Evidence of Gas Phase Glucosyl Transfer and Glycation in the CID/HCD-Spectra of S-Glucosylated Peptides
Abstract
1. Introduction
2. Results
2.1. General Characteristics of S-Glucopeptides MS-Fragmentation
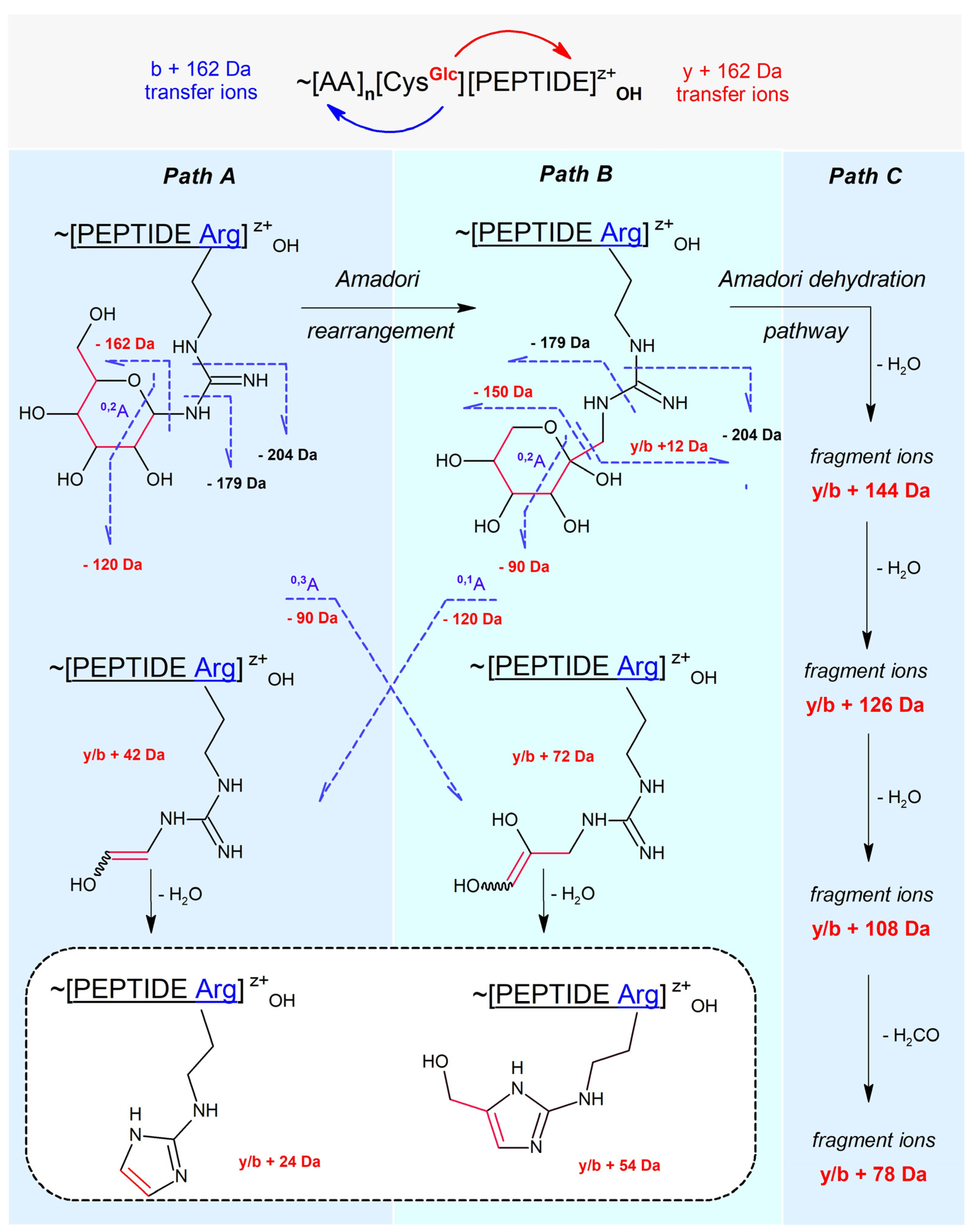
2.2. Literature-Based Characteristics of Peptidyl N-Glucosylated Arg and Lys MS-Fragmentation
- Path A: peptidyl N-glucoside fragmentation (Schiff base fragmentation path)
