Metabolic and Transcriptomic Profile Revealing the Differential Accumulating Mechanism in Different Parts of Dendrobium nobile
Abstract
1. Introduction
2. Results
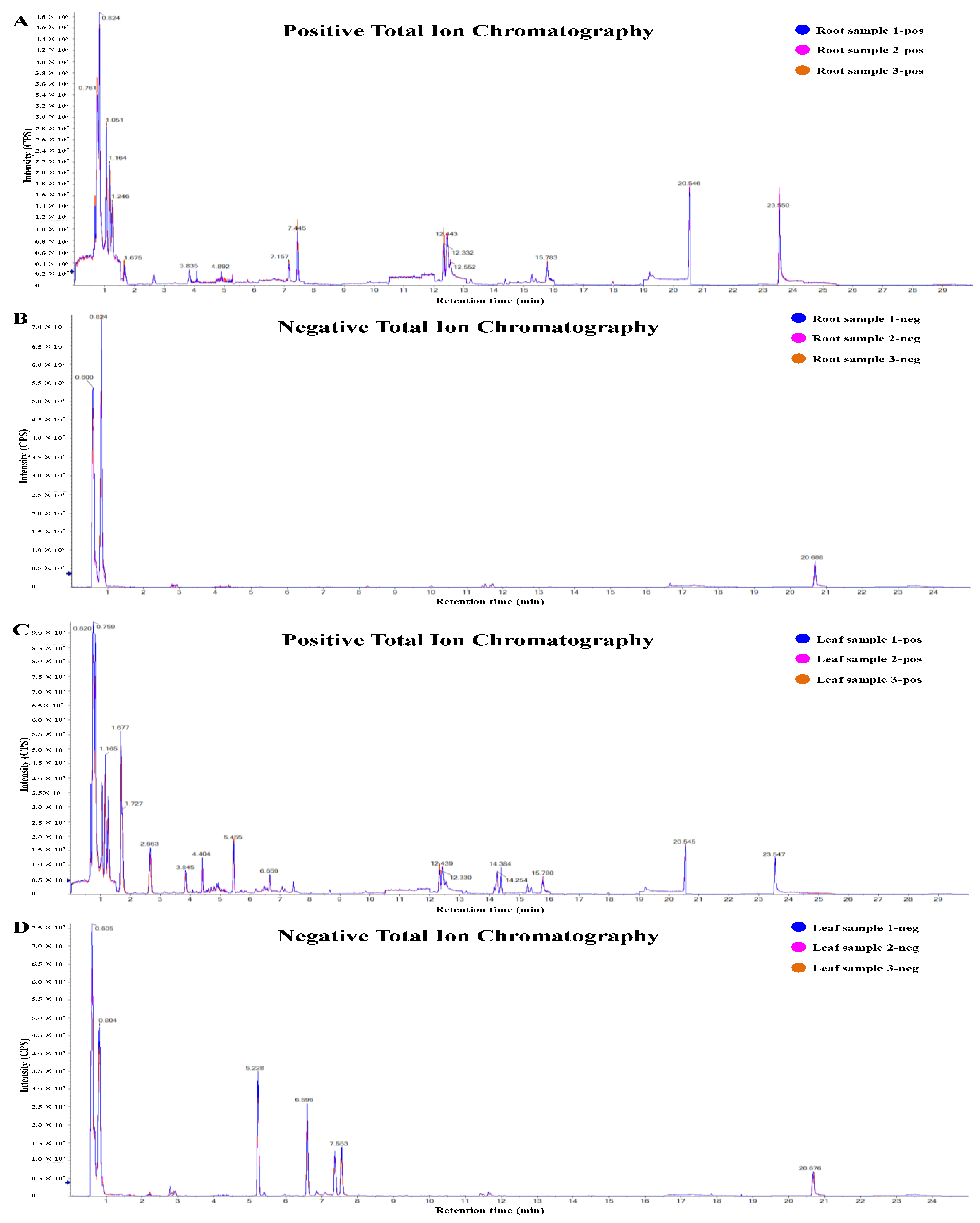
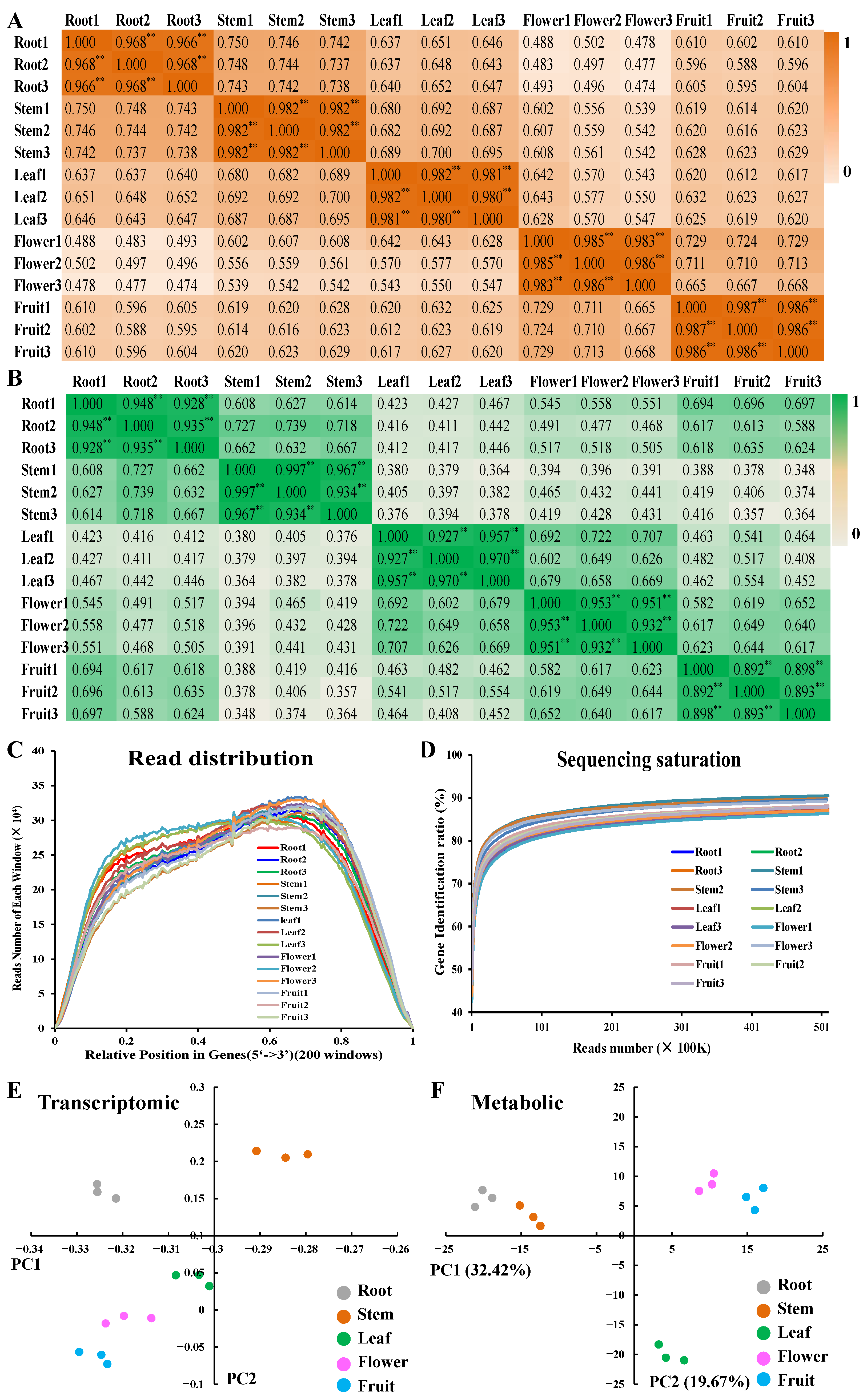
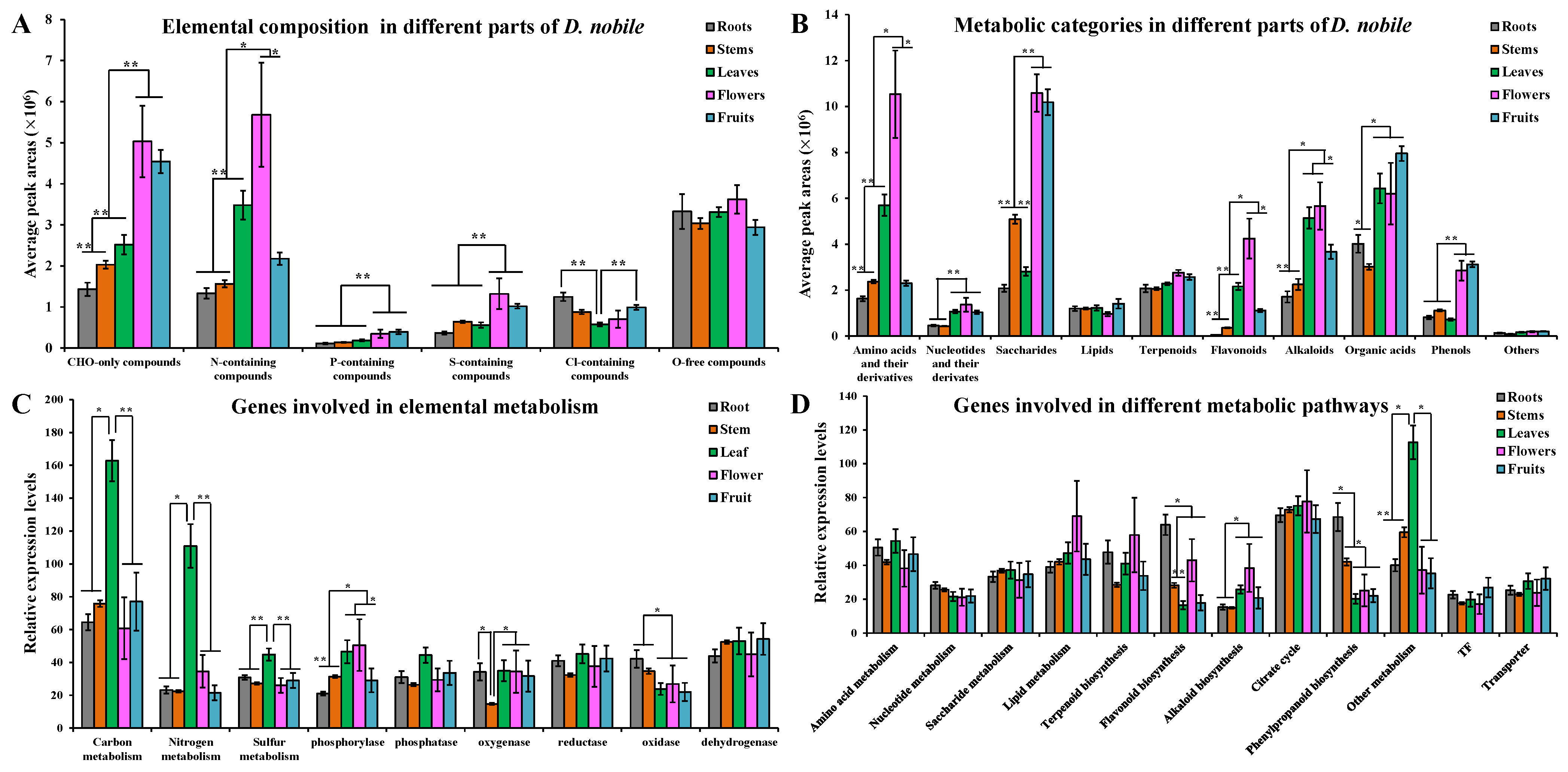
2.1. Overview of Metabolics and Transcriptomics in Different Parts of D. nobile
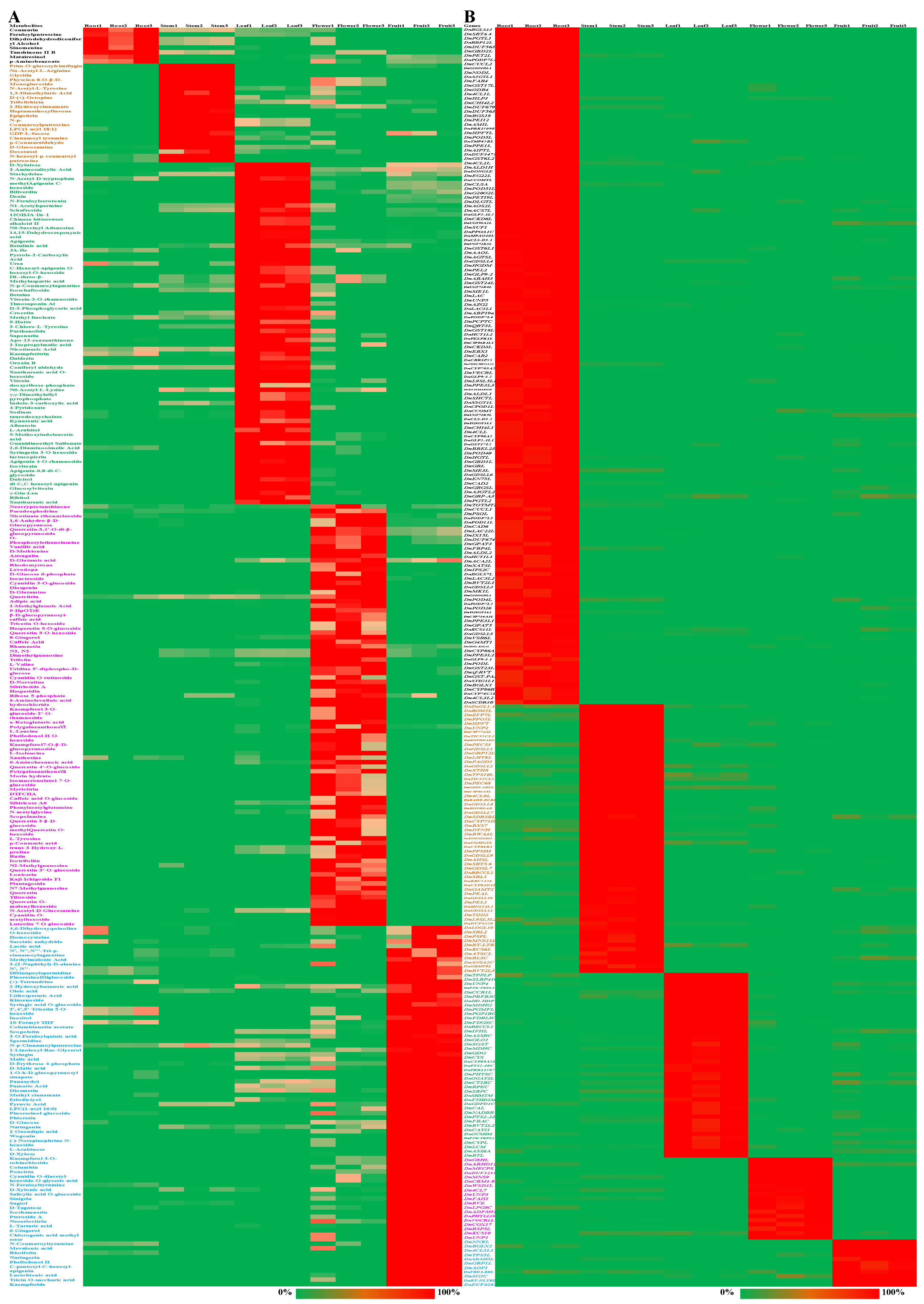
2.2. Differentially Accumulated Metabolites and Differentially Expressed Genes in Each Part of D. nobile
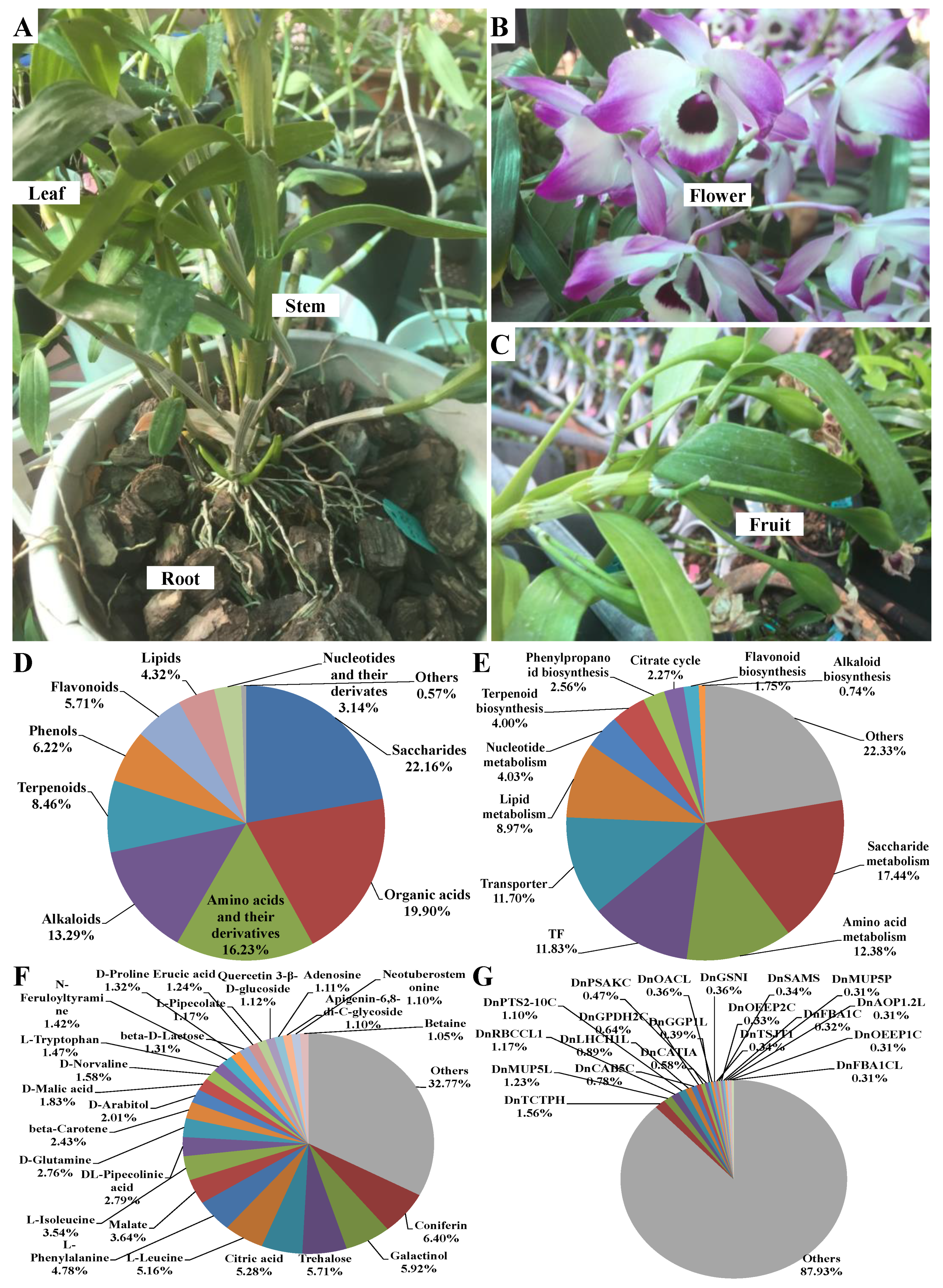
2.3. Distribution of Different Metabolic Categories and Expression of Genes Involved in Different Metabolic Pathways
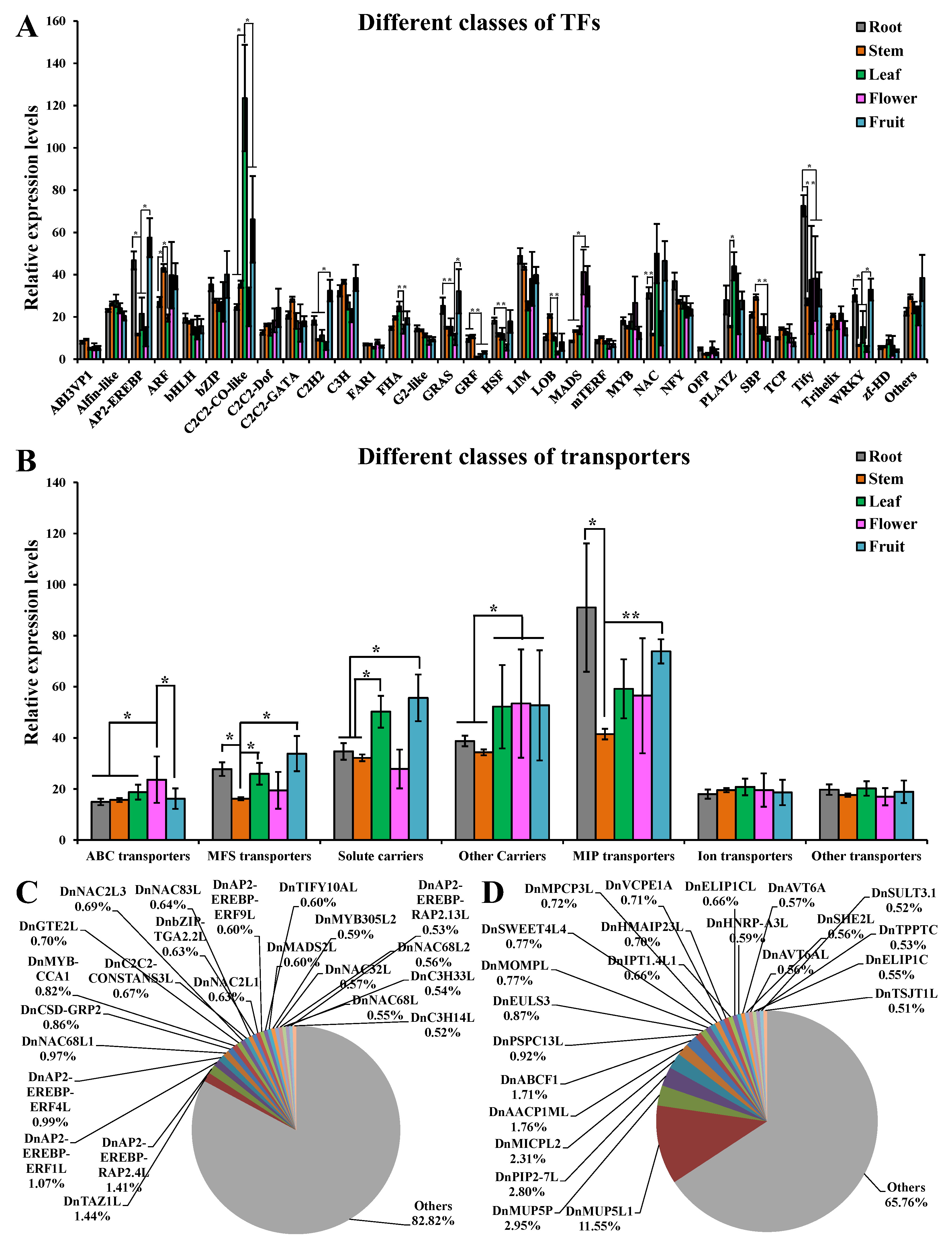
2.4. Expression of TFs and Transporters in Each Part of D. nobile
2.5. Extremely Significant Associations between Different Metabolic Categories and Gene Classes
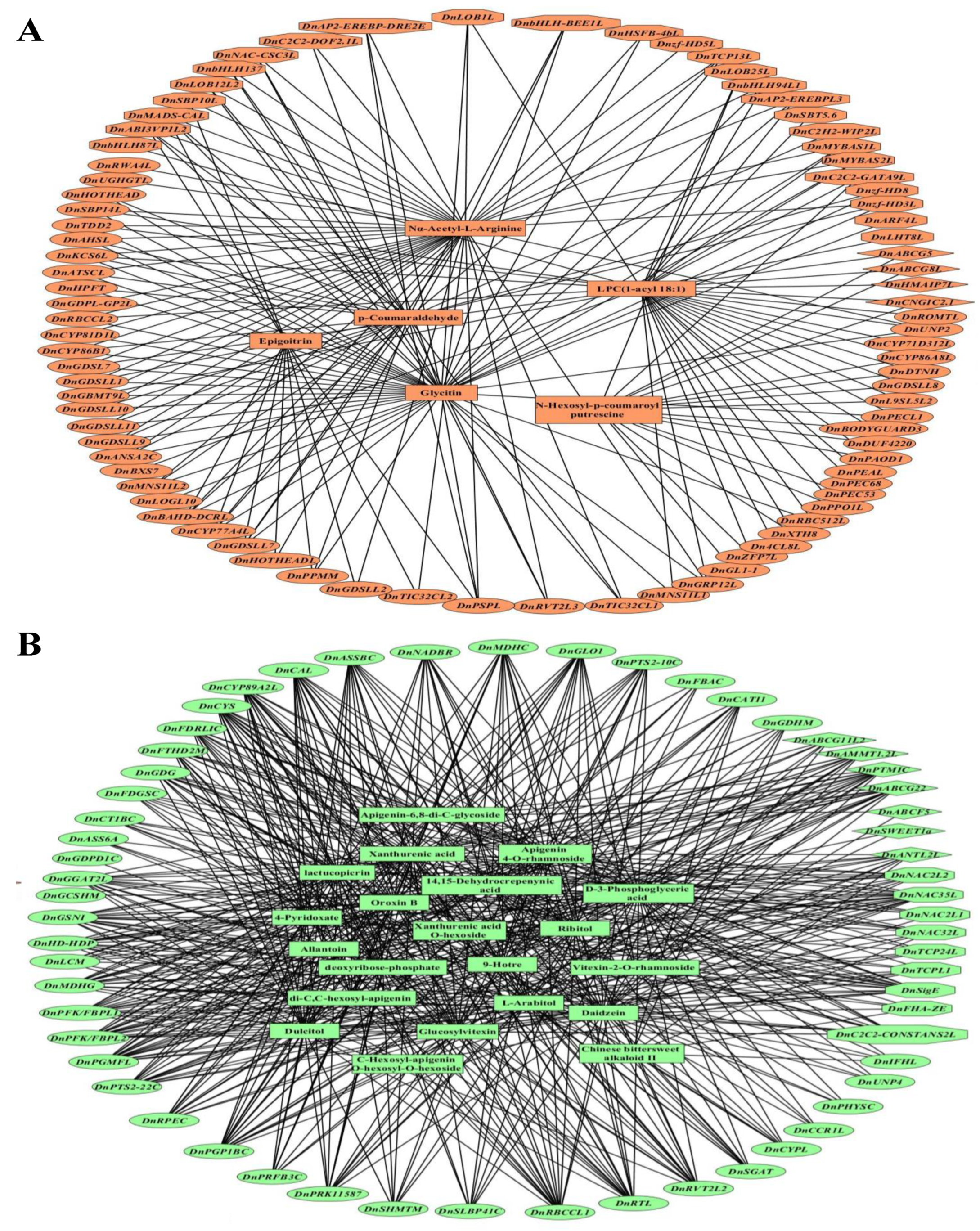
2.6. Extremely Significant Associations between DAMs and DEGs
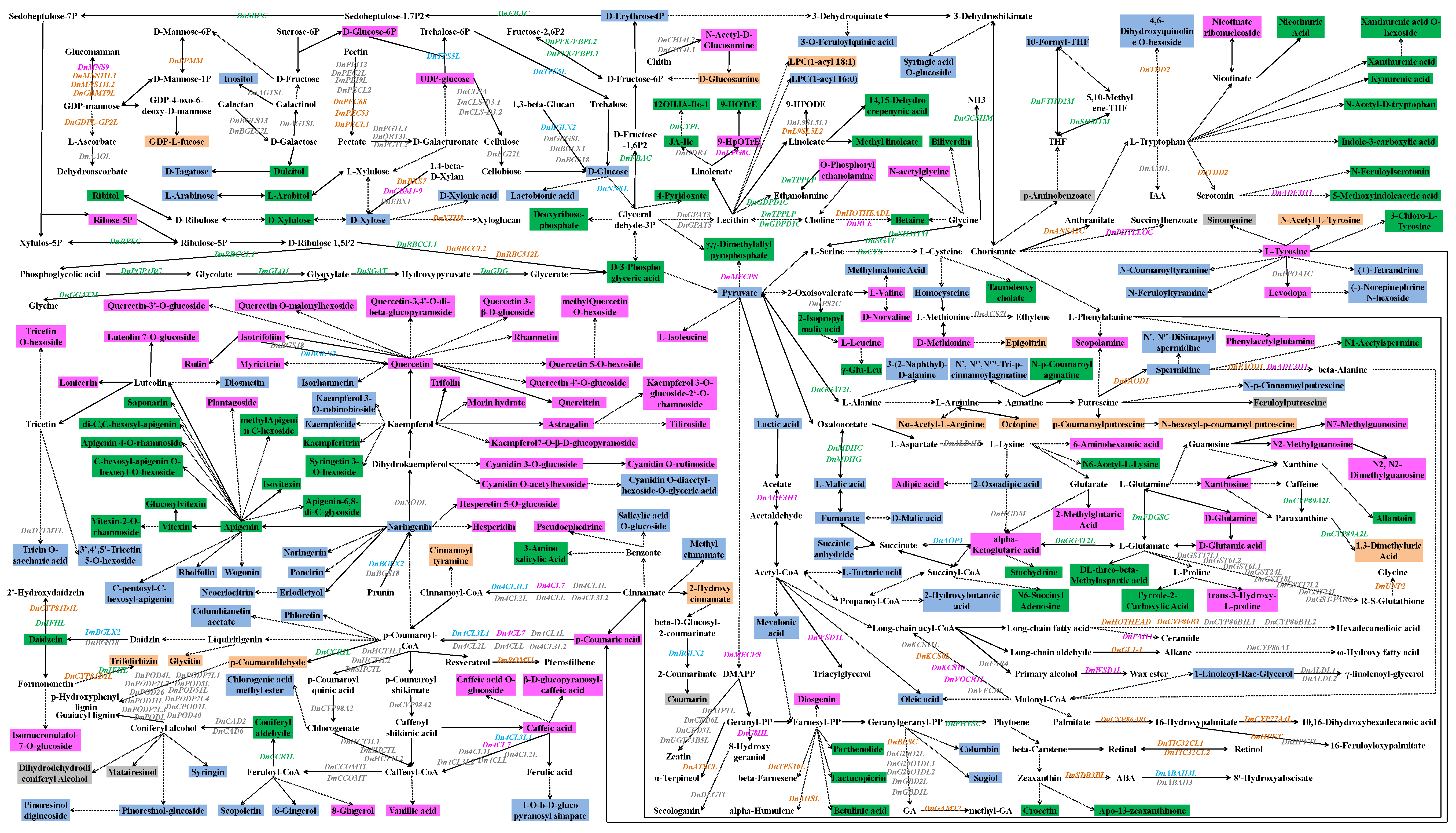
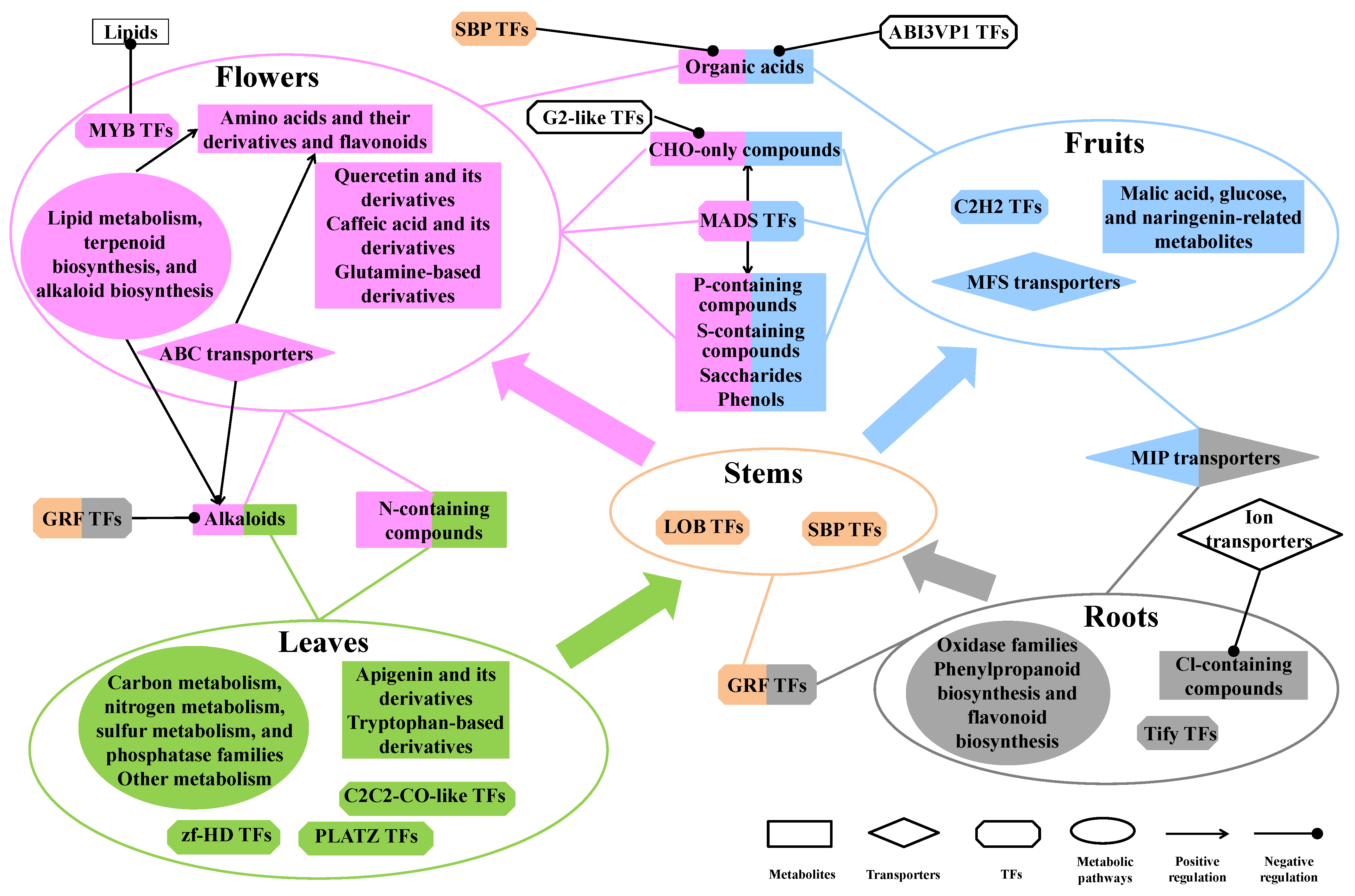
2.7. Biosynthesis, Regulation, and Transporting of DAMs in Different Parts of D. nobile
3. Discussion
3.1. Different Members of Metabolic Enzyme Families Showed Differential Effects on Tissue-Specific Metabolic Accumulation
3.2. Many TF Families Participated Widely in Tissue-Specific Metabolic Accumulation
3.3. ABC Transporters and Some Other Transporters also Showed Positive Effects on Tissue-Specific Metabolic Accumulation
4. Materials and Methods
4.1. Plant Materials
4.2. Metabolite Extraction for HPLC-MS/MS
4.3. HPLC-MS/MS Analysis
4.4. novoDB Database of Standards
4.5. Metabolites Identification by Multiple Reaction Monitoring
4.6. Metabolite Quantification
4.7. Metabolite Annotation and Classification
4.8. Total RNA Extraction
4.9. RNA-Seq
4.10. Genes Annotation
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheng, J.; Dang, P.P.; Zhao, Z.; Yuan, L.C.; Zhou, Z.H.; Wolf, D.; Luo, Y.B. An assessment of the Chinese medicinal Dendrobium industry: Supply.; demand and sustainability. J. Ethnopharmacol. 2019, 229, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Lin, Y.; Farag, M.A.; Li, Z.; Shao, P. Dendrobium as a new natural source of bioactive for the prevention and treatment of digestive tract diseases: A comprehensive review with future perspectives. Phytomedicine 2023, 114, 154784. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Sun, X.; Wang, X.; Yu, H. Therapeutic potential of the chemical composition of Dendrobium nobile Lindl. Front. Pharmacol. 2023, 14, 1163830. