Identification and Functional Analysis of circRNAs during Goat Follicular Development
Abstract
1. Introduction
2. Results
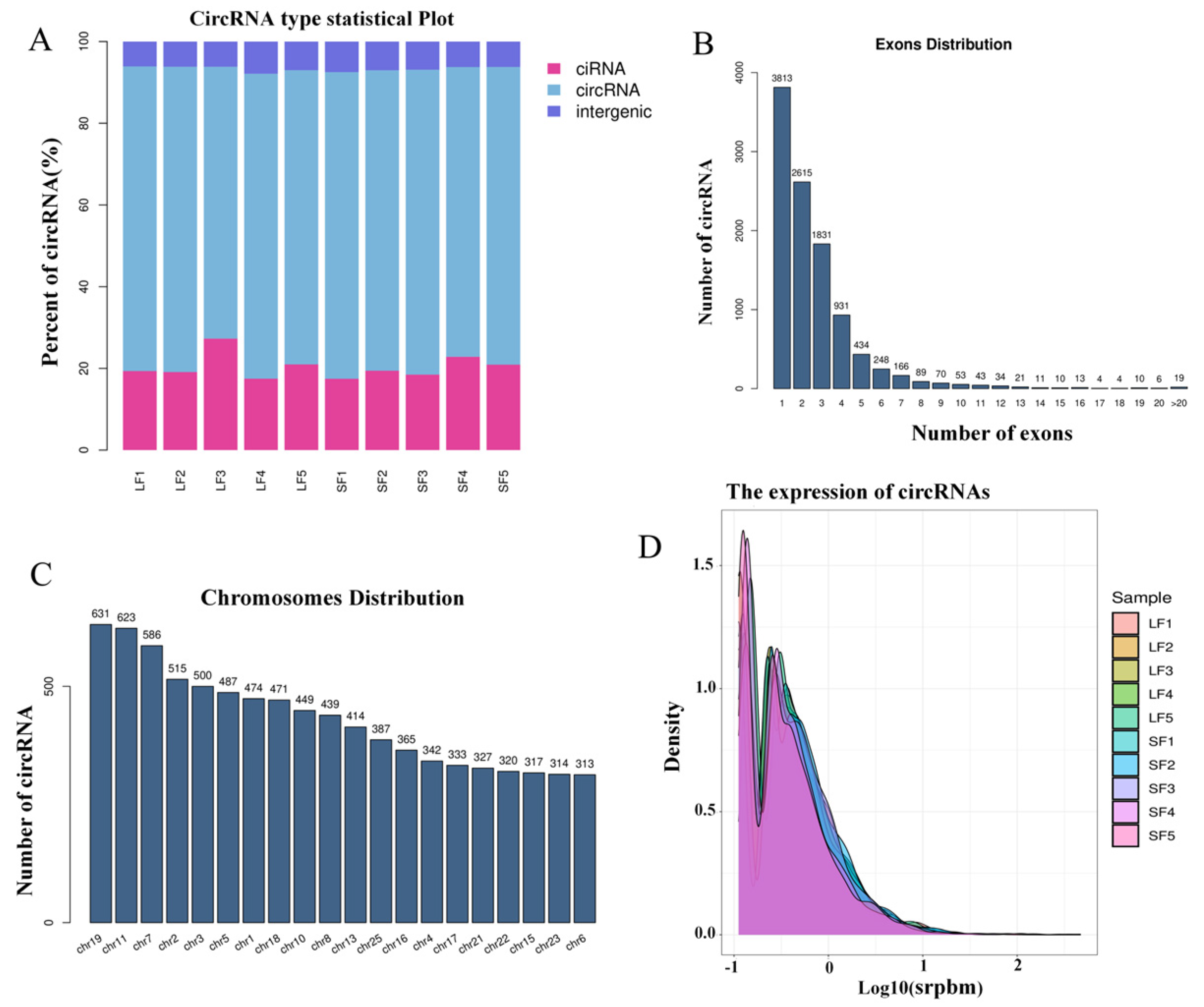
2.1. Identification of Follicular circRNA
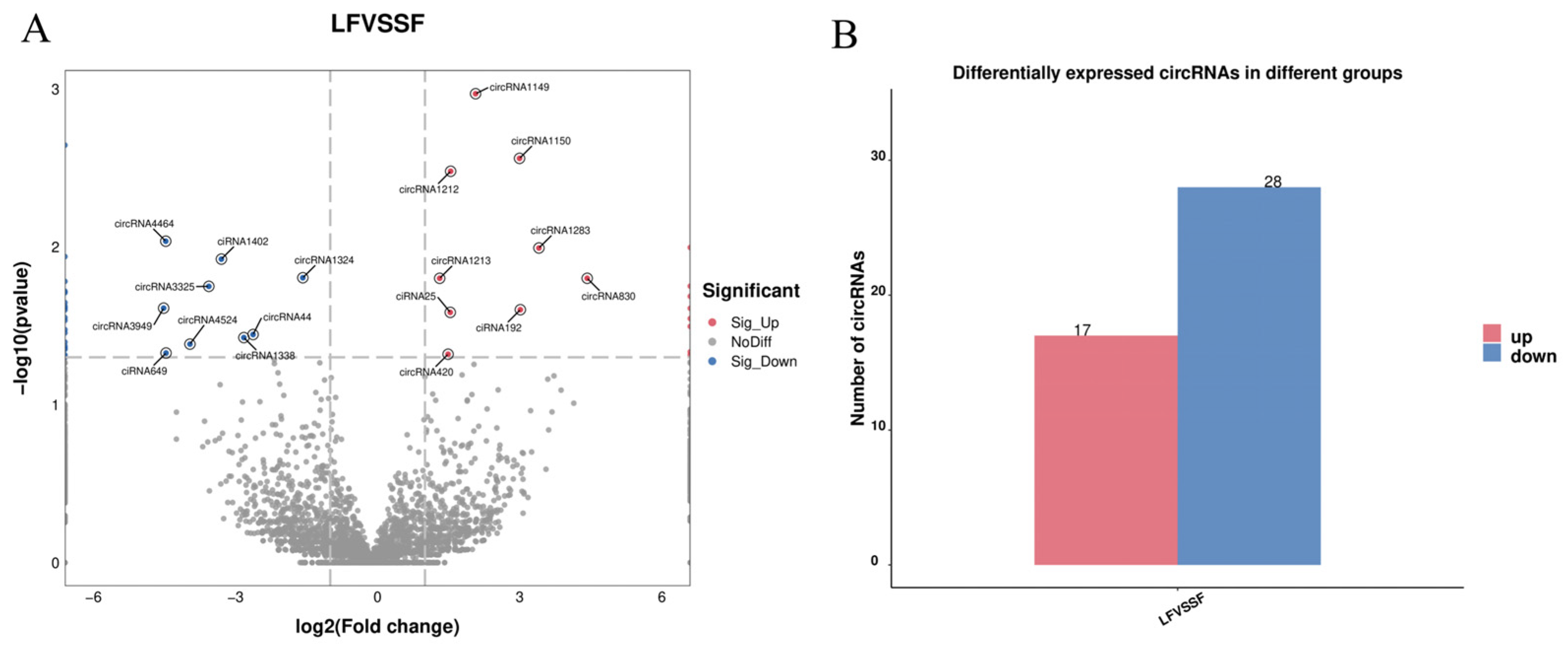
2.2. Differential Expression Analysis of Follicular circRNAs
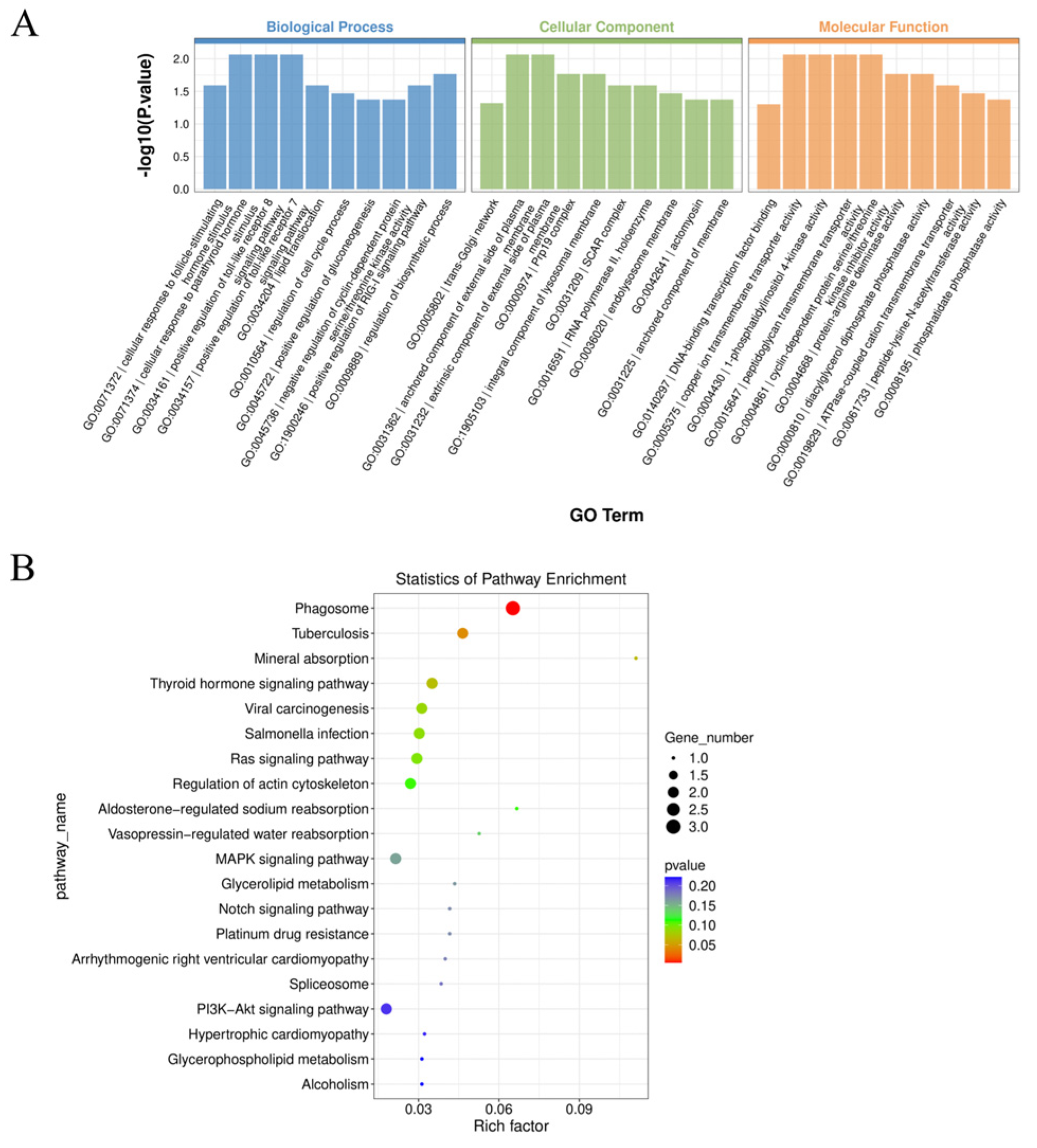
2.3. Functional Enrichment Analysis of Differential circRNA Parental Genes
2.4. Interaction Network Analysis of Differential circRNAs with miRNAs
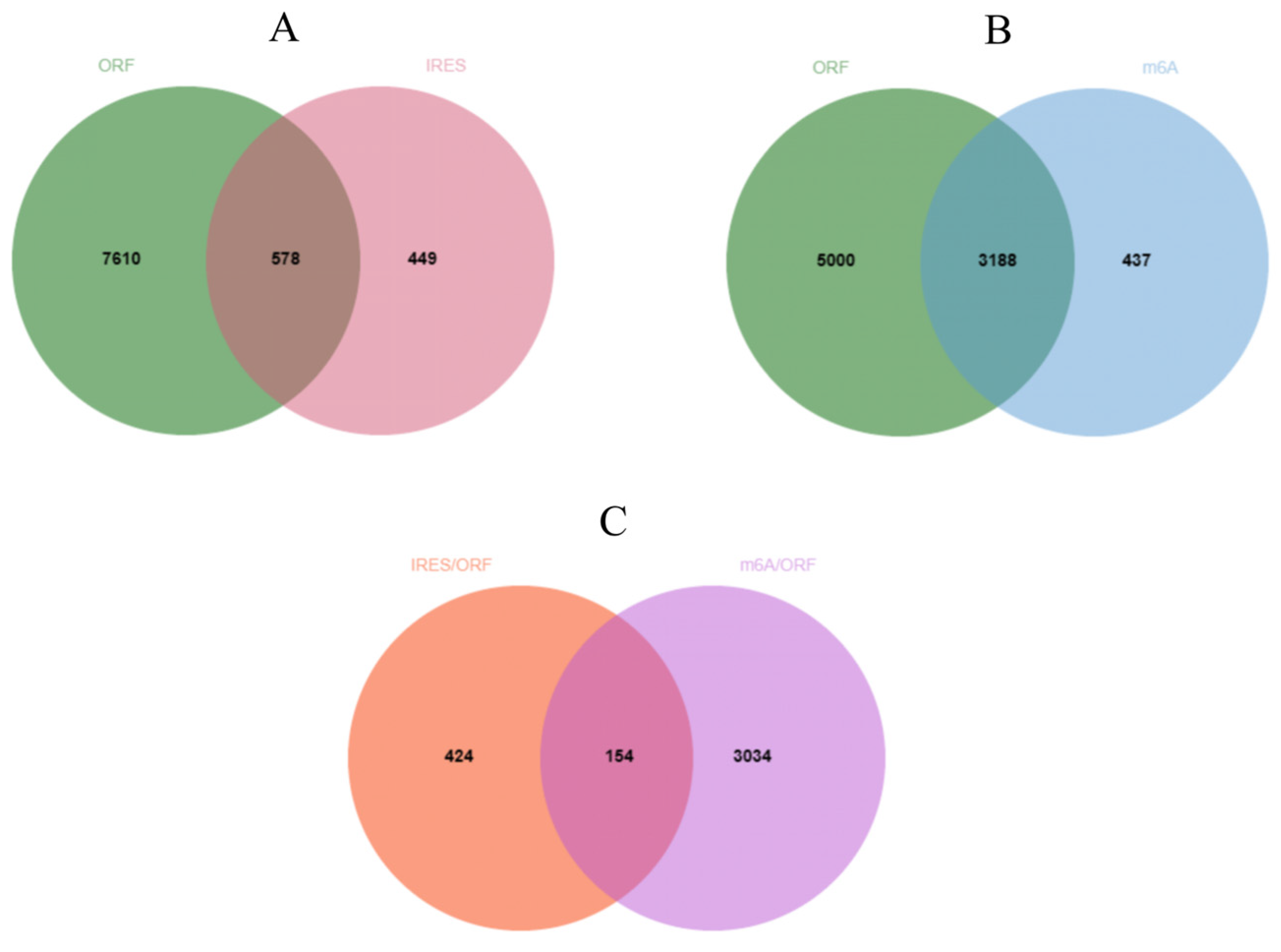
2.5. Prediction of the Translational Potential of circRNA
2.6. Conservativeness Analysis of circRNA
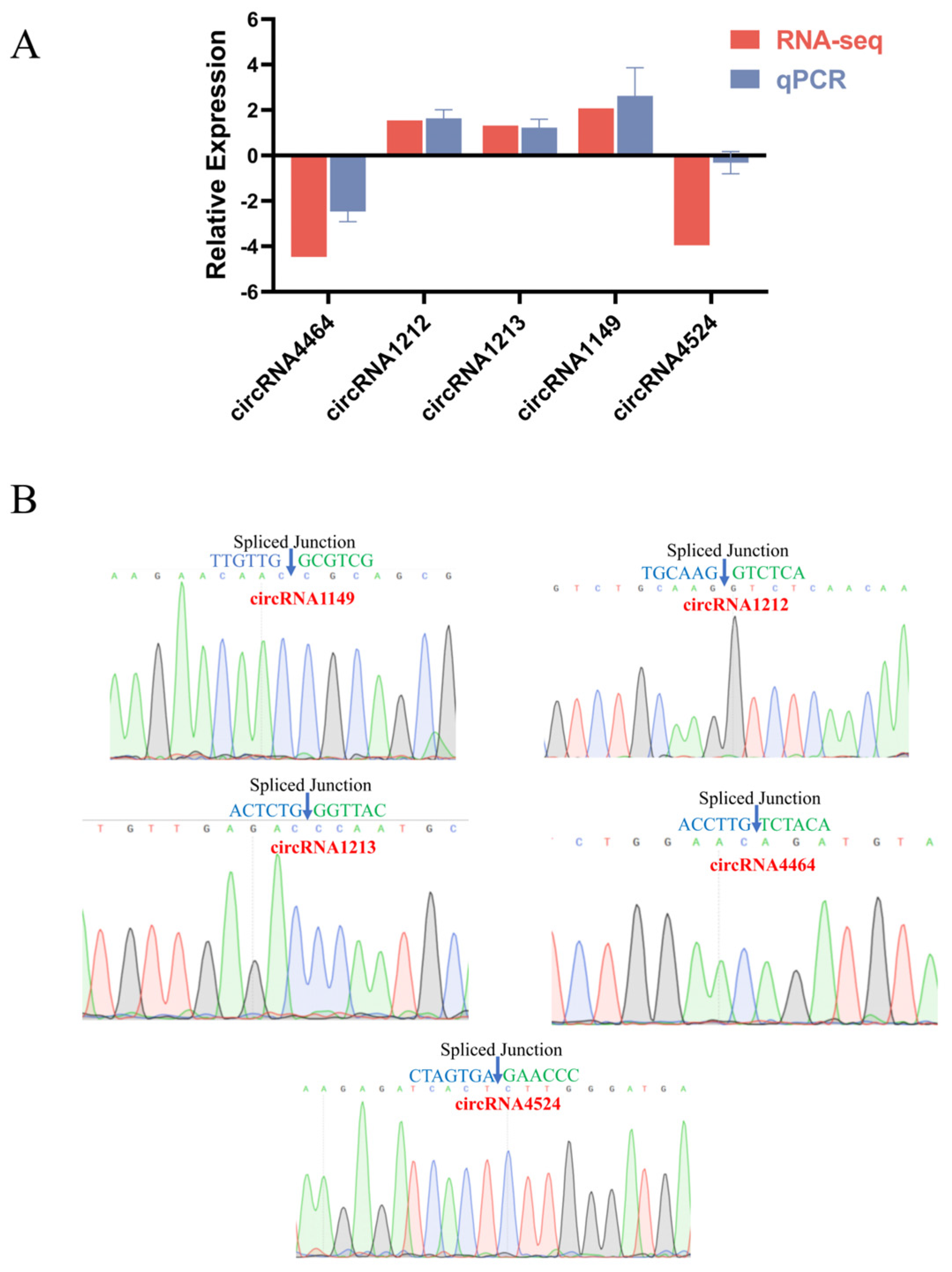
2.7. RT-qPCR and DNA Sequencing Validation of Differential circRNAs
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Total RNA Extraction, Library Construction and Sequencing
4.3. Analysis of circRNA Sequencing Data
4.4. Differential Expression Analysis of circRNA
4.5. GO and KEGG Analysis of Differential circRNA Host Genes
4.6. Prediction of Target miRNAs for circRNAs
4.7. Prediction of the Translational Potential of circRNA
4.8. Conservativeness Analysis of circRNA
4.9. RT-qPCR and DNA Sequencing Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jia, W.; Shi, Q.; Shi, L. Effect of irradiation treatment on the lipid composition and nutritional quality of goat meat. Food Chem. 2021, 351, 129295. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Peng, X.; Mei, S. Autophagy in Ovarian Follicular Development and Atresia. Int. J. Biol. Sci. 2019, 15, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, E.; Kamili, G.; Ou, S.; Yang, D. Effect of resveratrol on superovulation in mice. Biomed. Pharmacother. 2022, 146, 112565. [Google Scholar] [CrossRef] [PubMed]