- Path B: peptidyl N-fructoside fragmentation (Amadori product fragmentation path)
- Path C: peptidyl N-fructoside dehydration (Amadori product dehydration path)
2.3. Detailed Analysis of MS-Fragmentation Spectra for S-Glucopeptides
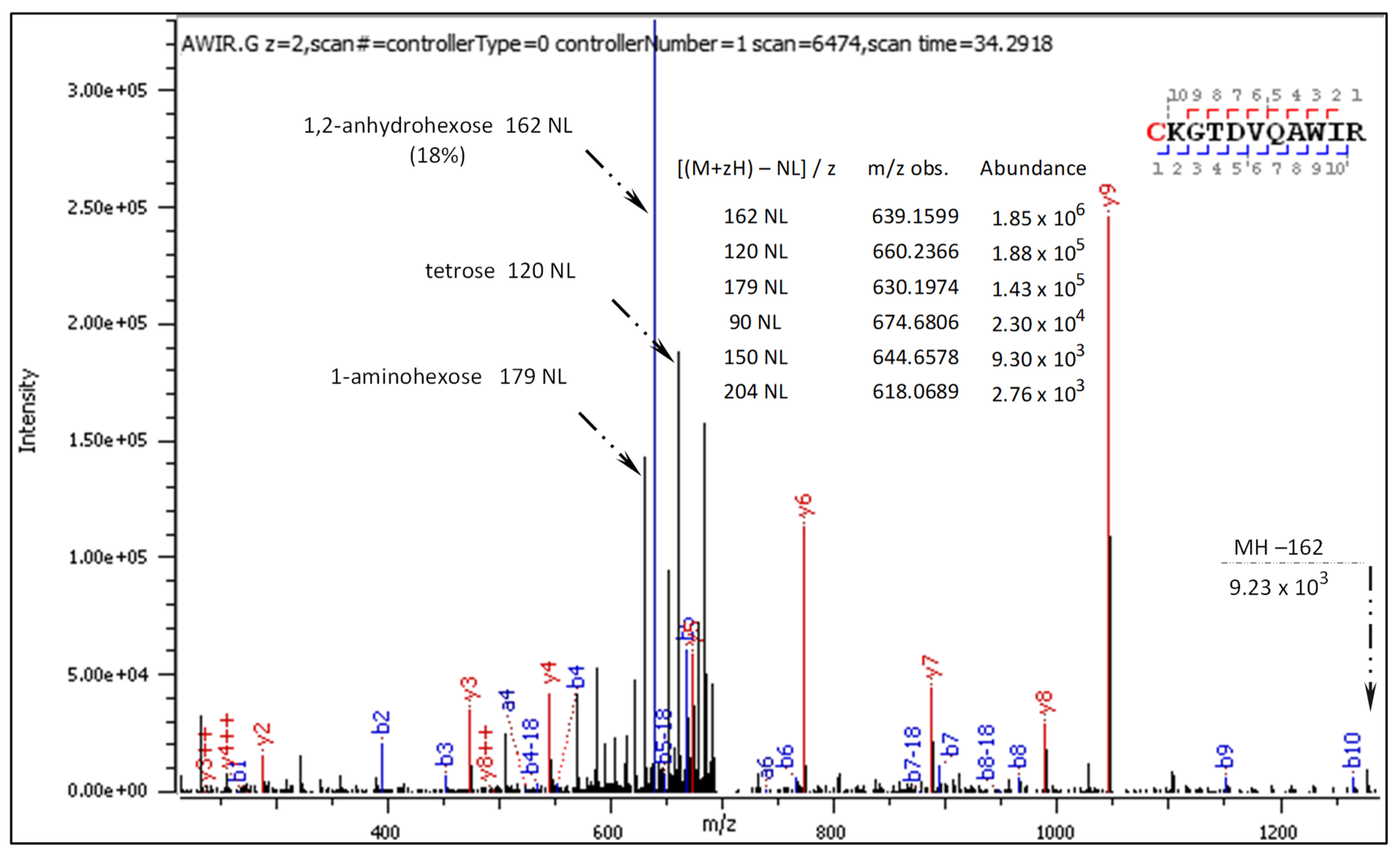
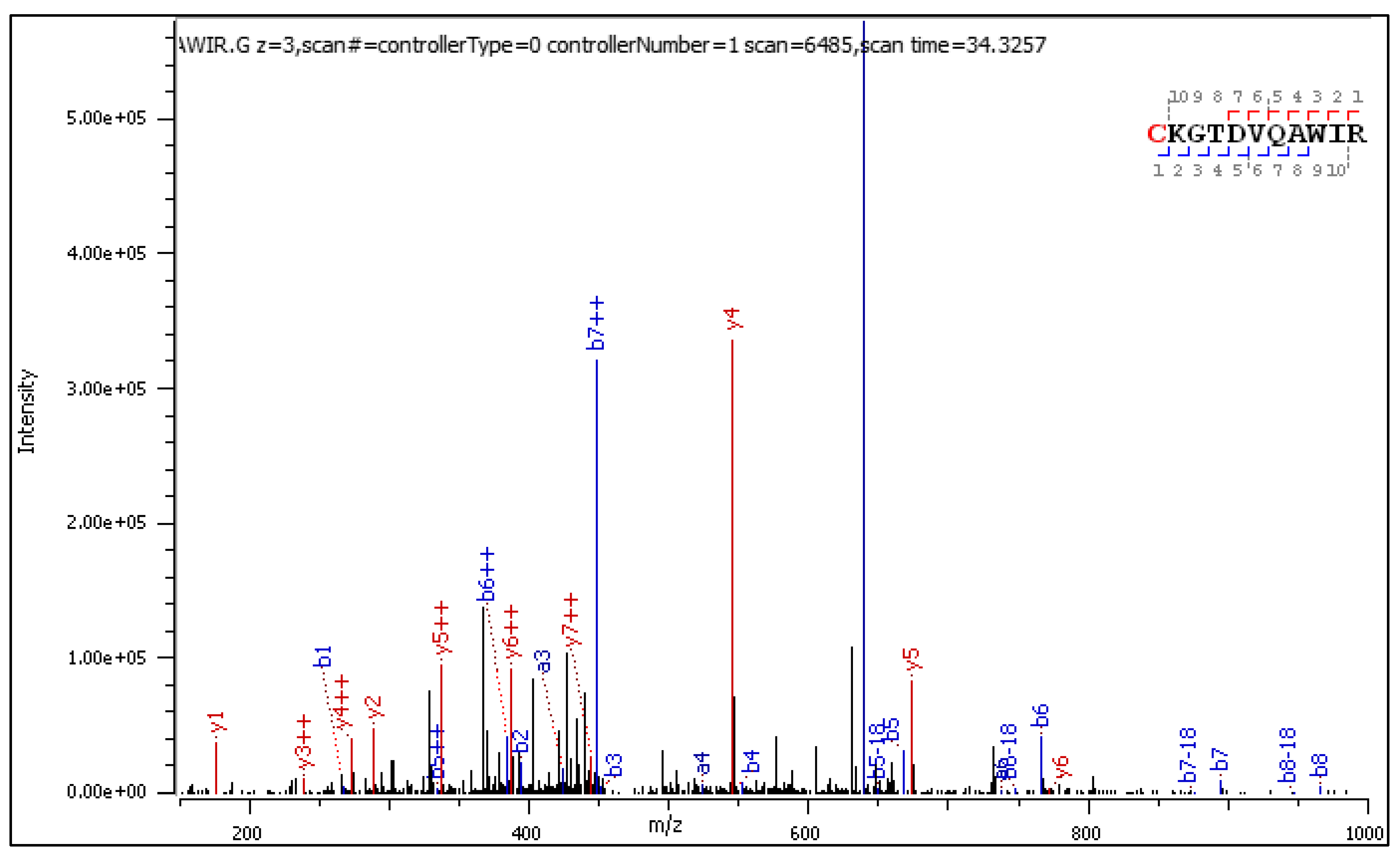
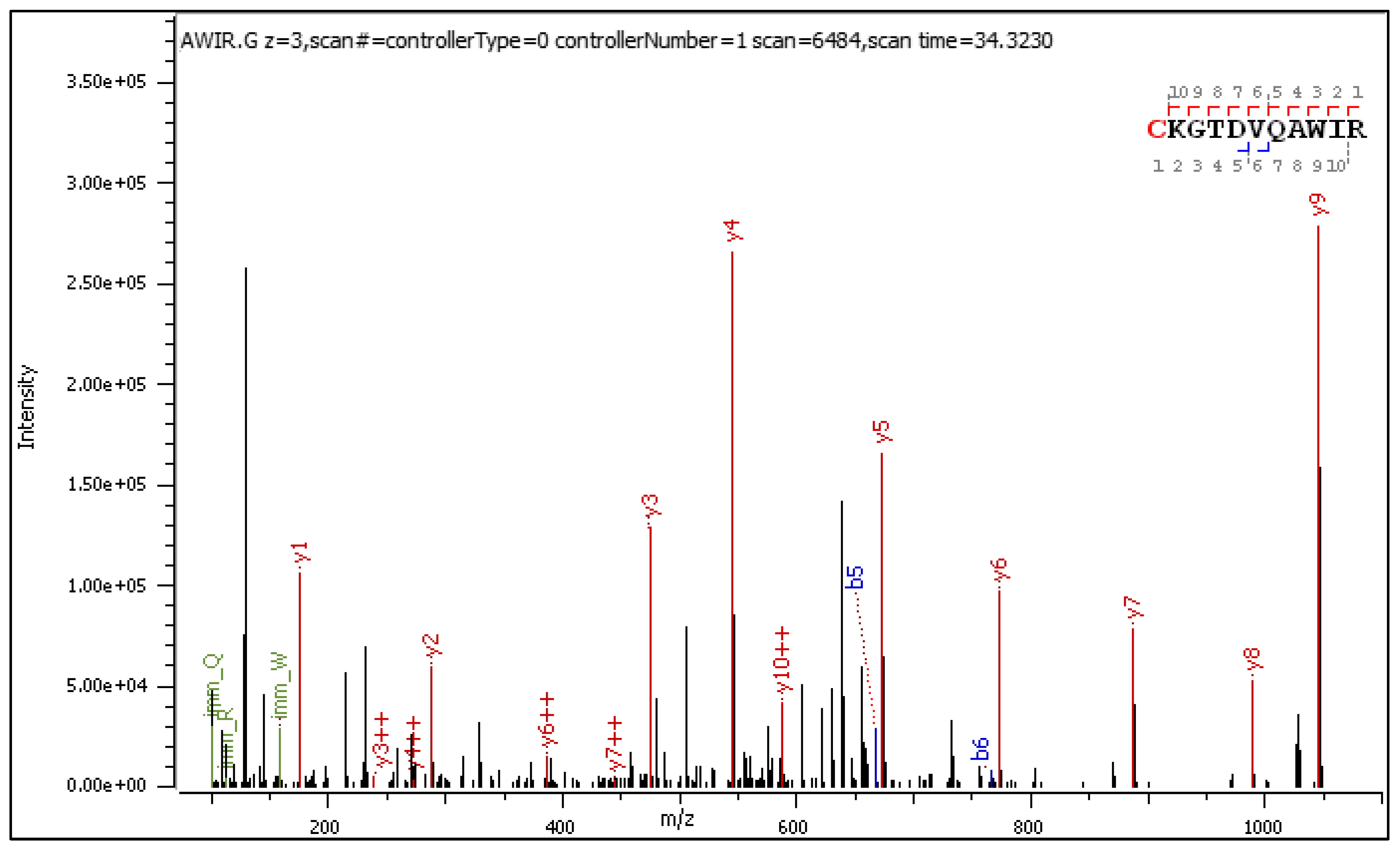
- C[+162.]KGTDVQAWIR
- C[+162]ELAAAMK (Tables S1 and S2; Figures S5 and S6)
- RC[+162]ELAAAMK (Table S3; Figures S7 and S8)