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, L.; Chang, H.; Cai, B.; Gao, H.; Chen, G.; Hou, W.; Jappar, Z.; Yan, Y. Research progress on extraction, purification, structure and biological activity of Dendrobium officinale polysaccharides. Front. Nutr. 2022, 9, 965073. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Er-Bu, A.; Dongzhi, Z.; Xie, H.; Ye, B.; He, J. Alkaloids from Dendrobium and their biosynthetic pathway, biological activity and total synthesis. Phytomedicine 2022, 102, 154132. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.; Hu, Y.; Zhao, R.; Li, H.; Chun, Z.; Zheng, S. Quantitative identification of antioxidant basis for Dendrobium nobile flower by high performance liquid chromatography-tandem mass spectrometry. Int. J. Anal. Chem. 2022, 2022, 9510598. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.; Zhao, R.; Hu, Y.; Li, H.; Chun, Z.; Zheng, S. Revealing of intracellular antioxidants in Dendrobium nobile by high performance liquid chromatography-tandem mass spectrometry. Metabolites 2023, 13, 702. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.G.; Hu, Y.D.; Zhao, R.X.; Yan, S.; Zhang, X.Q.; Zhao, T.M.; Chun, Z. Genome-wide researches and applications on Dendrobium. Planta 2018, 248, 769–784. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Wang, G.; Ma, J.; Liu, Y.; Xu, E.; Luo, A. Transcriptome analysis reveals the role of polysaccharide biosynthesis in the detoxification of Dendrobium nobile under zinc stress. Int. J. Biol. Macromol. 2023, 252, 126406. [Google Scholar] [CrossRef]
- Xu, Q.; Niu, S.C.; Li, K.L.; Zheng, P.J.; Zhang, X.J.; Jia, Y.; Liu, Y.; Niu, Y.X.; Yu, L.H.; Chen, D.F.; et al. Chromosome-Scale assembly of the Dendrobium nobile genome provides insights into the molecular mechanism of the biosynthesis of the medicinal active ingredient of Dendrobium. Front. Genet. 2022, 13, 844622. [Google Scholar] [CrossRef]
- Li, Q.; Ding, G.; Li, B.; Guo, S.X. Transcriptome analysis of genes involved in dendrobine biosynthesis in Dendrobium nobile Lindl. infected with Mycorrhizal fungus MF23 (Mycena sp.). Sci. Rep. 2017, 7, 316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Shi, Y.C.; Zhang, S.B. Metabolic and transcriptomic analyses elucidate a novel insight into the network for biosynthesis of carbohydrate and secondary metabolites in the stems of a medicinal orchid Dendrobium nobile. Plant Divers. 2022, 45, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Shi, M.; Zhang, Q.; Xie, W.; Chu, L.; Qiu, M.; Li, L.; Zeng, Z.; Han, L.; Sun, Z. Transcriptomic and metabolomic analyses reveal differences in flavonoid pathway gene expression profiles between two Dendrobium varieties during vernalization. Int. J. Mol. Sci. 2023, 24, 11039. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Cai, C.; Mo, X.; Zhao, X.; Wu, L.; Liu, F.; Li, R.; Liu, C.; Chen, J.; Tian, M. Transcriptome and metabolome analysis reveals the effect of flavonoids on flower color variation in Dendrobium nobile Lindl. Front. Plant Sci. 2023, 14, 1220507. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.; Li, B.; Wu, B.; Fu, D.; Li, Z.; Wei, H.; Guo, S.; Ding, G.; Wang, B. The integration of the metabolome and transcriptome for Dendrobium nobile Lindl. in response to methyl jasmonate. Molecules 2023, 28, 7892. [Google Scholar] [CrossRef] [PubMed]
- Li, K.L.; Liang, Y.M.; Chen, Z.; Zheng, P.J.; Zhang, G.Q.; Yan, B.; Elshikh, M.S.; Rizwana, H.; Chen, B.; Xu, Q. Genome-wide identification of the alkaloid synthesis gene family CYP450, gives new insights into alkaloid resource utilization in medicinal Dendrobium. Int. J. Biol. Macromol. 2024, 259, 129229. [Google Scholar] [CrossRef] [PubMed]
- Lavhale, S.G.; Kalunke, R.M.; Giri, A.P. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 2018, 248, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhang, S.; Zheng, Z.; Maoz, I.; Zhang, L.; Kai, G. Molecular regulation of the key specialized metabolism pathways in medicinal plants. J. Integr. Plant Biol. 2024, 66, 510–531. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, W.; Sabir, I.A.; Jiao, C.; Li, G.; Wang, Y.; Zhu, F.; Dai, J.; Liu, L.; Chen, C.; et al. The spatiotemporal profile of Dendrobium huoshanense and functional identification of bHLH genes under exogenous MeJA using comparative transcriptomics and genomics. Front. Plant Sci. 2023, 14, 1169386. [Google Scholar] [CrossRef]
- Li, D.; Liu, L.; Li, X.; Wei, G.; Cai, Y.; Sun, X.; Fan, H. DoAP2/ERF89 activated the terpene synthase gene DoPAES in Dendrobium officinale and participated in the synthesis of beta-patchoulene. PeerJ 2024, 12, e16760. [Google Scholar] [CrossRef]
- Zhao, M.; Zhao, Y.; Yang, Z.; Ming, F.; Li, J.; Kong, D.; Wang, Y.; Chen, P.; Wang, M.; Wang, Z. Metabolic pathway engineering improves dendrobine production in Dendrobium catenatum. Int. J. Mol. Sci. 2023, 25, 397. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Hou, Y.; Wu, M.; Pan, Q.; Xie, Y.; Zhang, Y.; Sun, F.; Zhang, Z.; Wu, J. DoMYB5 and DobHLH24, transcription factors involved in regulating anthocyanin accumulation in Dendrobium officinale. Int. J. Mol. Sci. 2023, 24, 7552. [Google Scholar] [CrossRef]