- Soede, N.M.; Langendijk, P.; Kemp, B. Reproductive cycles in pigs. Anim. Reprod. Sci. 2011, 124, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Jamal, M.A.; Khan, I.M.; Ullah, I.; Jabbar, A.; Khan, N.M.; Liu, Y. Factors affecting superovulation induction in goats (Capra hericus): An analysis of various approaches. Front. Vet. Sci. 2023, 10, 1152103. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; He, Y.; Pan, X.; Quan, H.; He, B.; Li, Y.; Bai, G.; Li, N.; Zhang, Z.; Zhang, H.; et al. DNMT1-mediated lncRNA IFFD controls the follicular development via targeting GLI1 by sponging miR-370. Cell Death Differ. 2023, 30, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Altesha, M.A.; Ni, T.; Khan, A.; Liu, K.; Zheng, X. Circular RNA in cardiovascular disease. J. Cell. Physiol. 2019, 234, 5588–5600. [Google Scholar] [CrossRef]
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Misir, S.; Wu, N.; Yang, B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022, 29, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Zhang, J.; Du, X.; Li, Q.; Pan, Z. circSLC41A1 Resists Porcine Granulosa Cell Apoptosis and Follicular Atresia by Promoting SRSF1 through miR-9820-5p Sponging. Int. J. Mol. Sci. 2022, 23, 1509. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Wei, Y.; Chen, Y.; Zhao, X.; Shen, X.; Zhu, Q.; Yin, H. High expression circRALGPS2 in atretic follicle induces chicken granulosa cell apoptosis and autophagy via encoding a new protein. J. Anim. Sci. Biotechnol. 2024, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xiao, H.; Pi, J.; Zhang, H.; Pan, A.; Pu, Y.; Liang, Z.; Shen, J.; Du, J. The circular RNA aplacirc_13267 upregulates duck granulosa cell apoptosis by the apla-miR-1-13/THBS1 signaling pathway. J. Cell. Physiol. 2020, 235, 5750–5763. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Lu, T.; Zhao, Z.; Liu, G.; Lian, Z.; Guo, Y.; Sun, B.; Liu, D.; Li, Y. Comprehensive analysis of mRNAs and miRNAs in the ovarian follicles of uniparous and multiple goats at estrus phase. BMC Genom. 2020, 21, 267. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, Z.; Qin, Q.; Nisenblat, V.; Chang, H.M.; Yu, Y.; Wang, T.; Lu, C.; Yang, M.; Yang, S.; et al. Transcriptome Landscape of Human Folliculogenesis Reveals Oocyte and Granulosa Cell Interactions. Mol. Cell 2018, 72, 1021–1034. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Li, B.; Zhang, Y.; Song, S.; Ding, W.; Xu, D.; Zhao, Z. BMP6 regulates AMH expression via SMAD1/5/8 in goat ovarian granulosa cells. Theriogenology 2023, 197, 167–176. [Google Scholar] [CrossRef]
- Wang, P.; Li, W.; Liu, Z.; He, X.; Hong, Q.; Lan, R.; Liu, Y.; Chu, M. Identification of WNT4 alternative splicing patterns and effects on proliferation of granulosa cells in goat. Int. J. Biol. Macromol. 2022, 223, 1230–1242. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zeng, C.; Wang, F.; Zhang, Z.; Yang, F.; Liu, Z.P.; Li, K.; Zhang, G.M. Neuromedin S Regulates Steroidogenesis through Maintaining Mitochondrial Morphology and Function via NMUR2 in Goat Ovarian Granulosa Cells. Int. J. Mol. Sci. 2022, 23, 13402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Jing, J.; Qin, S.; Zheng, Q.; Lu, J.; Zhu, C.; Liu, Y.; Fang, F.; Li, Y.; Ling, Y. miR-130a-3p regulates steroid hormone synthesis in goat ovarian granulosa cells by targeting the PMEPA1 gene. Theriogenology 2021, 165, 92–98. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Ma, H.; Liu, Y.; Li, F.; Song, Y.; Li, G.; Bai, Y.; Cao, B. Effects of miR-101-3p on goat granulosa cells in vitro and ovarian development in vivo via STC1. J. Anim. Sci. Biotechnol. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, L.; Liu, X.; Niu, M.; Cui, J.; Che, S.; Liu, Y.; An, X.; Cao, B. Analyses of circRNA profiling during the development from pre-receptive to receptive phases in the goat endometrium. J. Anim. Sci. Biotechnol. 2019, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, K.; Liu, J.; Zhang, H.; Fan, Y.; Chen, Y.; Han, H.; Yang, J.; Liu, Y. Expression Profile Analysis to Identify Circular RNA Expression Signatures in Muscle Development of Wu’an Goat Longissimus Dorsi Tissues. Front. Vet. Sci. 2022, 9, 833946. [Google Scholar] [CrossRef]
