Allied Sci. 1961, 16, 124–144. [Google Scholar] [CrossRef]
- Goodey, C.F. The Coining of a Developmental Theory: Leibniz to Bonnet. In Development: The History of a Psychological Concept; Cambridge University Press: Cambridge, UK, 2021; pp. 137–165. ISBN 9781108833479. [Google Scholar]
- Cohen, A.A. Kant on epigenesis, monogenesis and human nature: The biological premises of anthropology. Stud. Hist. Philos. Sci. Part C 2016, 37, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Lamarck, J.B. Philosophie Zoologique V2: Ou Exposition (1809) (French Edition); Kessinger Publishing: Whitefish, MT, USA, 2010. [Google Scholar]
- Driesch, H.; Ogden, C.K. The History & Theory of Vitalism; Macmillan and Co.: London, UK, 1941. [Google Scholar] [CrossRef]
- Darwin, C, Sir Wyville Thomson and natural selection. Nature 1880, 23, 32. [CrossRef]
- Darwin, C.R. The Variation of Animals and Plants under Domestication, 2nd ed.; John Murray: London, UK, 1875; Volume 1. [Google Scholar]
- Winston, M.L. The Biology of the Honey Bee; Harvard University Press: Cambridge, MA, USA, 1987; ISBN 9780674074095. [Google Scholar]
- Tian, L.; Hines, H.M. Morphological characterization and staging of Bumble bee pupae. PeerJ 2018, 6, e6089. [Google Scholar] [CrossRef] [PubMed]
- Simola, D.F.; Graham, R.J.; Brady, C.M.; Enzmann, B.L.; Desplan, C.; Ray, A.; Zwiebel, L.J.; Bonasio, R.; Reinberg, D.; Liebig, J.; et al. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science 2016, 351, aac6633. [Google Scholar] [CrossRef] [PubMed]
- Kováč, L. LaMarck and Darwin revisited. EMBO Rep. 2019, 20, e47922. [Google Scholar] [CrossRef] [PubMed]
- Dupras, C.; Saulnier, K.M.; Joly, Y. Epigenetics, ethics, law and society: A multidisciplinary review of descriptive, instru-mental, dialectical and reflexive analyses. Soc. Stud. Sci. 2019, 49, 785–810. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef]
- Waddington, C.H. An Introduction to Modern Genetics; George Allen and Unwin Ltd.: London, UK; The Macmillan Company: New York, NY, USA, 1939. [Google Scholar] [CrossRef]
- Waddington, C.H. The epigenotype. Int. J. Epidemiol. 1942, 41, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.F. Induction and the Origins of Developmental Genetics. In A Conceptual History of Modern Embryology; Springer: Boston, MA, USA, 1991; Volume 7, pp. 181–206. [Google Scholar] [CrossRef]
- Griffith, F. The Significance of Pneumococcal Types. J. Hyg. 1928, 27, 113–159. [Google Scholar] [CrossRef]
- Watson, J.; Crick, F. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Nanney, D.L. Epigenetic control systems. Proc. Natl. Acad. Sci. USA 1958, 44, 712–717. [Google Scholar] [CrossRef]
- Griffiths, B.B.; Hunter, R.G. Neuroepigenetics of stress. Neuroscience 2014, 275, 420–435. [Google Scholar] [CrossRef]
- Holliday, R. The inheritance of epigenetic defects. Science 1987, 238, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Balter, M. Was Lamarck Just a Little Bit Right? Science 2000, 288, 38. [Google Scholar] [CrossRef] [PubMed]
- Russo, V.; Martienssen, R.A.; Riggs, A.D. Epigenetic Mechanisms of Gene Regulation; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1996; Available online: http://ci.nii.ac.jp/ncid/BA29742198 (accessed on 14 May 2024).