- Yuan, Y.; Zuo, J.; Zhang, H.; Zu, M.; Liu, S. Analysis of the different growth years accumulation of flavonoids in Dendrobium moniliforme (L.) Sw. by the integration of metabolomic and transcriptomic approaches. Front. Nutr. 2022, 9, 928074. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Wei, M.; Fan, H.; Song, C.; Wang, Z.; Cai, Y.; Jin, Q. Transcriptomic analysis of genes related to alkaloid biosynthesis and the regulation mechanism under precursor and methyl jasmonate treatment in Dendrobium officinale. Front. Plant Sci. 2022, 13, 941231. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Jin, Y.; Wang, W.; Xia, K.; Chen, Z. Molecular and metabolic insights into floral scent biosynthesis during flowering in Dendrobium chrysotoxum. Front. Plant Sci. 2022, 13, 1030492. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Alonso, A.; Yepes-Molina, L.; Guarnizo, A.L.; Carvajal, M. Modification of gene expression of tomato plants through foliar flavonoid application in relation to enhanced growth. Genes 2023, 14, 2208. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ortiz, C.; Dutta, S.K.; Natarajan, P.; Peña-Garcia, Y.; Abburi, V.; Saminathan, T.; Nimmakayala, P.; Reddy, U.K. Genome-wide identification and gene expression pattern of ABC transporter gene family in Capsicum spp. PLoS ONE 2019, 14, e0215901. [Google Scholar] [CrossRef]
- Mi, Y.; Cao, X.; Zhu, X.; Chen, W.; Meng, X.; Wan, H.; Sun, W.; Wang, S.; Chen, S. Characterization and co-expression analysis of ATP-binding cassette transporters provide insight into genes related to cannabinoid transport in Cannabis sativa L. Int. J. Biol. Macromol. 2023, 242, 124934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, G.Q.; Zhang, D.; Liu, X.D.; Xu, X.Y.; Sun, W.H.; Yu, X.; Zhu, X.; Wang, Z.W.; Zhao, X.; et al. Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res. 2021, 8, 183. [Google Scholar] [CrossRef]
- Gao, J.N.; Li, Y.; Liang, J.; Chai, J.H.; Kuang, H.X.; Xia, Y.G. Direct acetylation for full analysis of polysaccharides in edible plants and fungi using reverse phase liquid chromatography-multiple reaction monitoring mass spectrometry. J. Pharm. Biomed. Anal. 2023, 222, 115083. [Google Scholar] [CrossRef]
- Luo, Z.; Liu, L.; Nie, Q.; Huang, M.; Luo, C.; Sun, Y.; Ma, Y.; Yu, J.; Du, F. HPLC-based metabolomics of Dendrobium officinale revealing its antioxidant ability. Front. Plant Sci. 2023, 14, 1060242. [Google Scholar] [CrossRef] [PubMed]


| Name | Amino Acids and Their Derivatives | Nucleotides and their Derivatives | Saccharides | Lipids | Terpenoids | Flavonoids | Alkaloids | Organic acids | Phenols | Other Categories | CHO-Only Compounds | N-Containing Compounds | P-Containing Compounds | S-Containing Compounds | Cl-Containing Compounds | O-Free Compounds |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | 0.016 | 0.082 | −0.623 | 0.174 | −0.301 | 0.012 | 0.291 | 0.126 | −0.624 | −0.066 | −0.389 | 0.025 | −0.386 | −0.504 | −0.507 | 0.069 |
| Metabolic genes | 0.145 | 0.043 | −0.658 | −0.068 | −0.348 | 0.094 | 0.303 | −0.070 | −0.707 | −0.215 | −0.441 | 0.119 | −0.502 | −0.488 | −0.587 | 0.222 |