- Shen, M.; Li, T.; Zhang, G.; Wu, P.; Chen, F.; Lou, Q.; Chen, L.; Yin, X.; Zhang, T.; Wang, J. Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken. BMC Genom. 2019, 20, 96. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, Y.; Wu, W.; Tan, X.; Wang, Z.; Irwin, D.M.; Wang, Z.; Zhang, S. Circular RNA Profiling Identifies Novel circPPARA that Promotes Intramuscular Fat Deposition in Pigs. J. Agric. Food Chem. 2022, 70, 4123–4137. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Z.; Zi, C.; Wu, P.; Lv, X.; Chen, L.; Chen, F.; Zhang, G.; Wang, J. CircRNA expression in chicken granulosa cells illuminated with red light. Poult. Sci. 2022, 101, 101734. [Google Scholar] [CrossRef]
- Zhou, C.; Cheng, X.; Meng, F.; Wang, Y.; Luo, W.; Zheng, E.; Cai, G.; Wu, Z.; Li, Z.; Hong, L. Identification and characterization of circRNAs in peri-implantation endometrium between Yorkshire and Erhualian pigs. BMC Genom. 2023, 24, 412. [Google Scholar] [CrossRef]
- Dewailly, D.; Robin, G.; Peigne, M.; Decanter, C.; Pigny, P.; Catteau-Jonard, S. Interactions between androgens, FSH, anti-Mullerian hormone and estradiol during folliculogenesis in the human normal and polycystic ovary. Hum. Reprod. Update 2016, 22, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Buensuceso, A.V.; Son, A.I.; Zhou, R.; Paquet, M.; Withers, B.M.; Deroo, B.J. Ephrin-A5 Is Required for Optimal Fertility and a Complete Ovulatory Response to Gonadotropins in the Female Mouse. Endocrinology 2016, 157, 942–955. [Google Scholar] [CrossRef][Green Version]
- Yuan, H.; Lu, J.; Xiao, S.Y.; Han, X.Y.; Song, X.T.; Qi, M.Y.; Liu, G.S.; Yang, C.X.; Yao, Y.C. miRNA expression analysis of the sheep follicle during the prerecruitment, dominant, and mature stages of development under FSH stimulation. Theriogenology 2022, 181, 161–169. [Google Scholar] [CrossRef]
