miRNA Expression Profiles in Isolated Ventricular Cardiomyocytes: Insights into Doxorubicin-Induced Cardiotoxicity
Abstract
1. Introduction
2. Results
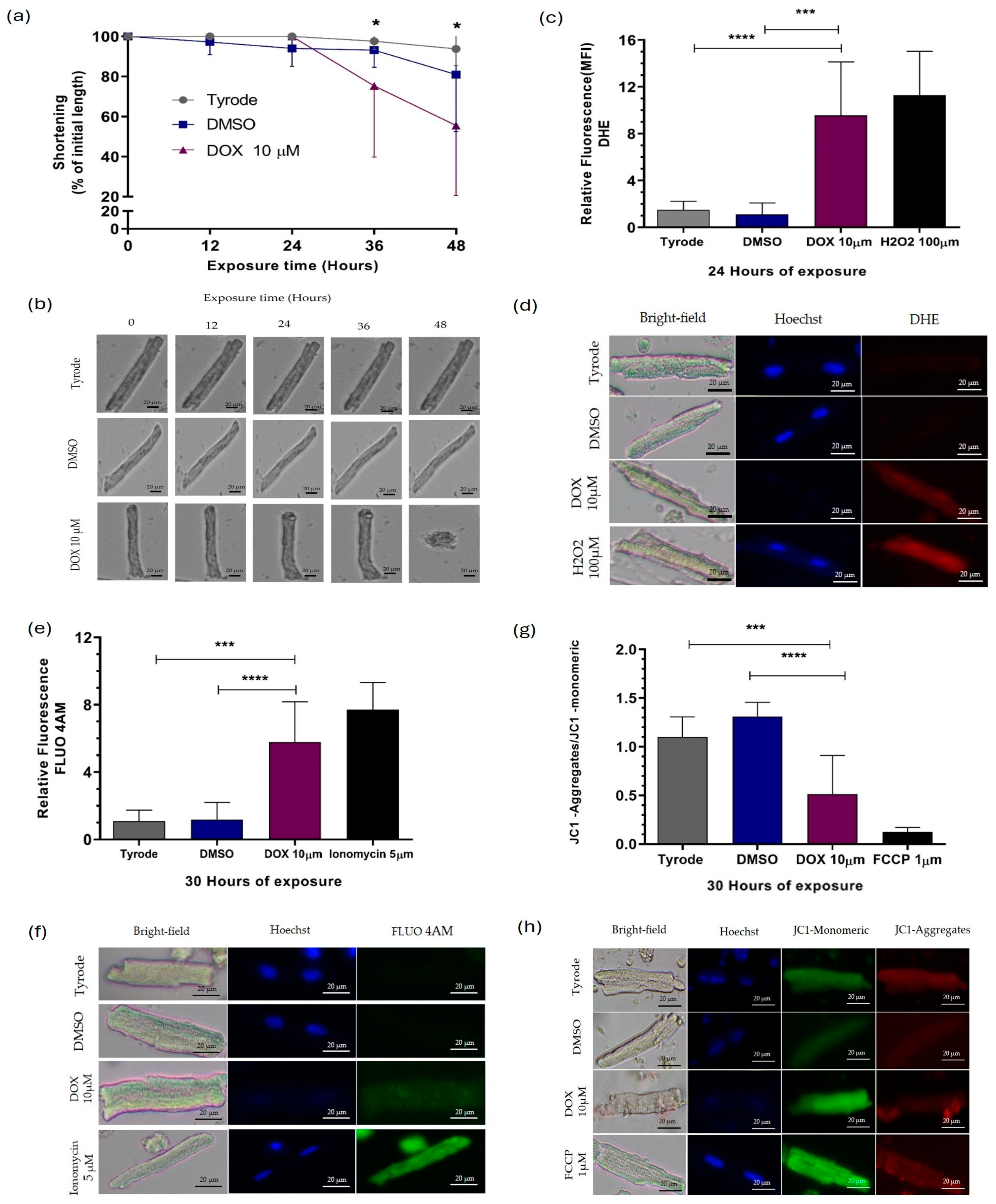
2.1. Doxorubicin Causes Damage to Guinea Pig Ventricular Cardiomyocytes
2.2. Small RNAseq and Differential Expression Analysis of miRNAs
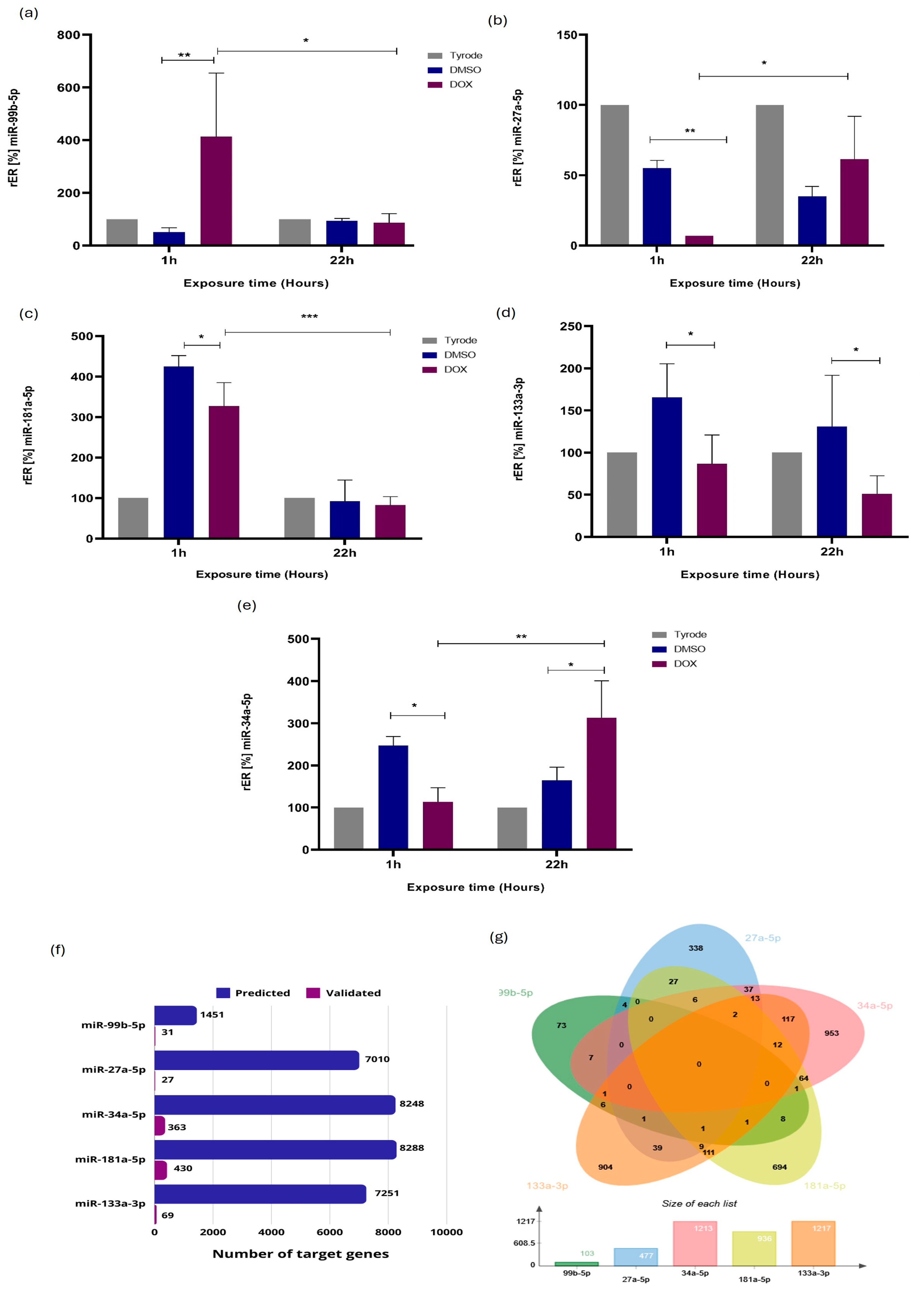
2.3. Validation of miRNA Differential Expression by RT-qPCR Stem-Loop
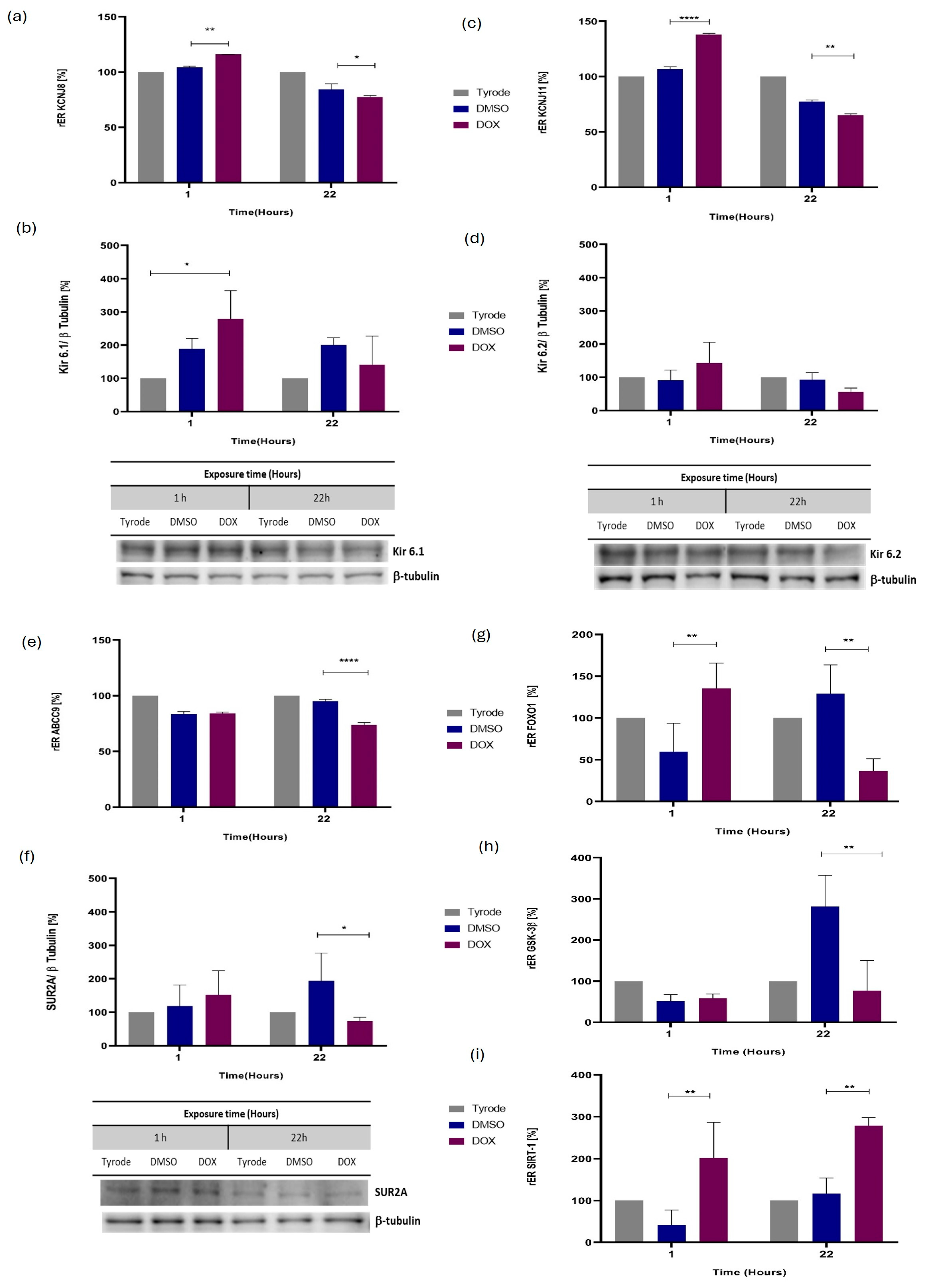
2.4. ATP-Sensitive Potassium Channel (KATP) Coding Genes Are Expressed Differently in Cardiomyocytes Injured by Direct Exposure to DOX
2.5. Differential mRNA Expression for Additional miRNA Molecular Targets, Including GSK3β, SIRT1, and FOXO1
2.6. miRNA Functional Analysis and Phenotype-Associated Molecules Network
3. Discussion
4. Materials and Methods
4.1. Isolation of Ventricular Cardiomyocytes from Guinea Pig (Cavia porcellus)
4.2. Evaluation of the Length and Percentage of Cell Shortening of Cardiomyocytes
4.3. Evaluation of ROS Levels, Intracellular Calcium, and Mitochondrial Membrane Potential
4.4. RNA Extraction, Small RNA Sequencing, and Analysis
4.5. RT-qPCR-Stem-Loop for miRNAs and RT-PCR for mRNAs
4.6. Prediction of Molecular Targets (mRNAs) of microRNAs by CSmiRTar
4.7. Protein Expression by Western Blot
4.8. miRNA Gene Regulatory Networks and Functional Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Müller, I.; Jenner, A.; Bruchelt, G.; Niethammer, D.; Halliwell, B. Effect of Concentration on the Cytotoxic Mechanism of Doxorubicin-Apoptosis and Oxidative DNA Damage. Biochem. Biophys. Res. Commun. 1997, 230, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.; Santos, R.; Cardoso, S.; Correia, S.; Oliveira, P.; Santos, M.; Moreira, P. Doxorubicin: The Good, the Bad and the Ugly Effect. Curr. Med. Chem. 2009, 16, 3267–3285. [Google Scholar] [CrossRef] [PubMed]
- Sheibani, M.; Azizi, Y.; Shayan, M.; Nezamoleslami, S.; Eslami, F.; Farjoo, M.H.; Dehpour, A.R. Doxorubicin-Induced Cardiotoxicity: An Overview on Pre-Clinical Therapeutic Approaches. Cardiovasc. Toxicol. 2022, 22, 292–310. [Google Scholar] [CrossRef]
- Octavia, Y.; Tocchetti, C.G.; Gabrielson, K.L.; Janssens, S.; Crijns, H.J.; Moens, A.L. Doxorubicin-Induced Cardiomyopathy: From Molecular Mechanisms to Therapeutic Strategies. J. Mol. Cell Cardiol. 2012, 52, 1213–1225. [Google Scholar] [CrossRef]
- Carvalho, F.S.; Burgeiro, A.; Garcia, R.; Moreno, A.J.; Carvalho, R.A.; Oliveira, P.J. Doxorubicin-Induced Cardiotoxicity: From Bioenergetic Failure and Cell Death to Cardiomyopathy. Med. Res. Rev. 2014, 34, 106–135. [Google Scholar] [CrossRef] [PubMed]