- WWC[+162]NDGR (Table S4; Figures S9 and S10)
- SLGNWVC[+162]AAK (Table S5)
2.3.1. CID-Fragmentation Analysis of C[+162.]KGTDVQAWIR Sequence
2.3.2. HCD-Fragmentation Analysis of the C[+162.]KGTDVQAWIR Sequence
3. Discussion
3.1. Reported Spectral Evidence of Gas Phase Glycosyl Transfer
3.2. Glucosyl Transfer Evidence in CID/HCD Spectra of S-Glucopeptides
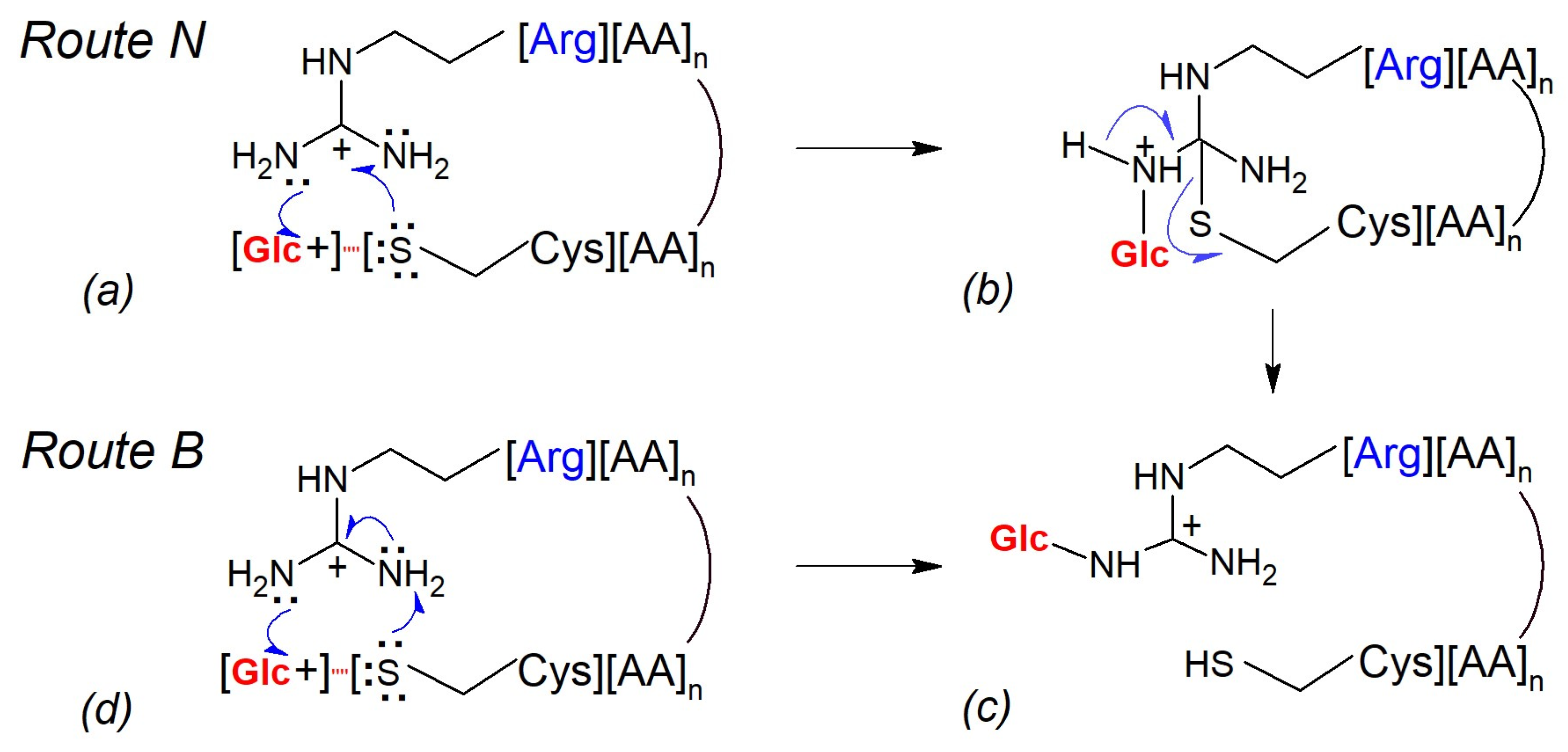
- Route N: Cysteine thiolate acts as a catalytic nucleophile
- 2.