- Ding, L.; Yang, L.; He, Y.; Zhu, B.; Ren, F.; Fan, X.; Wang, Y.; Li, M.; Li, J.; Kuang, Y.; et al. CREPT/RPRD1B associates with Aurora B to regulate Cyclin B1 expression for accelerating the G2/M transition in gastric cancer. Cell Death Dis. 2018, 9, 1172. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Chen, H.; Zuo, Q.; Huang, C.; Zhao, R.; Yu, X.; Wang, Y.; Zhang, Y.; Chang, Z.; Li, B. CREPT and p15RS regulate cell proliferation and cycling in chicken DF-1 cells through the Wnt/beta-catenin pathway. J. Cell. Biochem. 2018, 119, 1083–1092. [Google Scholar] [CrossRef]
- Costa, J.; Cezar, G.A.; Monteiro, P.; Silva, D.; Araujo, S.R.; Bartolomeu, C.C.; Santos, F.A.; Wischral, A.; Batista, A.M. Leptin improves in-vitro maturation of goat oocytes through MAPK and JAK2/STAT3 pathways and affects gene expression of cumulus cells. Reprod. Biol. 2022, 22, 100609. [Google Scholar] [CrossRef]
- Wang, P.; Liu, S.; Zhu, C.; Duan, Q.; Jiang, Y.; Gao, K.; Bu, Q.; Cao, B.; An, X. MiR-29 regulates the function of goat granulosa cell by targeting PTX3 via the PI3K/AKT/mTOR and Erk1/2 signaling pathways. J. Steroid Biochem. Mol. Biol. 2020, 202, 105722. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zou, P.; He, Y.; Meng, K.; Quan, F.; Zhang, Y. Effect of luteinizing hormone on goat theca cell apoptosis and steroidogenesis through activation of the PI3K/AKT pathway. Anim. Reprod. Sci. 2018, 190, 108–118. [Google Scholar] [CrossRef]
- Trombly, D.J.; Woodruff, T.K.; Mayo, K.E. Suppression of Notch signaling in the neonatal mouse ovary decreases primordial follicle formation. Endocrinology 2009, 150, 1014–1024. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Zhou, Z.; He, X.; Tao, L.; Jiang, Y.; Lan, R.; Hong, Q.; Chu, M. chi-miR-324-3p Regulates Goat Granulosa Cell Proliferation by Targeting DENND1A. Front. Vet. Sci. 2021, 8, 732440. [Google Scholar] [CrossRef]