- Mitry, M.A.; Edwards, J.G. Doxorubicin Induced Heart Failure: Phenotype and Molecular Mechanisms. IJC Heart Vasc. 2016, 10, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Whaley, F.S.; Ewer, M.S. Congestive Heart Failure in Patients Treated with Doxorubicin: A Retrospective Analysis of Three Trials. Cancer 2003, 97, 2869–2879. [Google Scholar] [CrossRef] [PubMed]
- Agunbiade, T.A.; Zaghlol, R.Y.; Barac, A. Heart Failure in Relation to Anthracyclines and Other Chemotherapies. Methodist. Debakey Cardiovasc. J. 2019, 15, 243. [Google Scholar] [CrossRef]
- Maillet, A.; Tan, K.; Chai, X.; Sadananda, S.N.; Mehta, A.; Ooi, J.; Hayden, M.R.; Pouladi, M.A.; Ghosh, S.; Shim, W.; et al. Modeling Doxorubicin-Induced Cardiotoxicity in Human Pluripotent Stem Cell Derived-Cardiomyocytes. Sci. Rep. 2016, 6, 25333. [Google Scholar] [CrossRef]
- Ichikawa, Y.; Ghanefar, M.; Bayeva, M.; Wu, R.; Khechaduri, A.; Naga Prasad, S.V.; Mutharasan, R.K.; Jairaj Naik, T.; Ardehali, H. Cardiotoxicity of Doxorubicin Is Mediated through Mitochondrial Iron Accumulation. J. Clin. Investig. 2014, 124, 617–630. [Google Scholar] [CrossRef]
- Yin, J.; Guo, J.; Zhang, Q.; Cui, L.; Zhang, L.; Zhang, T.; Zhao, J.; Li, J.; Middleton, A.; Carmichael, P.L.; et al. Doxorubicin-Induced Mitophagy and Mitochondrial Damage Is Associated with Dysregulation of the PINK1/Parkin Pathway. Toxicol. In Vitro 2018, 51, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.B.; Katugampola, S.; Able, S.; Napier, C.; Harding, S.E. Profiling of CAMP and CGMP Phosphodiesterases in Isolated Ventricular Cardiomyocytes from Human Hearts: Comparison with Rat and Guinea Pig. Life Sci. 2012, 90, 328–336. [Google Scholar] [CrossRef]
- Lookin, O.; Khokhlova, A.; Myachina, T.; Butova, X.; Cazorla, O.; de Tombe, P. Contractile State Dependent Sarcomere Length Variability in Isolated Guinea-Pig Cardiomyocytes. Front. Physiol. 2022, 13, 857471. [Google Scholar] [CrossRef]
- Soltysinska, E.; Olesen, S.P.; Osadchii, O.E. Myocardial Structural, Contractile and Electrophysiological Changes in the Guinea-Pig Heart Failure Model Induced by Chronic Sympathetic Activation. Exp. Physiol. 2011, 96, 647–663. [Google Scholar] [CrossRef]
- Brian Foster, D.; Liu, T.; Kammers, K.; O’Meally, R.; Yang, N.; Papanicolaou, K.N.; Conover Talbot, C.; Cole, R.N.; O’Rourke, B. Integrated Omic Analysis of a Guinea Pig Model of Heart Failure and Sudden Cardiac Death. J. Proteome Res. 2016, 15, 3009–3028. [Google Scholar] [CrossRef] [PubMed]
- Shiotani, M.; Harada, T.; Abe, J.; Hamada, Y.; Horii, I. Aging-Related Changes of QT and RR Intervals in Conscious Guinea Pigs. J. Pharmacol. Toxicol. Methods 2008, 57, 23–29. [Google Scholar] [CrossRef]