- Route B: Cysteine thiolate behaves as a catalytic base
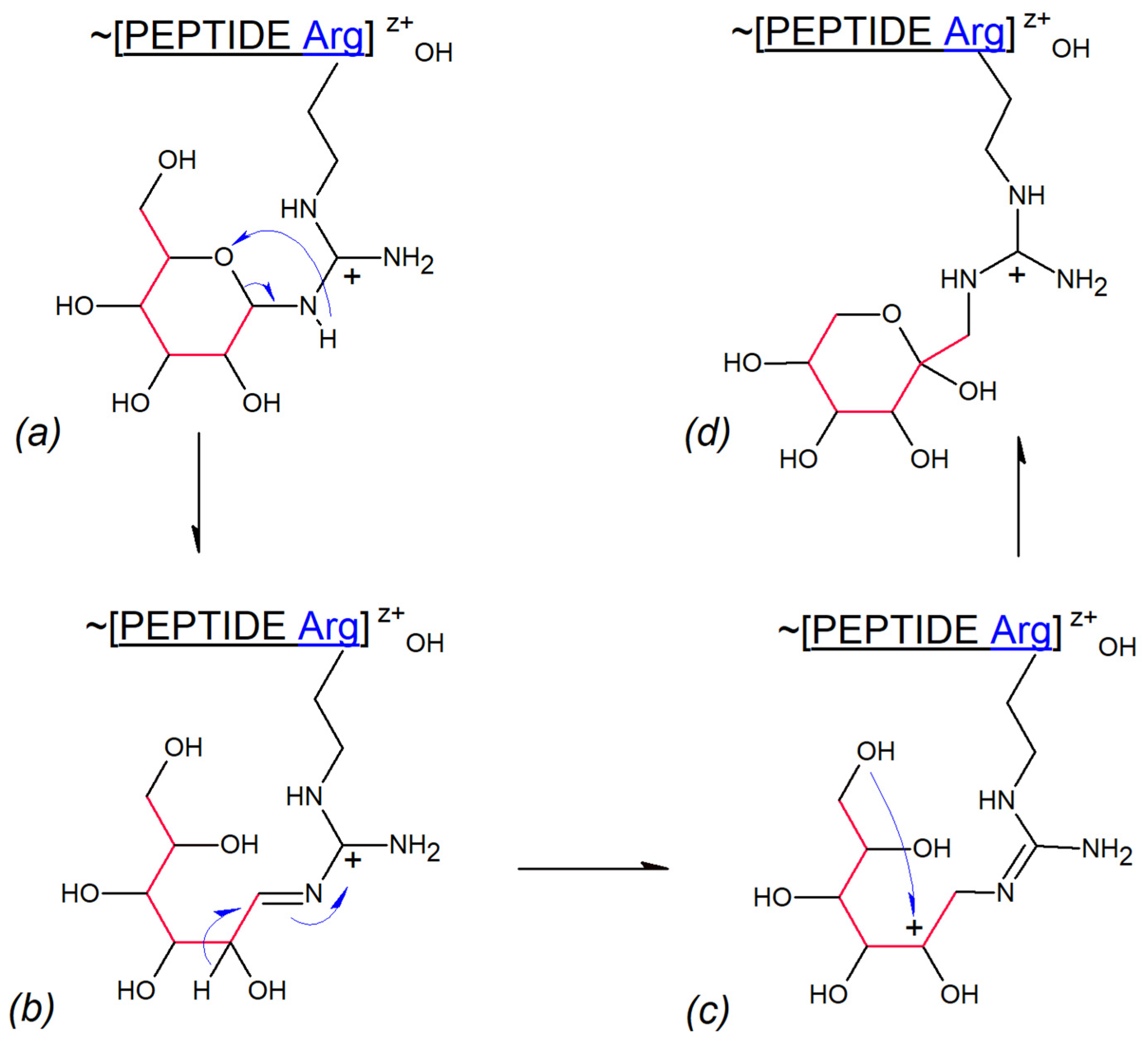
3.3. Gas Phase Amadori Rearrangement
4. Materials and Methods
4.1. MS/MS Spectra Acquisition
4.2. Manual Curation of Diagnostic Fragment Ions
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
List of Abbreviations
| Abbreviation | Meaning |
| anh-Glc | 1,2-anhydroglucose |
| anh-Glc+ | protonated 1,2-anhydroglucose |
| Arg- | arginyl residue |
| Arg+ | charged peptidyl arginine |
| ArgH+ | protonated peptidyl arginine |
| Asn- | asparaginyl residue |
| -C[S-Glc] | S-glucosylated peptidyl cysteine residue |
| CID | collision induced dissociation |
| Cys- | peptidyl cysteine |
| ESI | electrospray ionization |
| Fru- | fructosyl residue |
| Glc- | glucosyl residue |
| Glc+ | glucose oxocarbenium ion |
| GlcNAc- | N-Acetylglucosamine residue |
| HCD | higher energy collisional dissociation |
| LC | liquid chromatography |
| Mono-ADP-ribosyl | mono(adenosine diphosphate [ADP]–ribosyl residue |
| MS | mass spectrometry |
| MS/MS | tandem mass spectrometry |
| NG | neutral gain |
| NL | neutral loss |
| Rha- | rhamnosyl residue |
References
- Bagdonaite, I.; Malakar, S.A.; Polasky, D.A.; Riley, N.M.; Schjoldager, K.; Vakhrushev, S.Y.; Halim, A.; Aoki-Kinoshita, K.F.; Nesvizhskii, A.I.; Bertozzi, C.R.; et al. Glycoproteomics. Nat. Rev. Methods Primers 2022, 2, 48. [Google Scholar] [CrossRef]
- Grabarics, M.; Lettow, M.; Kirschbaum, C.; Greis, K.; Manz, C.; Pagel, K. Mass Spectrometry-Based Techniques to Elucidate the Sugar Code. Chem. Rev. 2022, 122, 7840–7908. [Google Scholar] [CrossRef] [PubMed]
- Illiano, A.; Pinto, G.; Melchiorre, C.; Carpentieri, A.; Faraco, V.; Amoresano, A. Protein Glycosylation Investigated by Mass Spectrometry: An Overview. Cells 2020, 9, 1986. [Google Scholar] [CrossRef] [PubMed]
- Krasnova, L.; Wong, C.H. Oligosaccharide Synthesis and Translational Innovation. J. Am. Chem. Soc. 2019, 141, 3735–3754. [Google Scholar] [CrossRef] [PubMed]
- Doelman, W.; van Kasteren, S.I. Synthesis of Glycopeptides and Glycopeptide Conjugates. Org. Biomol. Chem. 2022, 20, 6487–6507. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.G.; Demchenko, A.V. Intramolecular Glycosylation. Beilstein J. Org. Chem. 2017, 13, 2028–2048. [Google Scholar] [CrossRef] [PubMed]
- Escopy, S.; Demchenko, A.V. Transition-Metal-Mediated Glycosylation with Thioglycosides. Chem. A Eur. J. 2022, 28, e202103747. [Google Scholar] [CrossRef] [PubMed]
- Buch-Larsen, S.C.; Hendriks, I.A.; Lodge, J.M.; Rykær, M.; Furtwängler, B.; Shishkova, E.; Westphall, M.S.; Coon, J.J.; Nielsen, M.L. Mapping Physiological ADP-Ribosylation Using Activated Ion Electron Transfer Dissociation. Cell Rep. 2020, 32, 108176. [Google Scholar] [CrossRef] [PubMed]
- Maynard, J.C.; Burlingame, A.L.; Medzihradszky, K.F. Cysteine S-Linked N-Acetylglucosamine (S-GlcNAcylation), a New Post-Translational Modification in Mammals. Mol. Cell. Proteom. 2016, 15, 3405–3411. [Google Scholar] [CrossRef]
- Xiao, H.; Wu, R. Global and Site-Specific Analysis Revealing Unexpected and Extensive Protein S-GlcNAcylation in Human Cells. Anal. Chem. 2017, 89, 3656–3663. [Google Scholar] [CrossRef]
- Sharma, Y.; Ahlawat, S.; Rao, A. Biochemical Characterization of an Inverting S/O-HexNAc-Transferase and Evidence of S-Linked Glycosylation in Actinobacteria. Glycobiology 2022, 32, 148–161. [Google Scholar] [CrossRef]
- Wan, L.Q.; Zhang, X.; Zou, Y.; Shi, R.; Cao, J.G.; Xu, S.Y.; Deng, L.F.; Zhou, L.; Gong, Y.; Shu, X.; et al. Nonenzymatic Stereoselective S-Glycosylation of Polypeptides and Proteins. J. Am. Chem. Soc. 2021, 143, 11919–11926. [Google Scholar] [CrossRef]
- Soya, N.; Fang, Y.; Palcic, M.M.; Klassen, J.S. Trapping and Characterization of Covalent Intermediates of Mutant Retaining Glycosyltransferases. Glycobiology 2011, 21, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Buchowiecka, A.K. Protein Cysteine S-Glycosylation: Oxidative Hydrolysis of Protein S-Glycosidic Bonds in Aqueous Alka-line Environments. Amino Acids 2023, 55, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Moises, J.E.; Regl, C.; Hinterholzer, A.; Huber, C.G.; Schubert, M. Unambiguous Identification of Glucose-Induced Glycation in MAbs and Other Proteins by NMR Spectroscopy. Pharm. Res. 2023, 40, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Mossine, V.V.; Yaylayan, V. Identification of MS/MS Diagnostic Ions to Distinguish Schiff Bases of Nα- or Nε-Mono-Glycated and Nα,Nε-Di-Glycated Lysines from Their Amadori Isomers. Eur. Food Res. Technol. 2022, 248, 2753–2763. [Google Scholar] [CrossRef]