- Feng, G.; Liu, J.; Lu, Z.; Li, Y.; Deng, M.; Liu, G.; Sun, B.; Guo, Y.; Zou, X.; Liu, D. miR-450-5p and miR-202-5p Synergistically Regulate Follicle Development in Black Goat. Int. J. Mol. Sci. 2022, 24, 401. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Z.; Guo, S.; Li, K.; Wang, P.; Fan, Y.; He, X.; Jiang, Y.; Lan, R.; Chen, S.; et al. Transcriptome Analysis Reveals Key miRNA-mRNA Pathways in Ovarian Tissues of Yunshang Black Goats with Different Kidding Numbers. Front. Endocrinol. 2022, 13, 883663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, L.; Wang, L.; Chen, Y.; Li, F. miR-17-5p affects porcine granulosa cell growth and oestradiol synthesis by targeting E2F1 gene. Reprod. Domest. Anim. 2019, 54, 1459–1469. [Google Scholar] [CrossRef]
- Sun, X.; Klinger, F.G.; Liu, J.; De Felici, M.; Shen, W.; Sun, X. miR-378-3p maintains the size of mouse primordial follicle pool by regulating cell autophagy and apoptosis. Cell Death Dis. 2020, 11, 737. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cao, M.; Yuan, C.; Luo, Y.; Wang, N.; Liu, K.; Chen, T.; Chen, L.; Zhang, B.; Li, C.; et al. Follicular fluid exosomes inhibit expression of BTG2 and promote glucose uptake in granulosa cells by delivering miR-21-5p. Theriogenology 2024, 218, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Z.; Su, P.; Liang, Y.; Li, Z.; Zhang, H.; Song, X.; Han, D.; Wang, X.; Liu, Y.; et al. circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/beta-catenin pathway. Mol. Ther. 2022, 30, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Li, Z.; Zhong, D.; Yang, Y.; Yan, X.; Feng, T.; Wang, X.; Zhang, L.; Shen, X.; Chen, M.; et al. A Circular RNA Generated from Nebulin (NEB) Gene Splicing Promotes Skeletal Muscle Myogenesis in Cattle as Detected by a Multi-Omics Approach. Adv. Sci. 2024, 11, e2300702. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Huang, X.; Li, L.; Huang, M.; Cai, R.; Liao, X. A Regulatory Axis of circ_0008193/miR-1180-3p/TRIM62 Suppresses Proliferation, Migration, Invasion, and Warburg Effect in Lung Adenocarcinoma Cells under Hypoxia. Med. Sci. Monitor 2020, 26, e922900. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wang, Z.; Pang, W.; Zhou, J.; Liang, Y.; Yang, J.; Yang, L.; Zhang, Q. Upregulated hsa_circ_0004458 Contributes to Progression of Papillary Thyroid Carcinoma by Inhibition of miR-885-5p and Activation of RAC1. Med. Sci. Monitor 2018, 24, 5488–5500. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, Y.; Xu, D.; Cao, L. hsa_circ_0023409 Accelerates Gastric Cancer Cell Growth and Metastasis through Regulating the IRS4/PI3K/AKT Pathway. Cell Transplant. 2021, 30, 2138968094. [Google Scholar] [CrossRef]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S.A. ORF-FINDER: A vector for high-throughput gene identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, J.; Xu, T.; Yang, Q.; He, J.; Song, X. IRESfinder: Identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J. Genet. Genomics 2018, 45, 403–406. [Google Scholar] [CrossRef]


| Sample | Raw Data Read | Valid Data Read | Valid Ratio (Reads) | Q20% | Q30% |
|---|---|---|---|---|---|
| LF1 | 92957118 | 83771828 | 90.12 | 99.20 | 95.20 |
| LF2 | 94341552 | 84923578 | 90.02 | 99.09 | 95.16 |
| LF3 | 94068076 | 84851900 | 90.20 | 99.19 | 95.35 |
| LF4 | 82973554 | 76122068 | 91.74 | 99.58 | 95.33 |
| LF5 | 75735446 | 67783970 | 89.50 | 98.81 | 93.54 |
| SF1 | 93375512 | 84413204 | 90.40 | 99.12 | 95.34 |
| SF2 | 90519034 | 81379192 | 89.90 | 99.19 | 95.36 |
| SF3 | 77917452 | 70803598 | 90.87 | 99.64 | 95.89 |
| SF4 | 80346710 | 71427864 | 88.90 | 98.78 | 93.69 |
| SF5 | 90400602 | 82115126 | 90.83 | 99.18 | 94.48 |
| Accession | Parent Gene | Conservative_circRNA/Parent Gene |
|---|---|---|
| circRNA1150 | SERPINE2 | hsa_circ_0005773/SERPINE2 |