- Wang, G.X.; Wang, Y.X.; Zhou, X.B.; Korth, M. Effects of Doxorubicinol on Excitation–Contraction Coupling in Guinea Pig Ventricular Myocytes. Eur. J. Pharmacol. 2001, 423, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Willis, M.S.; Parry, T.L.; Brown, D.I.; Mota, R.I.; Huang, W.; Beak, J.Y.; Sola, M.; Zhou, C.; Hicks, S.T.; Caughey, M.C.; et al. Doxorubicin Exposure Causes Subacute Cardiac Atrophy Dependent upon the Striated Muscle-Specific Ubiquitin Ligase Muscle Ring Finger-1. Circ. Heart Fail. 2019, 12, e005234. [Google Scholar] [CrossRef]
- Rodrigues, P.G.; Miranda-Silva, D.; Costa, S.M.; Barros, C.; Hamdani, N.; Moura, C.; Mendes, M.J.; Sousa-Mendes, C.; Trindade, F.; Fontoura, D.; et al. Early Myocardial Changes Induced by Doxorubicin in the Nonfailing Dilated Ventricle. Am. J. Physiol. Heart Circ. Physiol. 2019, 316, H459–H475. [Google Scholar] [CrossRef]
- Abdullah, C.S.; Alam, S.; Aishwarya, R.; Miriyala, S.; Bhuiyan, M.A.N.; Panchatcharam, M.; Pattillo, C.B.; Orr, A.W.; Sadoshima, J.; Hill, J.A.; et al. Doxorubicin-Induced Cardiomyopathy Associated with Inhibition of Autophagic Degradation Process and Defects in Mitochondrial Respiration. Sci. Rep. 2019, 9, 2002. [Google Scholar] [CrossRef]
- Abe, K.; Ikeda, M.; Ide, T.; Tadokoro, T.; Miyamoto, H.D.; Furusawa, S.; Tsutsui, Y.; Miyake, R.; Ishimaru, K.; Watanabe, M.; et al. Doxorubicin Causes Ferroptosis and Cardiotoxicity by Intercalating into Mitochondrial DNA and Disrupting Alas1-Dependent Heme Synthesis. Sci. Signal 2022, 15, eabn8017. [Google Scholar] [CrossRef] [PubMed]
- Villani, F.; Meazza, R.; Materazzo, C. Non-Invasive Monitoring of Cardiac Hemodynamic Parameters in Doxorubicin-Treated Patients: Comparison with Echocardiography. Anticancer Res. 2006, 26, 797–801. [Google Scholar] [PubMed]
- Desai, V.G.; Kwekel, J.C.; Vijay, V.; Moland, C.L.; Herman, E.H.; Lee, T.; Han, T.; Lewis, S.M.; Davis, K.J.; Muskhelishvili, L.; et al. Early Biomarkers of Doxorubicin-Induced Heart Injury in a Mouse Model. Toxicol. Appl. Pharmacol. 2014, 281, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Gomez, L.A.; Alekseev, A.E.; Aleksandrova, L.A.; Brady, P.A.; Terzic, A. Use of the MTT Assay in Adult Ventricular Cardiomyocytes to Assess Viability: Effects of Adenosine and Potassium on Cellular Survival. J. Mol. Cell Cardiol. 1997, 29, 1255–1266. [Google Scholar] [CrossRef]