| Carbon metabolism | 0.068 | 0.208 | −0.449 | 0.236 | −0.165 | 0.093 | 0.407 | 0.263 | −0.468 | 0.037 | −0.222 | 0.095 | −0.212 | −0.344 | −0.633 | −0.028 |
| Nitrogen metabolism | 0.310 | 0.358 | −0.400 | −0.018 | −0.030 | 0.315 | 0.565 | 0.277 | −0.429 | 0.130 | −0.129 | 0.325 | −0.176 | −0.229 | −0.719 | 0.256 |
| Sulfur metabolism | −0.009 | 0.126 | −0.586 | 0.246 | −0.245 | 0.007 | 0.301 | 0.228 | −0.560 | 0.035 | −0.338 | 0.017 | −0.312 | −0.483 | −0.455 | 0.050 |
| Phosphorylase | 0.913 * | 0.802 | 0.337 | −0.585 | 0.598 | 0.912 * | 0.936 ** | 0.374 | 0.219 | 0.421 | 0.533 | 0.914 * | 0.358 | 0.565 | −0.931 * | 0.606 |
| Phosphatase | 0.098 | 0.373 | −0.331 | 0.308 | 0.038 | 0.165 | 0.476 | 0.524 | −0.276 | 0.344 | −0.056 | 0.161 | −0.006 | −0.245 | −0.486 | 0.059 |
| Oxygenase | 0.384 | 0.588 | 0.065 | −0.085 | 0.479 | 0.446 | 0.492 | 0.634 | 0.197 | 0.735 | 0.318 | 0.449 | 0.331 | 0.159 | −0.059 | 0.544 |
| Reductase | 0.003 | 0.394 | −0.177 | 0.401 | 0.191 | 0.104 | 0.349 | 0.680 | −0.033 | 0.582 | 0.078 | 0.094 | 0.186 | −0.156 | −0.107 | 0.079 |
| Oxidase | −0.428 | −0.812 * | −0.590 | −0.209 | −0.708 | −0.570 | −0.788 | −0.858 * | −0.571 | −0.729 | −0.735 | −0.528 | −0.759 | −0.614 | 0.688 | 0.093 |
| Dehydrogenase | −0.330 | 0.034 | 0.085 | 0.694 | −0.059 | −0.224 | 0.084 | 0.326 | 0.053 | 0.016 | 0.068 | −0.264 | 0.190 | −0.029 | −0.351 | −0.761 |
| Amino acid metabolism | −0.440 | −0.204 | −0.709 | 0.546 | −0.457 | −0.410 | −0.137 | 0.142 | −0.593 | −0.046 | −0.538 | −0.402 | −0.411 | −0.724 | 0.080 | −0.190 |
| Nucleotide metabolism | −0.666 | −0.923 * | −0.641 | 0.073 | −0.812* | −0.778 | −0.920 * | −0.806 * | −0.595 | −0.756 | −0.809 * | −0.745 | −0.766 | −0.731 | 0.786 | −0.172 |
| Saccharide metabolism | −0.500 | −0.381 | −0.501 | 0.563 | −0.587 | −0.490 | −0.202 | −0.163 | −0.550 | −0.475 | −0.507 | −0.499 | −0.424 | −0.557 | −0.256 | −0.640 |
| Lipid metabolism | 0.973 ** | 0.802 | 0.624 | −0.774 | 0.799 | 0.966 ** | 0.804 | 0.331 | 0.538 | 0.539 | 0.734 | 0.968 ** | 0.548 | 0.815 * | −0.582 | 0.752 |
| Terpenoid biosynthesis | 0.724 | 0.513 | 0.168 | −0.738 | 0.491 | 0.679 | 0.474 | 0.164 | 0.193 | 0.446 | 0.330 | 0.706 | 0.185 | 0.357 | −0.108 | 0.941 ** |
| Flavonoid biosynthesis | −0.017 | −0.357 | −0.258 | −0.469 | −0.217 | −0.129 | −0.424 | −0.504 | −0.200 | −0.238 | −0.313 | −0.089 | −0.376 | −0.221 | 0.626 | 0.503 |
| Alkaloid biosynthesis | 0.975 ** | 0.911 * | 0.578 | −0.654 | 0.843 * | 0.996 ** | 0.913 * | 0.512 | 0.520 | 0.679 | 0.755 | 0.995 ** | 0.596 | 0.775 | −0.656 | 0.745 |