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef]
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2013, 9, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Morris, J.R. Genes, Genetics, and Epigenetics: A Correspondence. Science 2001, 293, 1103–1105. [Google Scholar] [CrossRef]
- Van Otterdijk, S.D.; Michels, K.B. Transgenerational epigenetic inheritance in mammals: How good is the evidence? FASEB J. 2016, 30, 2457–2465. [Google Scholar] [CrossRef]
- Pang, T.Y.; Short, A.K.; Bredy, T.W.; Hannan, A.J. Transgenerational paternal transmission of acquired traits: Stress-induced modification of the sperm regulatory transcriptome and offspring phenotypes. Curr. Opin. Behav. Sci. 2017, 14, 140–147. [Google Scholar] [CrossRef]
- Rodgers, A.B.; Morgan, C.P.; Bronson, S.L.; Revello, S.D.; Bale, T.L. Paternal Stress Exposure Alters Sperm MicroRNA Content and Reprograms Offspring HPA Stress Axis Regulation. J. Neurosci. 2013, 33, 9003–9012. [Google Scholar] [CrossRef]
- Gapp, K.; Von Ziegler, L.; Tweedie-Cullen, R.Y.; Mansuy, I.M. Early life epigenetic programming and transmission of stress-induced traits in mammals. BioEssays 2014, 36, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Yeshurun, S.; Hannan, A.J. Transgenerational epigenetic influences of paternal environmental exposures on brain function and predisposition to psychiatric disorders. Mol. Psychiatry 2018, 24, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Bakusic, J.; Schaufeli, W.B.; Claes, S.; Godderis, L. Stress, burnout and depression: A systematic review on DNA methylation mechanisms. J. Psychosom. Res. 2017, 92, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Houri-Zeevi, L.; Rechavi, O. A Matter of Time: Small RNAs Regulate the Duration of Epigenetic Inheritance. Trends Genet. 2017, 33, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. What is an epigenetic transgenerational phenotype? Reprod. Toxicol. 2008, 25, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Jackson, F.; Niculescu, M.D.; Jackson, R.T. Conceptual Shifts Needed to Understand the Dynamic Interactions of Genes, Environment, Epigenetics, Social Processes, and Behavioral Choices. Am. J. Public Health 2013, 103 (Suppl. S1), S33–S42. [Google Scholar] [CrossRef] [PubMed]
- Flores, K.; Wolschin, F.; Amdam, G.V. The role of methylation of DNA in environmental adaptation. Integr. Comp. Biol. 2013, 53, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J. Multi-omics integration Strategies for animal Epigenetic studies. Anim. Biosci. 2021, 34, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Costello, J. DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 2017, 49, e322. [Google Scholar] [CrossRef]
- Melamed, P.; Yosefzon, Y.; David, C.; Tsukerman, A.; Pnueli, L. TET Enzymes, variants, and differential effects on function. Front. Cell Dev. Biol. 2018, 6, 22. [Google Scholar] [CrossRef]
- Slesarev, A.I.; Belova, G.I.; Kozyavkin, S.A.; Lake, J.A. Evidence for an early prokaryotic origin of histones H2A and H4 prior to the emergence of eukaryotes. Nucleic Acids Res. 1998, 26, 427–430. [Google Scholar] [CrossRef] [PubMed]
- McGinty, R.K.; Tan, S. Nucleosome structure and function. Chem. Rev. 2014, 115, 2255–2273. [Google Scholar] [CrossRef]
- Chen, Z.; Li, S.; Subramaniam, S.; Shyy, J.Y.; Chien, S. Epigenetic regulation: A new frontier for biomedical engineers. Annu. Rev. Biomed. Eng. 2017, 19, 195–219. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Rothenberg, M.E.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The human transcription factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Xiong, H.; Fang, J. Natural antisense transcripts regulate gene expression in an epigenetic manner. Biochem. Biophys. Res. Commun. 2010, 396, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.; Fried, M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3′ ends. Nature 1986, 322, 275–279. [Google Scholar] [CrossRef]