- Chazotte, B. Labeling Nuclear DNA with Hoechst 33342. Cold Spring Harb Protoc 2011, 2011, pdb-prot5557. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.G. Adenosine Triphosphate-Sensitive Potassium Currents in Heart Disease and Cardioprotection. Card. Electrophysiol. Clin. 2016, 8, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Kraljevic, J.; Høydal, M.A.; Ljubkovic, M.; Moreira, J.B.N.; Jørgensen, K.; Ness, H.O.; Bækkerud, F.H.; Dujic, Z.; Wisløff, U.; Marinovic, J. Role of KATP Channels in Beneficial Effects of Exercise in Ischemic Heart Failure. Med. Sci. Sports Exerc. 2015, 47, 2504–2512. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bian, W.; Yan, Y.; Zhang, D.M. Functional Regulation of KATP Channels and Mutant Insight Into Clinical Therapeutic Strategies in Cardiovascular Diseases. Front. Pharmacol. 2022, 13, 868401. [Google Scholar] [CrossRef] [PubMed]
- Storz, P. Forkhead Homeobox Type O Transcription Factors in the Responses to Oxidative Stress. Antioxid. Redox Signal. 2011, 14, 593. [Google Scholar] [CrossRef]
- Yan, D.; Cai, Y.; Luo, J.; Liu, J.; Li, X.; Ying, F.; Xie, X.; Xu, A.; Ma, X.; Xia, Z. FOXO1 Contributes to Diabetic Cardiomyopathy via Inducing Imbalanced Oxidative Metabolism in Type 1 Diabetes. J. Cell Mol. Med. 2020, 24, 7850. [Google Scholar] [CrossRef]
- Philip-Couderc, P.; Tavares, N.I.; Roatti, A.; Lerch, R.; Montessuit, C.; Baertschi, A.J. Forkhead Transcription Factors Coordinate Expression of Myocardial KATP Channel Subunits and Energy Metabolism. Circ. Res. 2008, 102, e20–e35. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.H.; Zhang, Y.H.; Ding, Y.Q.; Bi, X.Y.; Yuan, J.; Zhou, H.; Wang, P.X.; Zhang, L.L.; Ye, J.T. MicroRNA-99b-3p Promotes Angiotensin II-Induced Cardiac Fibrosis in Mice by Targeting GSK-3β. Acta Pharmacol. Sin. 2021, 42, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Chen, Z.; Gao, J.; Shi, W.; Li, L.; Jiang, S.; Hu, H.; Liu, Z.; Xu, D.; Wu, L. The Key Roles of GSK-3β in Regulating Mitochondrial Activity. Cell Physiol. Biochem. 2017, 44, 1445–1459. [Google Scholar] [CrossRef] [PubMed]
- Van Le, T.N.; Zoungrana, L.I.; Wang, H.; Fatmi, M.K.; Ren, D.; Krause-Hauch, M.; Li, J. Sirtuin 1 Aggravates Hypertrophic Heart Failure Caused by Pressure Overload via Shifting Energy Metabolism. Biochem. Biophys. Res. Commun. 2022, 637, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape Plugin: Pathway Insights Using Integrated Experimental and in Silico Data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Panpan, T.; Yuchen, D.; Xianyong, S.; Meng, L.; Ruijuan, H.; Ranran, D.; Pengyan, Z.; Mingxi, L.; Rongrong, X. Cardiac Remodelling Following Cancer Therapy: A Review. Cardiovasc. Toxicol. 2022, 22, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Sarvazyan, N. Visualization of Doxorubicin-Induced Oxidative Stress in Isolated Cardiac Myocytes. Am. J. Physiol. Heart Circ. Physiol. 1996, 271, H2079–H2085. [Google Scholar] [CrossRef]