| circRNA1212 | ITGB5 | hsa_circ_0067080/ITGB5 |
| circRNA5092 | UBP1 | hsa_circ_0007147/UBP1 |
| circRNA1324 | FAM120A | hsa_circ_0008193/FAM120A |
| circRNA830 | MEF2C | hsa_circ_0129926/MEF2C |
| circRNA5493 | ZNF536 | hsa_circ_0109365/ZNF536 |
| circRNA5508 | intergenic_circRNA | hsa_circ_0004366/VPS13B |
| circRNA6643 | PUM2 | hsa_circ_0052867/PUM2 |
| circRNA5501 | intergenic_circRNA | hsa_circ_0064555/SATB1 |
| circRNA5025 | PLPP4 | hsa_circ_0020207/PPAPDC1A |
| circRNA5671 | MED13L | hsa_circ_0003059/MED13L |
| circRNA44 | RPRD1B | hsa_circ_0007365/RPRD1B |
| circRNA1338 | HSDL2 | hsa_circ_0088087/HSDL2 |
| circRNA5240 | R3HDM2 | hsa_circ_0002539/R3HDM2 |
| circRNA420 | RAB5C | hsa_circ_0106851/RAB5C |
| circRNA5490 | ATP9B | hsa_circ_0108910/ATP9B |
| circRNA1922 | LASP1 | hsa_circ_0043388/LASP1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Guo, C.; Fu, J.; Liu, D.; Liu, G.; Sun, B.; Deng, M.; Guo, Y.; Li, Y. Identification and Functional Analysis of circRNAs during Goat Follicular Development. Int. J. Mol. Sci. 2024, 25, 7548. https://doi.org/10.3390/ijms25147548
Liu J, Guo C, Fu J, Liu D, Liu G, Sun B, Deng M, Guo Y, Li Y. Identification and Functional Analysis of circRNAs during Goat Follicular Development. International Journal of Molecular Sciences. 2024; 25(14):7548. https://doi.org/10.3390/ijms25147548
Chicago/Turabian StyleLiu, Jie, Conghui Guo, Junjie Fu, Dewu Liu, Guangbin Liu, Baoli Sun, Ming Deng, Yongqing Guo, and Yaokun Li. 2024. "Identification and Functional Analysis of circRNAs during Goat Follicular Development" International Journal of Molecular Sciences 25, no. 14: 7548. https://doi.org/10.3390/ijms25147548
APA StyleLiu, J., Guo, C., Fu, J., Liu, D., Liu, G., Sun, B., Deng, M., Guo, Y., & Li, Y. (2024). Identification and Functional Analysis of circRNAs during Goat Follicular Development. International Journal of Molecular Sciences, 25(14), 7548. https://doi.org/10.3390/ijms25147548