- Xing, H.; Yaylayan, V. Insight into Isomeric Diversity of Glycated Amino Acids in Maillard Reaction Mixtures. Int. J. Mol. Sci. 2022, 23, 3430. [Google Scholar] [CrossRef] [PubMed]
- Giangrande, C.; Auberger, N.; Rentier, C.; Papini, A.M.; Mallet, J.M.; Lavielle, S.; Vinh, J. Multi-Stage Mass Spectrometry Analysis of Sugar-Conjugated β-Turn Structures to Be Used as Probes in Autoimmune Diseases. J. Am. Soc. Mass. Spectrom. 2016, 27, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Vakhrushev, S.Y.; Zamfir, A.; Peter-Katalinić, J. 0,2A n Cross-Ring Cleavage as a General Diagnostic Tool for Glycan Assignment in Glycoconjugate Mixtures. J. Am. Soc. Mass. Spectrom. 2004, 15, 1863–1868. [Google Scholar] [CrossRef][Green Version]
- Pan, X.; Luo, J.; Li, S. Bacteria-Catalyzed Arginine Glycosylation in Pathogens and Host. Front. Cell Infect. Microbiol. 2020, 10, 185. [Google Scholar] [CrossRef]
- Wang, S.; Corcilius, L.; Sharp, P.P.; Rajkovic, A.; Ibba, M.; Parker, B.L.; Payne, R.J. Synthesis of Rhamnosylated Arginine Glycopeptides and Determination of the Glycosidic Linkage in Bacterial Elongation Factor P. Chem. Sci. 2017, 8, 2296–2302. [Google Scholar] [CrossRef] [PubMed]
- Gehrig, P.M.; Nowak, K.; Panse, C.; Leutert, M.; Grossmann, J.; Schlapbach, R.; Hottiger, M.O. Gas-Phase Fragmentation of ADP-Ribosylated Peptides: Arginine-Specific Side-Chain Losses and Their Implication in Database Searches. J. Am. Soc. Mass. Spectrom. 2021, 32, 157–168. [Google Scholar] [CrossRef]
- Ren, G.-R.; Zhao, L.-J.; Sun, Q.; Xie, H.-J.; Lei, Q.-F.; Fang, W.-J. Explore the Reaction Mechanism of the Maillard Reaction: A Density Functional Theory Study. J. Mol. Model. 2015, 21, 132. [Google Scholar] [CrossRef] [PubMed]
- Korwar, A.M.; Vannuruswamy, G.; Jagadeeshaprasad, M.G.; Jayaramaiah, R.H.; Bhat, S.; Regin, B.S.; Ramaswamy, S.; Giri, A.P.; Mohan, V.; Balasubramanyam, M.; et al. Development of Diagnostic Fragment Ion Library for Glycated Peptides of Human Serum Albumin: Targeted Quantification in Prediabetic, Diabetic, and Microalbuminuria Plasma by Parallel Reaction Monitoring, SWATH, and MSE. Mol. Cell. Proteom. 2015, 14, 2150–2159. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.T.; Hemmler, D.; Diederich, P.; Rychlik, M.; Marshall, J.W.; Schmitt-Kopplin, P. Open Search of Peptide Glycation Products from Tandem Mass Spectra. Anal. Chem. 2022, 94, 5953–5961. [Google Scholar] [CrossRef] [PubMed]
- Soboleva, A.; Schmidt, R.; Vikhnina, M.; Grishina, T.; Frolov, A. Maillard Proteomics: Opening New Pages. Int. J. Mol. Sci. 2017, 18, 2677. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Hemmler, D.; Schmitt-Kopplin, P. Discovery of Glycation Products: Unraveling the Unknown Glycation Space Using a Mass Spectral Library from In Vitro Model Systems. Anal. Chem. 2024, 96, 3569–3577. [Google Scholar] [CrossRef] [PubMed]
- Keilhauer, E.C.; Geyer, P.E.; Mann, M. HCD Fragmentation of Glycated Peptides. J. Proteome Res. 2016, 15, 2881–2890. [Google Scholar] [CrossRef]
- Foreman, D.J.; Mcluckey, S.A. Recent Developments in Gas-Phase Ion/Ion Reactions for Analytical Mass Spectrometry. Anal. Chem. 2020, 92, 252–266. [Google Scholar] [CrossRef]
- Prentice, B.M.; Mc Luckey, S.A. Gas-Phase Ion/Ion Reactions of Peptides and Proteins: Acid/Base, Redox, and Covalent Chemistries. Chem. Commun. 2013, 49, 947–965. [Google Scholar] [CrossRef]
- Hecht, E.S.; Loziuk, P.L.; Muddiman, D.C. Xylose Migration During Tandem Mass Spectrometry of N-Linked Glycans. J. Am. Soc. Mass. Spectrom. 2017, 28, 729–732. [Google Scholar] [CrossRef] [PubMed]
- Lettow, M.; Mucha, E.; Manz, C.; Thomas, D.A.; Marianski, M.; Meijer, G.; von Helden, G.; Pagel, K. The Role of the Mobile Proton in Fucose Migration. Anal. Bioanal. Chem. 2019, 411, 4637–4645. [Google Scholar] [CrossRef]
- Banoub, J.; Boullanger, P.; Lafont, D.; Cohen, A.; El Aneed, A.; Rowlands, E. In Situ Formation of C-Glycosides during Electrospray Ionization Tandem Mass Spectrometry of a Series of Synthetic Amphiphilic Cholesteryl Polyethoxy Neoglycolipids Containing N-Acetyl-D-Glucosamine. J. Am. Soc. Mass. Spectrom. 2005, 16, 565–570. [Google Scholar] [CrossRef][Green Version]