- Sangweni, N.F.; Gabuza, K.; Huisamen, B.; Mabasa, L.; van Vuuren, D.; Johnson, R. Molecular Insights into the Pathophysiology of Doxorubicin-Induced Cardiotoxicity: A Graphical Representation. Arch. Toxicol. 2022, 96, 1541–1550. [Google Scholar] [CrossRef]
- Kalivendi, S.V.; Kotamraju, S.; Zhao, H.; Joseph, J.; Kalyanaraman, B. Doxorubicin-Induced Apoptosis Is Associated with Increased Transcription of Endothelial Nitric-Oxide Synthase: Effect of Antiapoptotic Antioxidants and Calcium. J. Biol. Chem. 2001, 276, 47266–47276. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yingjie, X.U.; Deng, Z.; Wang, Y.; Zheng, Y.; Jiang, W.; Jiang, L. MicroRNA Expression Profiling Involved in Doxorubicin-Induced Cardiotoxicity Using High-Throughput Deep-Sequencing Analysis. Oncol. Lett. 2021, 22, 1–9. [Google Scholar] [CrossRef]
- Gómez-Grosso, L.A. Preacondicionamiento Isquémico En Cardiomiocitos Ventriculares Aislados. Identificación y Expresión de Algunos MicroRNAs Asociados. Rev. Acad. Colomb. Cienc. Exactas Fis. Nat. 2013, 37, 433–447. [Google Scholar] [CrossRef]
- Xia, P.; Chen, J.; Liu, Y.; Fletcher, M.; Jensen, B.C.; Cheng, Z. Doxorubicin Induces Cardiomyocyte Apoptosis and Atrophy through Cyclin-Dependent Kinase 2-Mediated Activation of Forkhead Box O1. J. Biol. Chem. 2020, 295, 4265–4276. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, S.J.; Kim, B.J.; Rah, S.Y.; Sung, M.C.; Im, M.J.; Kim, U.H. Doxorubicin-Induced Reactive Oxygen Species Generation and Intracellular Ca2+ Increase Are Reciprocally Modulated in Rat Cardiomyocytes. Exp. Mol. Med. 2006, 38, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Bers, D.M. Calcium Cycling and Signaling in Cardiac Myocytes. Annu. Rev. Physiol. 2008, 70, 23–49. [Google Scholar] [CrossRef]
- Songbo, M.; Lang, H.; Xinyong, C.; Bin, X.; Ping, Z.; Liang, S. Oxidative Stress Injury in Doxorubicin-Induced Cardiotoxicity. Toxicol. Lett. 2019, 307, 41–48. [Google Scholar] [CrossRef]
- Fox, C.A.; Romenskaia, I.; Dagda, R.K.; Ryan, R.O. Cardiolipin Nanodisks Confer Protection against Doxorubicin-Induced Mitochondrial Dysfunction. Biochim. Biophys. Acta (BBA)-Biomembr. 2022, 1864, 183984. [Google Scholar] [CrossRef]
- Parker, M.A.; King, V.; Howard, K.P. Nuclear Magnetic Resonance Study of Doxorubicin Binding to Cardiolipin Containing Magnetically Oriented Phospholipid Bilayers. Biochim. Biophys. Acta Biomembr. 2001, 1514, 206–216. [Google Scholar] [CrossRef]
- Ikon, N.; Ryan, R.O. Cardiolipin and Mitochondrial Cristae Organization. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1156–1163. [Google Scholar] [CrossRef]