- Goodrich, J.A.; Kugel, J.F. Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell Biol. 2006, 7, 612–616. [Google Scholar] [CrossRef]
- He, Y.; Vogelstein, B.; Velculescu, V.E.; Papadopoulos, N.; Kinzler, K.W. The Antisense Transcriptomes of Human Cells. Science 2008, 322, 1855–1857. [Google Scholar] [CrossRef]
- Huang, S.; Yoshitake, K.; Asakawa, S. A Review of Discovery Profiling of PIWI-Interacting RNAs and Their Diverse Functions in Metazoans. Int. J. Mol. Sci. 2021, 22, 11166. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P. The long and short of RNAs. Nature 2009, 457, 974–975. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, Y.; Cao, X.; Tan, W.; Yu, J.; Lü, Y.; Kang, R.; Wang, X.; Li, E. The epigenetic regulatory mechanism of PIWI/piRNAs in human cancers. Mol. Cancer 2023, 22, 45. [Google Scholar] [CrossRef] [PubMed]
- Özata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2018, 20, 89–108. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Kang, L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics 2011, 27, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Verdel, A.; Vavasseur, A.; Le Gorrec, M.; Touat-Todeschini, L. Common themes in siRNA-mediated epigenetic silencing pathways. Int. J. Dev. Biol. 2009, 53, 245–257. [Google Scholar] [CrossRef]
- Hu, X.; Xiao, T.; Chen, L.; Huang, F. Dynamic supramolecular complexes constructed by Orthogonal Self-Assembly. Acc. Chem. Res. 2014, 47, 2041–2051. [Google Scholar] [CrossRef]
- Zeng, X.; Yuan, X.; Cai, Q.; Tang, C.; Gao, J. Circular RNA as an epigenetic regulator in chronic liver diseases. Cells 2021, 10, 1945. [Google Scholar] [CrossRef]
- Gu, A.; Jaijyan, D.K.; Yang, S.; Zeng, M.; Pei, S.; Zhu, H. Functions of circular RNA in human diseases and illnesses. Non-Coding RNA 2023, 9, 38. [Google Scholar] [CrossRef]
- Rahnamoun, H.; Orozco, P.; Lauberth, S. The role of enhancer RNAs in epigenetic regulation of gene expression. Transcription 2019, 11, 19–25. [Google Scholar] [CrossRef]
- Kim, Y.; Xie, P.; Cao, L.F.; Zhang, M.Q.; Kim, T.H. Global transcriptional activity dynamics reveal functional enhancer RNAs. Genome Res. 2018, 28, 1799–1811. [Google Scholar] [CrossRef] [PubMed]
- Bose, D.; Donahue, G.; Reinberg, D.; Shiekhattar, R.; Bonasio, R.; Berger, S.L. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 2017, 168, 135–149.e22. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Cao, C.; Xue, Y. Enhancer RNA: Biogenesis, function, and regulation. Essays Biochem. 2020, 64, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Veneziano, D.; Nigita, G.; Nana-Sinkam, S.P. RNA Methylation in ncRNA: Classes, Detection, and Molecular Associations. Front. Genet. 2018, 9, 243. [Google Scholar] [CrossRef] [PubMed]
- Beck, D.; Ben Maamar, M.; Skinner, M.K. Integration of sperm ncRNA-directed DNA methylation and DNA methylation-directed histone retention in epigenetic transgenerational inheritance. Epigenetics Chromatin 2021, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, C.; Liu, W.; Zhang, J.; Ohgi, K.A.; Grinstein, J.D.; Dorrestein, P.C.; Rosenfeld, M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2013, 147, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Keller, C.; Bühler, M. Chromatin-associated ncRNA activities. Chromosome Res. 2013, 21, 627–641. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ho, L.; Crabtree, G. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, Q.; Chang, C. Epigenetics in health and disease. In Epigenetics in Allergy and Autoimmunity; Springer: Singapure, 2020; Volume 1253, pp. 3–55. [Google Scholar] [CrossRef]