| Citrate cycle | 0.866 * | 0.482 | 0.079 | −0.851 * | 0.296 | 0.777 | 0.676 | −0.091 | −0.074 | 0.031 | 0.215 | 0.803 | −0.022 | 0.343 | −0.776 | 0.760 |
| Phenylpropanoid biosynthesis | −0.528 | −0.807 * | −0.582 | −0.060 | −0.685 | −0.643 | −0.832 * | −0.742 | −0.518 | −0.626 | −0.717 | −0.606 | −0.695 | −0.642 | 0.807 * | 0.020 |
| Other metabolic genes | 0.077 | 0.063 | −0.562 | 0.083 | −0.318 | 0.055 | 0.318 | 0.012 | −0.623 | −0.195 | −0.369 | 0.068 | −0.403 | −0.430 | −0.651 | 0.027 |
| TFs | −0.592 | −0.060 | 0.106 | 0.838 * | 0.060 | −0.450 | −0.286 | 0.514 | 0.290 | 0.399 | 0.093 | −0.485 | 0.329 | −0.111 | 0.550 | −0.574 |
| ABI3VP1 | −0.490 | −0.872 * | −0.394 | −0.118 | −0.720 | −0.621 | −0.813 * | −0.940 ** | −0.456 | −0.895 * | −0.645 | −0.595 | −0.672 | −0.466 | 0.519 | −0.231 |
| Alfin-like | 0.255 | −0.098 | −0.535 | −0.382 | −0.401 | 0.142 | 0.211 | −0.434 | −0.684 | −0.517 | −0.440 | 0.174 | −0.588 | −0.346 | −0.642 | 0.260 |
| AP2-EREBP | −0.618 | −0.158 | 0.014 | 0.755 | −0.019 | −0.501 | −0.391 | 0.394 | 0.212 | 0.327 | −0.007 | −0.527 | 0.218 | −0.195 | 0.685 | −0.479 |
| ARF | 0.040 | −0.015 | 0.703 | −0.117 | 0.304 | 0.055 | −0.135 | −0.112 | 0.622 | −0.030 | 0.453 | 0.031 | 0.430 | 0.593 | 0.142 | −0.268 |
| bHLH | −0.692 | −0.907 * | −0.631 | 0.123 | −0.790 | −0.792 | −0.925 * | −0.751 | −0.567 | −0.703 | −0.791 | −0.762 | −0.732 | −0.732 | 0.828* | −0.187 |
| bZIP | −0.592 | −0.172 | 0.230 | 0.679 | 0.076 | −0.477 | −0.446 | 0.333 | 0.404 | 0.310 | 0.128 | −0.508 | 0.340 | −0.009 | 0.756 | −0.524 |
| C2C2-CO-like | 0.006 | 0.313 | −0.257 | 0.433 | −0.009 | 0.085 | 0.440 | 0.490 | −0.244 | 0.235 | −0.040 | 0.070 | 0.024 | −0.209 | −0.583 | −0.189 |
| C2C2-Dof | 0.044 | 0.468 | 0.869 * | 0.385 | 0.679 | 0.200 | 0.257 | 0.680 | 0.899 * | 0.633 | 0.794 | 0.145 | 0.896 * | 0.720 | −0.028 | −0.424 |
| C2C2-GATA | −0.513 | −0.810 * | −0.361 | 0.049 | −0.727 | −0.617 | −0.693 | −0.845 * | −0.466 | −0.918 * | −0.606 | −0.603 | −0.618 | −0.442 | 0.249 | −0.456 |
| C2H2 | −0.593 | −0.053 | 0.219 | 0.849 * | 0.106 | −0.443 | −0.290 | 0.514 | 0.385 | 0.394 | 0.169 | −0.484 | 0.405 | −0.018 | 0.536 | −0.651 |
| C3H | −0.917 * | −0.641 | −0.117 | 0.833 * | −0.476 | −0.858 * | −0.739 | −0.154 | −0.055 | −0.350 | −0.328 | −0.885 * | −0.124 | −0.381 | 0.582 | −0.956 ** |
| FAR1 | 0.471 | 0.074 | 0.397 | −0.785 | 0.298 | 0.381 | 0.011 | −0.340 | 0.330 | −0.034 | 0.281 | 0.401 | 0.121 | 0.474 | 0.157 | 0.564 |
| FHA | −0.172 | 0.068 | −0.311 | 0.498 | −0.220 | −0.115 | 0.235 | 0.246 | −0.345 | −0.060 | −0.189 | −0.131 | −0.123 | −0.303 | −0.554 | −0.423 |
| G2-like | −0.668 | −0.946 ** | −0.842 * | 0.112 | −0.963 ** | −0.792 | −0.841 * | −0.853 * | −0.832 * | −0.888 * | −0.963 ** | −0.756 | −0.934 * | −0.892 * | 0.547 | −0.214 |