- Silveira, J.A.; Fort, K.L.; Kim, D.; Servage, K.A.; Pierson, N.A.; Clemmer, D.E.; Russell, D.H. From Solution to the Gas Phase: Stepwise Dehydration and Kinetic Trapping of Substance p Reveals the Origin of Peptide Conformations. J. Am. Chem. Soc. 2013, 135, 19147–19153. [Google Scholar] [CrossRef] [PubMed]
- Meroueh, O.; Hase, W.L. Collisional Activation of Small Peptides. J. Phys. Chem. A 1999, 103, 3981–3990. [Google Scholar] [CrossRef]
- Huang, F.; Nau, W.M. A Conformational Flexibility Scale for Amino Acids in Peptides. Angew. Chem. Int. Ed. 2003, 42, 2269–2272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bordas-Nagy, J. Peptide Conformation in Gas Phase Probed by Collision-Induced Dissociation and Its Correlation to Conformation in Condensed Phases. J. Am. Soc. Mass. Spectrom. 2006, 17, 786–794. [Google Scholar] [CrossRef][Green Version]
- Huang, F.; Hudgins, R.R.; Nau, W.M. Primary and Secondary Structure Dependence of Peptide Flexibility Assessed by Fluorescence-Based Measurement of End-to-End Collision Rates. J. Am. Chem. Soc. 2004, 126, 16665–16675. [Google Scholar] [CrossRef] [PubMed]
- Brintaki, A.N.; Lai-Yuen, S.K. EBGF: An Enhanced Geometric Hierarchical Representation for Protein Modeling and Rapid Self-Collision Detection. Comput. Aided Des. Appl. 2009, 6, 625–638. [Google Scholar] [CrossRef]
- Franconetti, A.; Ardá, A.; Asensio, J.L.; Blériot, Y.; Thibaudeau, S.; Jiménez-Barbero, J. Glycosyl Oxocarbenium Ions: Structure, Conformation, Reactivity, and Interactions. Acc. Chem. Res. 2021, 54, 2552–2564. [Google Scholar] [CrossRef]
- Moriarty, N.W.; Liebschner, D.; Tronrud, D.E.; Adams, P.D. Arginine Off-Kilter: Guanidinium Is Not as Planar as Restraints Denote. Acta Crystallogr. D Struct. Biol. 2020, 76, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Falcioni, F.; Molt, R.W.; Jin, Y.; Waltho, J.P.; Hay, S.; Richards, N.G.J.; Blackburn, G.M. Arginine Kinase Activates Arginine for Phosphorylation by Pyramidalization and Polarization. ACS Catal. 2024, 14, 6650–6658. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Galkin, A.; Herzberg, O.; Dunaway-Mariano, D. Arginine Deiminase Uses an Active-Site Cysteine in Nucleophilic Catalysis of L-Arginine Hydrolysis. J. Am. Chem. Soc. 2004, 126, 5374–5375. [Google Scholar] [CrossRef] [PubMed]
- Novichkov, A.I.; Hanopolskyi, A.I.; Miao, X.; Shimon, L.J.W.; Diskin-Posner, Y.; Semenov, S.N. Autocatalytic and Oscillatory Reaction Networks That Form Guanidines and Products of Their Cyclization. Nat. Commun. 2021, 12, 2994. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.; Mauguen, Y.; Prangé, T. Revisiting Glutaraldehyde Cross-Linking: The Case of the Arg-Lys Intermolecular Doublet. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 225–228. [Google Scholar] [CrossRef]
- Fitch, C.A.; Platzer, G.; Okon, M.; Garcia-Moreno, B.E.; McIntosh, L.P. Arginine: Its pKa Value Revisited. Protein Sci. 2015, 24, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Grundler, V.; Gademann, K. Direct Arginine Modification in Native Peptides and Application to Chemical Probe Development. ACS Med. Chem. Lett. 2014, 5, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Vazdar, K.; Margetić, D.; Kovačević, B.; Sundermeyer, J.; Leito, I.; Jahn, U. Design of Novel Uncharged Organic Superbases: Merging Basicity and Functionality. Acc. Chem. Res. 2021, 54, 3108–3123. [Google Scholar] [CrossRef] [PubMed]
- McGee, W.M.; Mentinova, M.; McLuckey, S.A. Gas-Phase Conjugation to Arginine Residues in Polypeptide Ions via N-Hydroxysuccinimide Ester-Based Reagent Ions. J. Am. Chem. Soc. 2012, 134, 11412–11414. [Google Scholar] [CrossRef]
- Awoonor-Williams, E.; Rowley, C.N. How Reactive Are Druggable Cysteines in Protein Kinases? J. Chem. Inf. Model. 2018, 58, 1935–1946. [Google Scholar] [CrossRef]
- Liu, R.; Zhan, S.; Che, Y.; Shen, J. Reactivities of the Front Pocket N-Terminal Cap Cysteines in Human Kinases. J. Med. Chem. 2022, 65, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Varanasi, L.; Hosler, J. Alternative Initial Proton Acceptors for the D Pathway of Rhodobacter sphaeroides Cytochrome c Oxidase. Biochemistry 2011, 50, 2820–2828. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, H.; Yang, S.; Saravanamurugan, S.; Riisager, A. Glucose Isomerization by Enzymes and Chemo-Catalysts: Status and Current Advances. ACS Catal. 2017, 7, 3010–3029. [Google Scholar] [CrossRef]