- Haghani, A.; Li, C.Z.; Robeck, T.R.; Zhang, J.; Lu, A.T.; Ablaeva, J.; Acosta-Rodríguez, V.A.; Adams, D.M.; Alagaili, A.N.; Almunia, J.; et al. DNA methylation networks underlying mammalian traits. Science 2023, 381, 1–15. [Google Scholar] [CrossRef]
- Stylianou, E. Epigenetics of chronic inflammatory diseases. J. Inflamm. Res. 2018, 12, 1–14. [Google Scholar] [CrossRef]
- Lacagnina, S. The Developmental Origins of Health and Disease (DOHAD). Am. J. Lifestyle Med. 2019, 14, 47–50. [Google Scholar] [CrossRef]
- Maruyama, R.; Choudhury, S.; Kowalczyk, A.; Bessarabova, M.; Beresford-Smith, B.; Conway, T.; Kaspi, A.; Wu, Z.J.; Nikolskaya, T.; Merino, V.F.; et al. Epigenetic regulation of cell Type–Specific expression patterns in the human mammary epithelium. PLOS Genet. 2011, 7, e1001369. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.R.; Arumugam, T.; Chuturgoon, A.A.; An, P.; Ramsuran, V. Editorial: Epigenetics of Infectious Diseases. Front. Immunol. 2022, 13, 1054151. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, R.; Zwergel, C.; Artico, M.; Taurone, S.; Ralli, M.; Greco, A.; Mai, A. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenetics 2019, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Sethi, G. Role of Epigenetics in Inflammation-Associated diseases. In Epigenetics: Development and Disease; Sub-cellular biochemistry; Springer: Dordrecht, The Netherlands, 2012; Volume 61, pp. 627–657. [Google Scholar] [CrossRef]
- Netea, M.G.; Joosten, L.A.B.; Latz, E.; Mills, K.H.G.; Natoli, G.; Stunnenberg, H.G.; O’Neill, L.A.; Xavier, R.J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Calzari, L.; Zanotti, L.; Inglese, E.; Scaglione, F.; Cavagnola, R.; Ranucci, F.; Di Blasio, A.M.; Stefanini, G.; Gaetano, C.; Parati, G.; et al. Role of epigenetics in the clinical evolution of COVID-19 disease. Epigenome-wide Association study identifies markers of severe outcome. Eur. J. Med. Res. 2023, 28, 81. [Google Scholar] [CrossRef] [PubMed]
- Roshan, J.; Rougeulle, C. Developmental epigenetics: Phenotype and the flexible epigenome. Front. Cell Dev. Biol. 2018, 6, 130. [Google Scholar] [CrossRef]
- Lacagnina, A.F.; Brockway, E.T.; Crovetti, C.R.; Shue, F.; McCarty, M.J.; Sattler, K.P.; Lim, S.C.; Santos, S.M.A.V.; Denny, C.A.; Drew, M.R. Distinct hippocampal engrams control extinction and relapse of fear memory. Nat. Neurosci. 2019, 22, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Gluckman, P.D.; Hanson, M.A.; Cooper, C.; Thornburg, K.L. Effect of in utero and Early-Life conditions on adult health and disease. New Engl. J. Med. 2008, 359, 61–73. [Google Scholar] [CrossRef]
- Kumar, S.V.; Wigge, P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef]
- Seong, K.H.; Li, D.; Shimizu, H.; Nakamura, R.; Ishii, S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 2011, 145, 1049–1061. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Paro, R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 1998, 93, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Rechavi, O.; Minevich, G.; Hobert, O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 2011, 147, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Rechavi, O.; Houri-Ze’evi, L.; Anava, S.; Goh, W.S.S.; Kerk, S.Y.; Hannon, G.J.; Hobert, O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 2014, 158, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Klosin, A.; Casas, E.; Hidalgo-Carcedo, C.; Vavouri, T.; Lehner, B. Transgenerational transmission of environmental information in C. elegans. Science 2017, 356, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Larriba, E.; del Mazo, J. Role of Non-Coding RNAs in the Transgenerational Epigenetic Transmission of the Effects of Reprotoxicants. Int. J. Mol. Sci. 2016, 17, 452. [Google Scholar] [CrossRef] [PubMed]