| GRAS | −0.738 | −0.270 | 0.010 | 0.855 * | −0.113 | −0.620 | −0.489 | 0.316 | 0.188 | 0.202 | −0.061 | −0.651 | 0.181 | −0.233 | 0.691 | −0.660 |
| GRF | −0.694 | −0.957 ** | −0.470 | 0.112 | −0.793 | −0.799 | −0.942 ** | −0.864 * | −0.479 | −0.855 * | −0.719 | −0.777 | −0.681 | −0.595 | 0.695 | −0.377 |
| HSF | −0.882 * | −0.535 | −0.355 | 0.833 * | −0.451 | −0.815 * | −0.676 | 0.037 | −0.185 | −0.099 | −0.421 | −0.829 * | −0.191 | −0.575 | 0.749 | −0.642 |
| LIM | −0.477 | −0.636 | −0.003 | 0.000 | −0.310 | −0.533 | −0.791 | −0.541 | 0.048 | −0.390 | −0.255 | −0.524 | −0.210 | −0.153 | 0.881 * | −0.183 |
| LOB | −0.650 | −0.812 * | −0.527 | 0.305 | −0.829 * | −0.724 | −0.663 | −0.712 | −0.608 | −0.893 * | −0.713 | −0.715 | −0.674 | −0.621 | 0.160 | −0.615 |
| MADS | 0.626 | 0.810 * | 0.968 ** | −0.233 | 0.968 ** | 0.727 | 0.643 | 0.675 | 0.956 ** | 0.801 | 0.991 ** | 0.693 | 0.949 ** | 0.983 ** | −0.292 | 0.231 |
| mTERF | −0.459 | −0.792 | −0.480 | 0.000 | −0.781 | −0.576 | −0.624 | −0.867 * | −0.597 | −0.956 ** | −0.682 | −0.557 | −0.714 | −0.522 | 0.133 | −0.379 |
| MYB | 0.879 * | 0.514 | 0.221 | −0.943 ** | 0.471 | 0.797 | 0.562 | −0.015 | 0.154 | 0.259 | 0.344 | 0.827 * | 0.127 | 0.455 | −0.366 | 0.967 ** |
| NAC | −0.268 | 0.226 | −0.191 | 0.697 | 0.045 | −0.141 | 0.181 | 0.655 | −0.053 | 0.453 | −0.001 | −0.164 | 0.166 | −0.250 | −0.057 | −0.292 |
| NFY | −0.439 | −0.678 | −0.749 | −0.056 | −0.679 | −0.548 | −0.664 | −0.604 | −0.661 | −0.510 | −0.771 | −0.506 | −0.752 | −0.754 | 0.670 | 0.173 |
| OFP | 0.492 | 0.314 | 0.335 | −0.617 | 0.474 | 0.458 | 0.167 | 0.071 | 0.388 | 0.392 | 0.367 | 0.476 | 0.271 | 0.419 | 0.268 | 0.728 |
| PLATZ | 0.236 | 0.461 | −0.295 | 0.139 | 0.144 | 0.291 | 0.537 | 0.547 | −0.222 | 0.449 | 0.009 | 0.295 | 0.031 | −0.175 | −0.432 | 0.286 |
| SBP | −0.321 | −0.784 | −0.449 | −0.261 | −0.720 | −0.473 | −0.643 | −0.973 ** | −0.560 | −0.946 ** | −0.658 | −0.441 | −0.736 | −0.455 | 0.252 | −0.117 |
| TCP | 0.319 | −0.133 | −0.300 | −0.540 | −0.314 | 0.189 | 0.146 | −0.571 | −0.492 | −0.591 | −0.305 | 0.217 | −0.488 | −0.130 | −0.586 | 0.246 |
| Tify | −0.245 | −0.360 | −0.512 | −0.084 | −0.337 | −0.308 | −0.430 | −0.264 | −0.375 | −0.113 | −0.467 | −0.273 | −0.437 | −0.505 | 0.660 | 0.343 |
| Trihelix | 0.602 | 0.163 | 0.339 | −0.780 | 0.215 | 0.504 | 0.278 | −0.374 | 0.151 | −0.226 | 0.251 | 0.518 | 0.039 | 0.477 | −0.444 | 0.405 |
| WRKY | −0.648 | −0.214 | −0.126 | 0.753 | −0.119 | −0.545 | −0.419 | 0.338 | 0.075 | 0.261 | −0.125 | −0.565 | 0.098 | −0.319 | 0.683 | −0.446 |
| zf-HD | 0.429 | 0.235 | −0.485 | −0.341 | −0.143 | 0.364 | 0.502 | −0.042 | −0.568 | −0.113 | −0.252 | 0.392 | −0.379 | −0.257 | −0.735 | 0.458 |