- Marcillat, O.; Zhang, Y.; Davies, K.J.A. Oxidative and Non-Oxidative Mechanisms in the Inactivation of Cardiac Mitochondrial Electron Transport Chain Components by Doxorubicin. Biochem. J. 1989, 259, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Asensio-López, M.C.; Soler, F.; Sánchez-Más, J.; Pascual-Figal, D.; Fernández-Belda, F.; Lax, A. Early Oxidative Damage Induced by Doxorubicin: Source of Production, Protection by GKT137831 and Effect on Ca2+transporters in HL-1 Cardiomyocytes. Arch. Biochem. Biophys. 2016, 594, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Hanna, A.D.; Lam, A.; Tham, S.; Dulhunty, A.F.; Beard, N.A. Adverse Effects of Doxorubicin and Its Metabolic Product on Cardiac RyR2 and SERCA2A. Mol. Pharmacol. 2014, 86, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Vercesi, A.E.; Castilho, R.F.; Kowaltowski, A.J.; de Oliveira, H.C.F.; de Souza-Pinto, N.C.; Figueira, T.R.; Busanello, E.N.B. Mitochondrial Calcium Transport and the Redox Nature of the Calcium-Induced Membrane Permeability Transition. Free Radic. Biol. Med. 2018, 129, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Lemasters, J.J.; Nieminen, A.L.; Qian, T.; Trost, L.C.; Elmore, S.P.; Nishimura, Y.; Crowe, R.A.; Cascio, W.E.; Bradham, C.A.; Brenner, D.A.; et al. The Mitochondrial Permeability Transition in Cell Death: A Common Mechanism in Necrosis, Apoptosis and Autophagy. Biochim. Biophys. Acta Bioenerg. 1998, 1366, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Tian, R. Mitochondrial Dysfunction in Pathophysiology of Heart Failure. J. Clin. Investig. 2018, 128, 3716–3726. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wang, Y.; Ayub, A.; Ashraf, M. Role of Mitochondrial Membrane Potential in Cardiac Protection against Ischemia. In Myocardial Ischemia and Preconditioning; Springer: Boston, MA, USA, 2003; pp. 205–218. [Google Scholar] [CrossRef]
- Sulkin, M.S.; Boukens, B.J.; Tetlow, M.; Gutbrod, S.R.; Ng, F.S.; Efimov, I.R. Mitochondrial Depolarization and Electrophysiological Changes during Ischemia in the Rabbit and Human Heart. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H1178. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.G.; Lee, P.; Gordon, R.E.; Sahoo, S.; Kho, C.; Jeong, D. Analysis of Extracellular Vesicle MiRNA Profiles in Heart Failure. J. Cell Mol. Med. 2020, 24, 7214. [Google Scholar] [CrossRef] [PubMed]
- Amara, V.R.; Surapaneni, S.K.; Tikoo, K. Metformin Attenuates Cardiovascular and Renal Injury in Uninephrectomized Rats on DOCA-Salt: Involvement of AMPK and MiRNAs in Cardioprotection. Toxicol. Appl. Pharmacol. 2019, 362, 95–104. [Google Scholar] [CrossRef]