- Dolinoy, D.C.; Huang, D.; Jirtle, R.L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. USA 2007, 104, 13056–13061. [Google Scholar] [CrossRef]
- Song, M.; Greenbaum, J.; Luttrell, J.; Zhou, W.; Wu, C.; Shen, H.; Gong, P.; Zhang, C.; Deng, H.W. A review of Integrative Imputation for Multi-Omics Datasets. Front. Genet. 2020, 11, 570255. [Google Scholar] [CrossRef]
- Orsini, A.; Diquigiovanni, C.; Bonora, E. Omics Technologies improving breast cancer research and diagnostics. Int. J. Mol. Sci. 2023, 24, 12690. [Google Scholar] [CrossRef]
- Grunau, C.; Luyer, J.L.; Laporte, M.; Joly, D. The epigenetics dilemma. Genes 2019, 11, 23. [Google Scholar] [CrossRef]
- Butler, R.N. The Longevity Revolution: The Benefits and Challenges of Living a Long Life; PublicAffairs: New York, NY, USA, 2008; ISBN 9781586488550. [Google Scholar]
- Shonkoff, J.P.; Phillips, D. From Neurons to Neighborhoods: The Science of Early Childhood Development; The National Academies Press: Washington, DC, USA, 2000. [Google Scholar] [CrossRef]
- Basta, J.; Rauchman, M. The nucleosome remodeling and deacetylase complex in development and disease. Transl. Res. (Online) 2015, 165, 36–47. [Google Scholar] [CrossRef]
- Choi, Y.; Mango, S.E. Hunting for Darwin’s Gemmules and Lamarck’s fluid: Transgenerational signaling and Histone methylation. Biochim. Et Biophys. Acta (BBA)—Gene Regul. Mech. 2014, 1839, 1440–1453. [Google Scholar] [CrossRef]
- Harvey, W. Exercitationes de Generatione Animalium: Quibus Accedunt Quaedam de Partu, de Membranis ac Humoribus vteri, et de Conceptione. Biblioteca Virtual Miguel de Cervantes. 1651. Available online: https://www.cervantesvirtual.com/obra/exercitationes-de-generatione-animalium--quibus-accedunt-quaedam-de-partu-de-membranis-ac-humoribus-vteri-et-de-conceptione/ (accessed on 16 February 2024).
- Descartes, R. Tractatus de homine: Et de formatione foetus. Amstelodami: Ex Typographia Blaviana, sumptibus Societatis; 1677. [Google Scholar]
- Needham, J.T.; Folkes, M. Observations upon the Generation, Composition, and Decomposition of Animal and Vegetable Substances: Communicated in a Letter to Martin Folkes, Esq.; President of the Royal Society: London, UK, 1749; pp. 1–53. [Google Scholar] [CrossRef]
- Bonnet, C. “Observations sur Quelques Auteurs D’histoire Naturelle” in Correspondance Litteraire, Philosophique et Critique par Grimm, Diderot, Meister, etc.; Tourneux, M., Ed.; Garnier Freres: Paris, France, 1878; Volume IV, p. 167. [Google Scholar]
- Gazzoli, I.; Loda, M.; Garber, J.E.; Syngal, S.; Kolodner, R.D. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002, 62, 3925–3928. [Google Scholar] [PubMed]
- Mattick, J.S. RNA regulation: A new genetics? Nat. Rev. Genet. 2004, 5, 316–323. [Google Scholar] [CrossRef]
- Meaney, M.J. Nature, Nurture, and the Disunity of Knowledge. Ann. N. Y. Acad. Sci. 2001, 935, 50–61. [Google Scholar] [CrossRef]
- Pauli, A.; Rinn, J.L.; Schier, A.F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011, 12, 136–149. [Google Scholar] [CrossRef]
- Skinner, M.K. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res. 2011, 93, 51–55. [Google Scholar] [CrossRef]
- Kuppusamy, S.P.; Kaiser, J.P.; Wesselkamper, S.C. Epigenetic Regulation in Environmental Chemical Carcinogenesis and its Applicability in Human Health Risk Assessment. Int. J. Toxicol. 2015, 34, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Zaina, S.; Lund, G. Atherosclerosis, cell biology and lipoproteins—Epigenetics and oxidation in atherosclerosis. Curr. Opin. Lipidol. 2014, 25, 235–236. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.; Qu, W.; Ho, T.; Clark, S.J.; Molloy, P. Conversion-specific detection of DNA methylation using real-time polymerase chain reaction (ConLight-MSP) to avoid false positives. Science 2002, 27, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Gnirke, A.; Bell, G.W.; Ramsahoye, B.; Lander, E.S.; Jaenisch, R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005, 33, 5868–5877. [Google Scholar] [CrossRef] [PubMed]