- Barnett, Z.M.E.; Feketeová, L.; O’Hair, R.A.J. The Major Product Ion of S-Adenosyl-L-Methionine Arises from a Neighboring Group Reaction. Rapid Commun. Mass. Spectrom. 2010, 24, 1387–1391. [Google Scholar] [CrossRef]
- Jiang, C.; Arthur, C.J.; Gates, P.J. A Computational and Experimental Study of the Fragmentation of L-Leucine, l-Isoleucine and l-Allo-Isoleucine under Collision-Induced Dissociation Tandem Mass Spectrometry. Analyst 2020, 145, 6632–6638. [Google Scholar] [CrossRef]

| Mark | Ion Type (Notation) | Moiety Lost or Gained |
|---|---|---|
| 162 NL | [M + zH+ − 162]/z | 1,2-anhydroglucose loss |
| NL ions | [M + zH+ − 180; −150; −120; −90]/z | hexose, pentose, tetrose, triose loss, respectively |
| 179 NL | [M + zH+ − 179]/z | aminoglucose/aminofructose loss |
| 204 NL | [M + zH+ − 204/z | 1,2-anhydroglucose loss + carbodiimide loss |
| NL ions | [M + zH+ − nH2O]/z, n = 1, 2, 3 | water loss, −18 Da, −36 Da, −54 Da |
| 162 NG | y+/b+ [+162] | glucosyl gain |
| 144 NGetc. | y+/b+ [+144; +126; +108] | loss of 1, 2, and 3 water molecules from 162 NG ions |
| 78 NGetc. | y+/b+ [+78; +72; +42] | loss of (3H2O + CH2O), triose, tetrose from 162 NG ions |
| [b-3H2O]+ | b1 | b2 | b3 | b4 | b5 | b6 | b7 | b8 | b9 | b10 | 0.20 | Path C | ||
| [b-2H2O]+ | b2 | b3 | b4 | b5 | b6 | b7 | b8 | b9 | b10 | 0.82 | ||||
| [b-H2O]+ | b2 | b3 | b10 | 0.03 | ||||||||||
| [b-150]+ | b3 | b4 | b5 | b6 | b8 | b9 | 0.004 | Path B | ||||||
| [b-90]+ | b2 | b3 | b4 | b5 | b6 | b9 | b10 | 0.22 | ||||||
| [b-120]+ | b2 | b3 | b5 | b6 | b7 | b9 | b10 | 0.24 | Path A | |||||
| [b]+ | b1 | b2 | b3 | b4 | b5 | b6 | b7 | b8 | b9 | b10 | # | 1.00 Total abundance | ||
| Total abundance 1.00 | CGlc | K | G | T | D | V | Q | A | W | I | R | |||
| # | y9 | y8 | y7 | y6 | y5 | y4 | y3 | y2 | [y]+ | |||||
| Path A | 0.02 | y9 | y7 | y5 | y3 | [y+162]+ | ||||||||
| 0.10 | y8 | y7 | y4 | y3 | y1 | [y+42]+ | ||||||||
| 0.04 | y7 | y6 | y4 | y3 | y2 | [y+24]+ | ||||||||
| Path B | 0.03 | y9 | y6 | y4 | y3 | y1 | [y+72]+ | |||||||
| 0.02 | y8 | y7 | y5 | y3 | y2 | y1 | [y+12]+ | |||||||
| 0.01 | y9 | y3 | y2 | y1 | [y+54]+ | |||||||||
| Path C | 0.03 | y7 | y4 | y3 | [y+144]+ | |||||||||
| 0.04 | y5 | y4 | y3 | y2 | y1 | [y+126]+ | ||||||||
| 0.03 | y10 | y9 | y7 | y4 | y3 | y2 | [y+108]+ | |||||||
| 0.01 | y4 | y1 | [y+78]+ | |||||||||||
| [b+108]+ | b1 | b3 | b4 | b5 | b9 | 4.37 | Path C | ||||||
| [b+126]+ | b1 | b3 | b6 | b7 | b8 | b9 | 0.28 | ||||||
| [b+144]+ | b1 | b3 | b5 | 0.48 | |||||||||
| [b+12]+ | b3 | b4 | b5 | b6 | b8 | b9 | 0.13 | Path B | |||||
| [b+72]+ | b2 | b3 | b4 | b5 | b6 | b9 | b10 | 0.33 | |||||
| [b+42]+ | b2 | b5 | b6 | b7 | b9 | b10 | 0.68 | Path A | |||||
| [b+162]+ | b2 | b3 | b8 | 1.00 Total abundance | |||||||||
| CGlc | K | G | T | D | V | Q | A | W | I | R | |||
| Transformations | Diagnostic Ion(s) | Path |
|---|---|---|
| N-glucosylation of Arg | [y+162]+ | A |
| N-glucosylation of Arg | [y+42; y+24]+ | A |
| N-fructosylation of Arg | [y+72; y+54; y+12]+ | B |
| N-fructosylation of Arg | [y+144; y+126; y+108; y+78]+ | C |
| S→N (Glc+) migration and tetrose loss | [b-120]+ | A |
| S→N (Glc+) migration and triose loss | [b-90]+ | B |
| S→N (Glc+) migration and pentose loss | [b-150]+ | B |
| [b-3H2O]++ | b2 | b3 | b4 | b5 | b6 | b7 | b10 | 0.15 | Path C | |||||
| [b-2H2O]++ | b4 | b6 | b7 | b10 | 0.10 | |||||||||
| [b-H2O]++ | b1 | b3 | b4 | b5 | b6 | b7 | b9 | 0.26 | ||||||
| [b-150]++ | b6 | b7 | b9 | b10 | 0.02 | Path B | ||||||||
| [b-90]++ | b2 | b3 | b5 | b6 | b7 | b9 | b10 | 0.29 | ||||||
| [b-120]++ | b4 | b5 | b6 | b7 | b10 | 0.14 | Path A | |||||||
| [b]++ | b5 | b6 | b7 | 1.00 | ||||||||||
| [b-3H2O]+ | b1 | b2 | b3 | b4 | b5 | b6 | b8 | 0.16 | Path C | |||||
| [b-2H2O]+ | b1 | b2 | b4 | b5 | b6 | 0.70 | ||||||||
| [b-H2O]+ | b1 | b2 | b5 | b6 | b7 | b8 | 0.06 | |||||||
| [b-150]+ | b2 | b3 | b4 | b5 | b6 | b7 | 0.81 | Path B | ||||||
| [b-90]+ | b1 | b2 | b3 | b5 | b6 | b7 | b8 | 0.13 | ||||||
| [b-120]+ | b1 | b2 | b3 | b4 | b5 | b6 | b7 | b8 | 0.41 | Path A | ||||
| [b]+ | b1 | b2 | b3 | b4 | b5 | b6 | b7 | b8 | # | 1.00 Total abundance | ||||
| Total abundance 1.00 | CGlc | K | G | T | D | V | Q | A | W | I | R | |||
| # | y7 | y6 | y5 | y4 | y2 | y1 | [y]+ | |||||||
| Path A | 0.02 | y3 | y2 | [y+162]+ | ||||||||||
| 0.06 | y8 | y5 | y4 | y3 | y2 | y1 | [y+42]+ | |||||||