- Zhang, J.; Qiu, W.; Ma, J.; Wang, Y.; Hu, Z.; Long, K.; Wang, X.; Jin, L.; Tang, Q.; Tang, G.; et al. MiR-27a-5p Attenuates Hypoxia-Induced Rat Cardiomyocyte Injury by Inhibiting Atg7. Int. J. Mol. Sci. 2019, 20, 2418. [Google Scholar] [CrossRef]
- Shyng, S.L. KATP Channel Function: More than Meets the Eye. Function 2022, 3, 70. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hu, D.; Huang, C.; Nichols, C.G. Genetic Discovery of KATP Channels in Cardiovascular Diseases. Circ. Arrhythm. Electrophysiol. 2019, 12, e007322. [Google Scholar] [CrossRef] [PubMed]
- Ronnebaum, S.M.; Patterson, C. The FoxO Family in Cardiac Function and Dysfunction. Annu. Rev. Physiol. 2010, 72, 81–94. [Google Scholar] [CrossRef]
- Qin, X.D.; Liu, L. Loss of MicroRNA-27a Induces Cardiac Dysfunction through Activating FoxO1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5941–5948. [Google Scholar] [CrossRef] [PubMed]
- Takahashi-Yanaga, F. Roles of Glycogen Synthase Kinase-3 (GSK-3) in Cardiac Development and Heart Disease. J. UOEH 2018, 40, 147–156. [Google Scholar] [CrossRef]
- Granchi, C.; Minutolo, F. Activators of Sirtuin-1 and Their Involvement in Cardioprotection. Curr. Med. Chem. 2018, 25, 4432–4456. [Google Scholar] [CrossRef]
- Packer, M. Cardioprotective Effects of Sirtuin-1 and Its Downstream Effectors. Circ. Heart Fail. 2020, 13, E007197. [Google Scholar] [CrossRef] [PubMed]
- Tuncay, E.; Gando, I.; Huo, J.Y.; Yepuri, G.; Sampler, N.; Turan, B.; Yang, H.Q.; Ramasamy, R.; Coetzee, W.A. The Cardioprotective Role of Sirtuins Is Mediated in Part by Regulating KATP Channel Surface Expression. Am. J. Physiol. Cell Physiol. 2023, 324, C1017–C1027. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.N.; Fu, Y.H.; Hu, Z.Q.; Li, W.Y.; Tang, C.M.; Fei, H.W.; Yang, H.; Lin, Q.X.; Gou, D.M.; Wu, S.L.; et al. Activation of MiR-34a-5p/Sirt1/P66shc Pathway Contributes to Doxorubicin-Induced Cardiotoxicity. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Perez, I.E.; Taveras Alam, S.; Hernandez, G.A.; Sancassani, R. Cancer Therapy-Related Cardiac Dysfunction: An Overview for the clinician. Clin. Med. Insights Cardiol. 2019, 13, 1179546819866445. [Google Scholar] [CrossRef]
- Schefe, J.H.; Lehmann, K.E.; Buschmann, I.R.; Unger, T.; Funke-Kaiser, H. Quantitative real-time RT-PCR data analysis: Current concepts and the novel “gene expression’s CT difference” formula. J. Mol. Med. 2006, 84, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-S.; Tu, B.-W.; Chen, T.-T.; Hou, S.-W.; Tseng, J.T. CSmiRTar: Condition-Specific MicroRNA Targets Database. PLoS ONE 2017, 12, e0181231. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING Database in 2017: Quality-Controlled Protein-Protein Association Networks, Made Broadly Accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