| Other TFs | −0.561 | −0.087 | 0.358 | 0.841 * | 0.065 | −0.420 | −0.232 | 0.375 | 0.410 | 0.161 | 0.223 | −0.471 | 0.421 | 0.116 | 0.208 | −0.905 * |
| Transporters | −0.200 | 0.367 | 0.080 | 0.735 | 0.243 | −0.040 | 0.284 | 0.799 | 0.201 | 0.588 | 0.238 | −0.076 | 0.412 | −0.007 | −0.136 | −0.391 |
| ABC transporters | 0.997 ** | 0.846 * | 0.526 | −0.747 | 0.769 | 0.992 ** | 0.883 * | 0.379 | 0.441 | 0.554 | 0.686 | 0.996 ** | 0.499 | 0.742 | −0.691 | 0.778 |
| MFS transporters | −0.395 | 0.143 | 0.055 | 0.733 | 0.167 | −0.250 | −0.053 | 0.659 | 0.247 | 0.557 | 0.144 | −0.280 | 0.353 | −0.093 | 0.371 | −0.351 |
| Solute carriers | −0.354 | 0.214 | 0.025 | 0.844 * | 0.104 | −0.197 | 0.147 | 0.692 | 0.131 | 0.434 | 0.134 | −0.237 | 0.327 | −0.098 | −0.093 | −0.571 |
| Other carriers | 0.617 | 0.951 ** | 0.557 | −0.022 | 0.850 * | 0.744 | 0.873 * | 0.936 ** | 0.599 | 0.943 ** | 0.784 | 0.717 | 0.780 | 0.640 | −0.541 | 0.314 |
| MIP transporters | −0.367 | −0.144 | −0.211 | 0.326 | −0.065 | −0.324 | −0.332 | 0.202 | −0.009 | 0.274 | −0.156 | −0.320 | −0.020 | −0.303 | 0.706 | 0.037 |
| Ion transporters | 0.505 | 0.427 | −0.079 | −0.233 | 0.113 | 0.493 | 0.669 | 0.153 | −0.216 | 0.021 | 0.091 | 0.495 | −0.026 | 0.102 | −0.955 ** | 0.203 |
| Other transporters | −0.477 | −0.214 | −0.676 | 0.581 | −0.434 | −0.439 | −0.179 | 0.169 | −0.541 | −0.006 | −0.514 | −0.434 | −0.372 | −0.709 | 0.170 | −0.207 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, R.; Yan, S.; Hu, Y.; Rao, D.; Li, H.; Chun, Z.; Zheng, S. Metabolic and Transcriptomic Profile Revealing the Differential Accumulating Mechanism in Different Parts of Dendrobium nobile. Int. J. Mol. Sci. 2024, 25, 5356. https://doi.org/10.3390/ijms25105356
Zhao R, Yan S, Hu Y, Rao D, Li H, Chun Z, Zheng S. Metabolic and Transcriptomic Profile Revealing the Differential Accumulating Mechanism in Different Parts of Dendrobium nobile. International Journal of Molecular Sciences. 2024; 25(10):5356. https://doi.org/10.3390/ijms25105356
Chicago/Turabian StyleZhao, Ruoxi, Shou Yan, Yadong Hu, Dan Rao, Hongjie Li, Ze Chun, and Shigang Zheng. 2024. "Metabolic and Transcriptomic Profile Revealing the Differential Accumulating Mechanism in Different Parts of Dendrobium nobile" International Journal of Molecular Sciences 25, no. 10: 5356. https://doi.org/10.3390/ijms25105356
APA StyleZhao, R., Yan, S., Hu, Y., Rao, D., Li, H., Chun, Z., & Zheng, S. (2024). Metabolic and Transcriptomic Profile Revealing the Differential Accumulating Mechanism in Different Parts of Dendrobium nobile. International Journal of Molecular Sciences, 25(10), 5356. https://doi.org/10.3390/ijms25105356