| 0.03 | y8 | y6 | y5 | y3 | y2 | y1 | [y+24]+ | |||||||
| Path B | 0.14 | y4 | y3 | [y+72]+ | ||||||||||
| 0.07 | y7 | y6 | y3 | y2 | y1 | [y+12]+ | ||||||||
| 0.02 | y5 | y1 | [y+54]+ | |||||||||||
| Path C | 0.02 | y3 | y2 | [y+144]+ | ||||||||||
| 0.04 | y3 | y2 | y1 | [y+126]+ | ||||||||||
| 0.03 | y3 | y2 | y1 | [y+108]+ | ||||||||||
| 0.01 | y3 | y1 | [y+78]+ | |||||||||||
| 1.00 | y10 | y7 | y6 | y5 | y4 | y3 | y2 | [y]++ | ||||||
| Path A | 0.20 | y10 | y9 | y8 | y7 | y5 | y4 | y2 | y1 | [y+162]++ | ||||
| 0.17 | y6 | y5 | y4 | y3 | y2 | [y+42]++ | ||||||||
| 0.12 | y9 | y8 | y6 | y5 | y4 | y3 | [y+24]++ | |||||||
| Path B | 0.10 | y9 | y6 | y5 | y4 | y3 | y1 | [y+72]++ | ||||||
| 0.19 | y10 | y9 | y8 | y7 | y6 | y5 | [y+12]++ | |||||||
| 0.08 | y9 | y6 | y4 | [y+54]++ | ||||||||||
| Path C | 0.11 | y8 | y7 | y4 | y3 | y2 | y1 | [y+144]++ | ||||||
| 0.17 | y10 | y9 | y8 | y7 | y5 | y3 | [y+126]++ | |||||||
| 0.04 | y10 | y9 | y7 | y4 | y2 | y1 | [y+108]++ | |||||||
| 0.14 | y9 | y8 | y6 | y5 | y4 | y2 | y1 | [y+78]++ | ||||||
| [b+108]++ | b1 | b4 | b5 | b6 | b7 | b8 | b9 | b10 | 13.23 | Path C | |||
| [b+126]++ | b2 | b3 | b4 | b5 | b6 | b7 | b8 | b10 | 7.82 | ||||
| [b+144]++ | b2 | b3 | b4 | b5 | b7 | b8 | b9 | b10 | 2.41 | ||||
| [b+12]++ | b2 | b3 | b4 | b5 | b8 | b9 | 2.45 | Path B | |||||
| [b+72]++ | b1 | b2 | b5 | b6 | b8 | b9 | b10 | 2.67 | |||||
| [b+42]++ | b4 | b9 | 0.33 | Path A | |||||||||
| [b+162]++ | b1 | b2 | b3 | b6 | b8 | b9 | 1.00 | ||||||
| [b+108]+ | b1 | b2 | b3 | b4 | b5 | 1.27 | Path C | ||||||
| [b+126]+ | b1 | b3 | 1.64 | ||||||||||
| [b+144]+ | b1 | b2 | b3 | b5 | 0.57 | ||||||||
| [b+12]+ | b1 | b2 | b3 | b4 | b6 | 1.15 | Path B | ||||||
| [b+72]+ | b1 | b4 | b7 | 0.70 | |||||||||
| [b+42]+ | b1 | b2 | b6 | 0.89 | Path A | ||||||||
| [b+162]+ | b1 | b2 | b3 | b4 | b5 | 1.00 Total abundance | |||||||
| CGlc | K | G | T | D | V | Q | A | W | I | R | |||
| Transformations | Diagnostic ion(s) | Path |
|---|---|---|
| N-glucosylation of Arg | [y+162]+/++ | A |
| N-glucosylation of Arg | [y+42; y+24]+/++ | A |
| N-fructosylation of Arg | [y+72; y+54; y+12]+/++ | B |
| N-fructosylation of Arg | y+144; y+126; y+108; y+78 | C |
| S→N (Glc+) migration and tetrose loss | [b-120]+/++ | A |
| S→N (Glc+) migration and triose loss | [b-90]+/++ | B |
| S→N (Glc+) migration and pentose loss | [b-150]+/++ | B |
| Entry | Notation | Monoisotopic Mass [Da] | Chemical Formula | Chemical Formula Calculation |
|---|---|---|---|---|
| 1 | 204 | 204.074623 | C7H12O5N2 | 1,2-anhydro-Glc + carbodiimide |
| 2 | 180 | 180.063390 | C6H12O6 | Glc |
| 3 | 179 | 179.079374 | C6H13O5N | 1-amino-Glc |
| 4 | 162 | 162.052824 | C6H10O5 | 1,2-anhydro-Glc |
| 5 | 150 | 150.052824 | C5H10O5 | pentose |
| 6 | 144 | 144.042260 | C6H8O4 | 162 − H2O |
| 7 | 126 | 126.031695 | C6H6O3 | 162 − 2H2O |
| 8 | 120 | 120.042260 | C4H8O4 | tetrose |
| 9 | 108 | 108.021130 | C6H4O2 | 162 − 3H2O |
| 10 | 90 | 90.031695 | C3H6O3 | triose |
| 11 | 78 | 78.010565 | C5H2O | 162 − 3H2O − CH2O |
| 12 | 72 | 72.021130 | C3H4O2 | 162 − triose |
| 13 | 54 | 54.031695 | H6O3 | 3H2O |
| 14 | 54 | 54.010565 | C3H2O | 162 − triose − H2O |
| 15 | 42 | 42.021798 | CH2N2 | carbodiimide |
| 16 | 42 | 42.010565 | C2H2O | 162 − tetrose |
| 17 | 36 | 36.021130 | H4O2 | 2H2O |
| 18 | 24 | 24.000000 | C2 | 162 − tetrose − H2O |
| 19 | 18 | 18.010565 | H2O | H2O |
| 20 | 12 | 12.00000 | C | C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buchowiecka, A.K. Evidence of Gas Phase Glucosyl Transfer and Glycation in the CID/HCD-Spectra of S-Glucosylated Peptides. Int. J. Mol. Sci. 2024, 25, 7483. https://doi.org/10.3390/ijms25137483
Buchowiecka AK. Evidence of Gas Phase Glucosyl Transfer and Glycation in the CID/HCD-Spectra of S-Glucosylated Peptides. International Journal of Molecular Sciences. 2024; 25(13):7483. https://doi.org/10.3390/ijms25137483
Chicago/Turabian StyleBuchowiecka, Alicja K. 2024. "Evidence of Gas Phase Glucosyl Transfer and Glycation in the CID/HCD-Spectra of S-Glucosylated Peptides" International Journal of Molecular Sciences 25, no. 13: 7483. https://doi.org/10.3390/ijms25137483
APA StyleBuchowiecka, A. K. (2024). Evidence of Gas Phase Glucosyl Transfer and Glycation in the CID/HCD-Spectra of S-Glucosylated Peptides. International Journal of Molecular Sciences, 25(13), 7483. https://doi.org/10.3390/ijms25137483