- Jacinto, F.V.; Ballestar, E.; Esteller, M. Methyl-DNA immunoprecipitation (MeDIP): Hunting down the DNA methylome. Biotechniques 2008, 44, 35–43. [Google Scholar] [CrossRef]
- Pillai, S.; Chellappan, S.P. ChIP on chip assays: Genome-wide analysis of transcription factor binding and histone modifications. Methods Mol. Biol. 2009, 523, 341–366. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tollefsbol, T.O. DNA methylation detection: Bisulfite genomic sequencing analysis. Methods Mol. Biol. 2011, 791, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.D.; Wang, C.I.; Kharchenko, P.V.; West, J.A.; Chapman, B.A.; Alekseyenko, A.A.; Borowsky, M.L.; Kuroda, M.I.; Kingston, R.E. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 20497–20502. [Google Scholar] [CrossRef] [PubMed]
- Gade, P.; Kalvakolanu, D.V. Chromatin immunoprecipitation assay as a tool for analyzing transcription factor activity. Methods Mol. Biol. 2012, 809, 85–104. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Furey, T.S. ChIP-seq and beyond: New and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012, 13, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Booth, M.J.; Ost, T.W.; Beraldi, D.; Bell, N.M.; Branco, M.R.; Reik, W.; Balasubramanian, S. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 2013, 8, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Laszlo, A.H.; Derrington, I.M.; Brinkerhoff, H.; Langford, K.W.; Nova, I.C.; Samson, J.M.; Bartlett, J.J.; Pavlenok, M.; Gundlach, J.H. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl. Acad. Sci. USA 2013, 110, 18904–18909. [Google Scholar] [CrossRef] [PubMed]
- Delaney, C.; Garg, S.K.; Yung, R. Analysis of DNA Methylation by Pyrosequencing. Methods Mol. Biol. 2015, 1343, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.; Lander, E.S.; Guttman, M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol. Biol. 2015, 1262, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Yong, W.S.; Hsu, F.M.; Chen, P.Y. Profiling genome-wide DNA methylation. Epigenet. Chromatin 2016, 9, 26. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meza-Menchaca, T.; Albores-Medina, A.; Heredia-Mendez, A.J.; Ruíz-May, E.; Ricaño-Rodríguez, J.; Gallegos-García, V.; Esquivel, A.; Vettoretti-Maldonado, G.; Campos-Parra, A.D. Revisiting Epigenetics Fundamentals and Its Biomedical Implications. Int. J. Mol. Sci. 2024, 25, 7927. https://doi.org/10.3390/ijms25147927
Meza-Menchaca T, Albores-Medina A, Heredia-Mendez AJ, Ruíz-May E, Ricaño-Rodríguez J, Gallegos-García V, Esquivel A, Vettoretti-Maldonado G, Campos-Parra AD. Revisiting Epigenetics Fundamentals and Its Biomedical Implications. International Journal of Molecular Sciences. 2024; 25(14):7927. https://doi.org/10.3390/ijms25147927
Chicago/Turabian StyleMeza-Menchaca, Thuluz, Arnulfo Albores-Medina, Alma Jaqueline Heredia-Mendez, Eliel Ruíz-May, Jorge Ricaño-Rodríguez, Verónica Gallegos-García, Adriana Esquivel, Giancarlo Vettoretti-Maldonado, and Alma D. Campos-Parra. 2024. "Revisiting Epigenetics Fundamentals and Its Biomedical Implications" International Journal of Molecular Sciences 25, no. 14: 7927. https://doi.org/10.3390/ijms25147927
APA StyleMeza-Menchaca, T., Albores-Medina, A., Heredia-Mendez, A. J., Ruíz-May, E., Ricaño-Rodríguez, J., Gallegos-García, V., Esquivel, A., Vettoretti-Maldonado, G., & Campos-Parra, A. D. (2024). Revisiting Epigenetics Fundamentals and Its Biomedical Implications. International Journal of Molecular Sciences, 25(14), 7927. https://doi.org/10.3390/ijms25147927