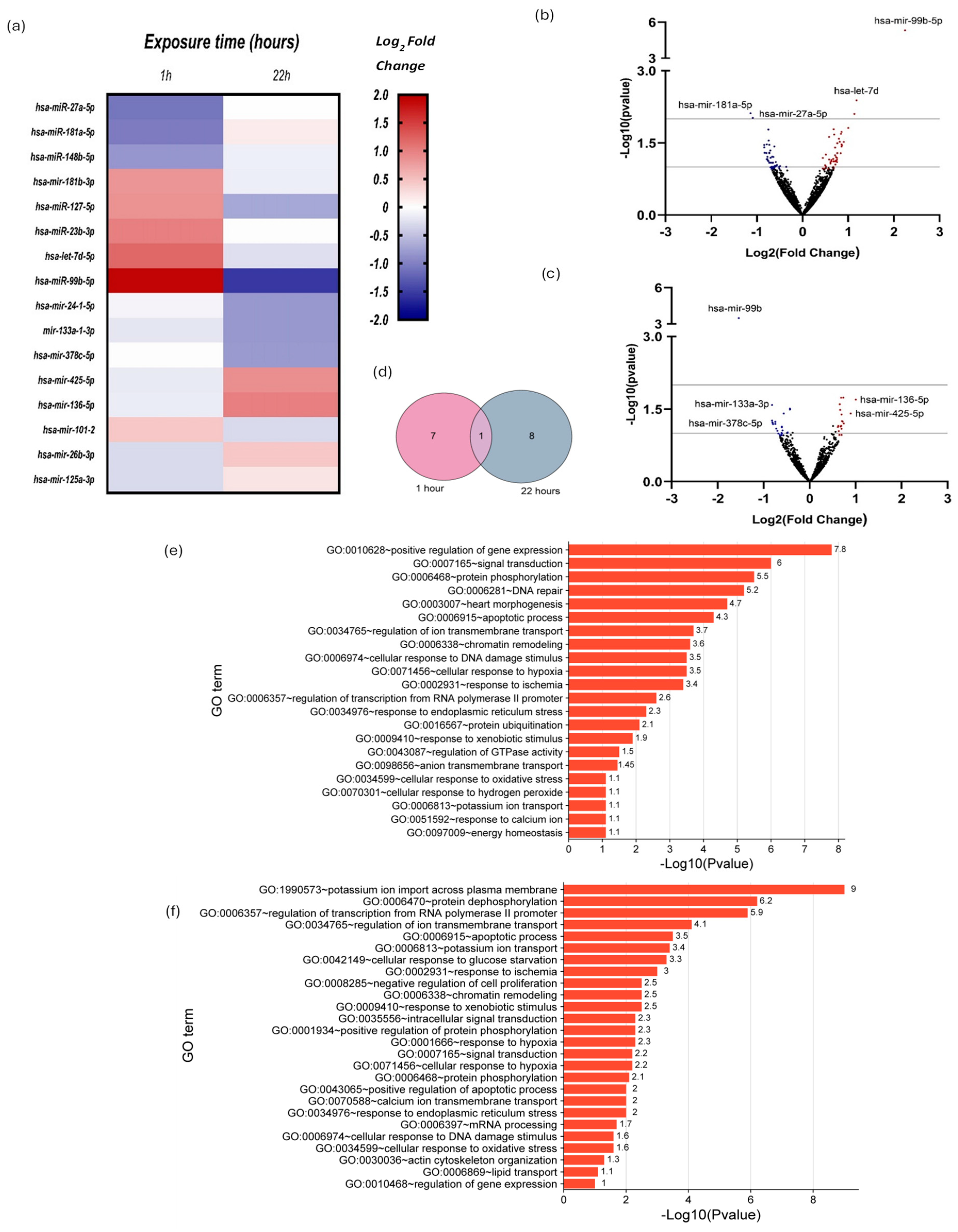

| Cardiomyocytes Exposed to 10 µM DOX for 1 h Versus Unexposed Cardiomyocytes | ||||
|---|---|---|---|---|
| miRNA ID | Expression Level | Sequence | log2 (Fold Change) | p-Value |
| miR-27a-5p | Downregulated | AGCTTAGCTGATTGGTGAACT | −1.09 | 9.53 × 10−3 |
| miR-181a-5p | CATTCAACGCTGTCGG | −1.05 | 1.44 × 10−2 | |
| miR-148b-5p | AGTTCTGTTATACACTCAG | −0.83 | 5.07 × 10−2 | |
| miR-181b-3p | Upreguladed | CACTGATCAATGAATGCAA | 0.83 | 5.17 × 10−2 |
| miR-127-5p | AGCTCAGAGGGCTCTGAT | 0.86 | 3.55 × 10−2 | |
| miR-23b-3p | CACATTGCCAGGGA | 1.00 | 1.54 × 10−2 | |
| let-7d-5p | AGGTAGTAGGTTGTATAGTTA | 1.18 | 4.11 × 10−2 | |
| miR-99b-5p | CCCGTAGAACCGATCTTGTG | 1.91 | 2.20 × 10−8 | |
| Cardiomyocytes Exposed to 10 µM DOX for 22 h Versus Unexposed Cardiomyocytes | ||||
|---|---|---|---|---|
| miRNA ID | Expression Level | Sequence | log2 (Fold Change) | p-Value |
| miR-99b-5p | Downregulated | CCCGTAGAACCGATCTTGTG | −1.46 | 4.49 × 10−4 |
| miR-24-1-5p | TGCCTACTGAGCTGATATC | −0.82 | 2.59 × 10−2 | |
| miR-133a-1-3p | GGTCCCCTTCAATCAGCTGTT | −0.82 | 5.47 × 10−2 | |
| miR-378c-5p | ACTGGACTTGGAGTCAGAA | −0.81 | 5.98 × 10−2 | |
| miR-425-5p | Upregulated | TGACACGATCACTCCCGTTGT | 0.89 | 3.90 × 10−2 |
| miR-136-5p | TCCATTTGTTTTGATGATGG | 1.00 | 2.01 × 10−2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domínguez Romero, Y.; Montoya Ortiz, G.; Novoa Herrán, S.; Osorio Mendez, J.; Gomez Grosso, L.A. miRNA Expression Profiles in Isolated Ventricular Cardiomyocytes: Insights into Doxorubicin-Induced Cardiotoxicity. Int. J. Mol. Sci. 2024, 25, 5272. https://doi.org/10.3390/ijms25105272
Domínguez Romero Y, Montoya Ortiz G, Novoa Herrán S, Osorio Mendez J, Gomez Grosso LA. miRNA Expression Profiles in Isolated Ventricular Cardiomyocytes: Insights into Doxorubicin-Induced Cardiotoxicity. International Journal of Molecular Sciences. 2024; 25(10):5272. https://doi.org/10.3390/ijms25105272
Chicago/Turabian StyleDomínguez Romero, Yohana, Gladis Montoya Ortiz, Susana Novoa Herrán, Jhon Osorio Mendez, and Luis A. Gomez Grosso. 2024. "miRNA Expression Profiles in Isolated Ventricular Cardiomyocytes: Insights into Doxorubicin-Induced Cardiotoxicity" International Journal of Molecular Sciences 25, no. 10: 5272. https://doi.org/10.3390/ijms25105272
APA StyleDomínguez Romero, Y., Montoya Ortiz, G., Novoa Herrán, S., Osorio Mendez, J., & Gomez Grosso, L. A. (2024). miRNA Expression Profiles in Isolated Ventricular Cardiomyocytes: Insights into Doxorubicin-Induced Cardiotoxicity. International Journal of Molecular Sciences, 25(10), 5272. https://doi.org/10.3390/ijms25105272







