The Current Situation and Development Prospect of Whole-Genome Screening
Abstract
1. Introduction
2. Traditional Screening Techniques
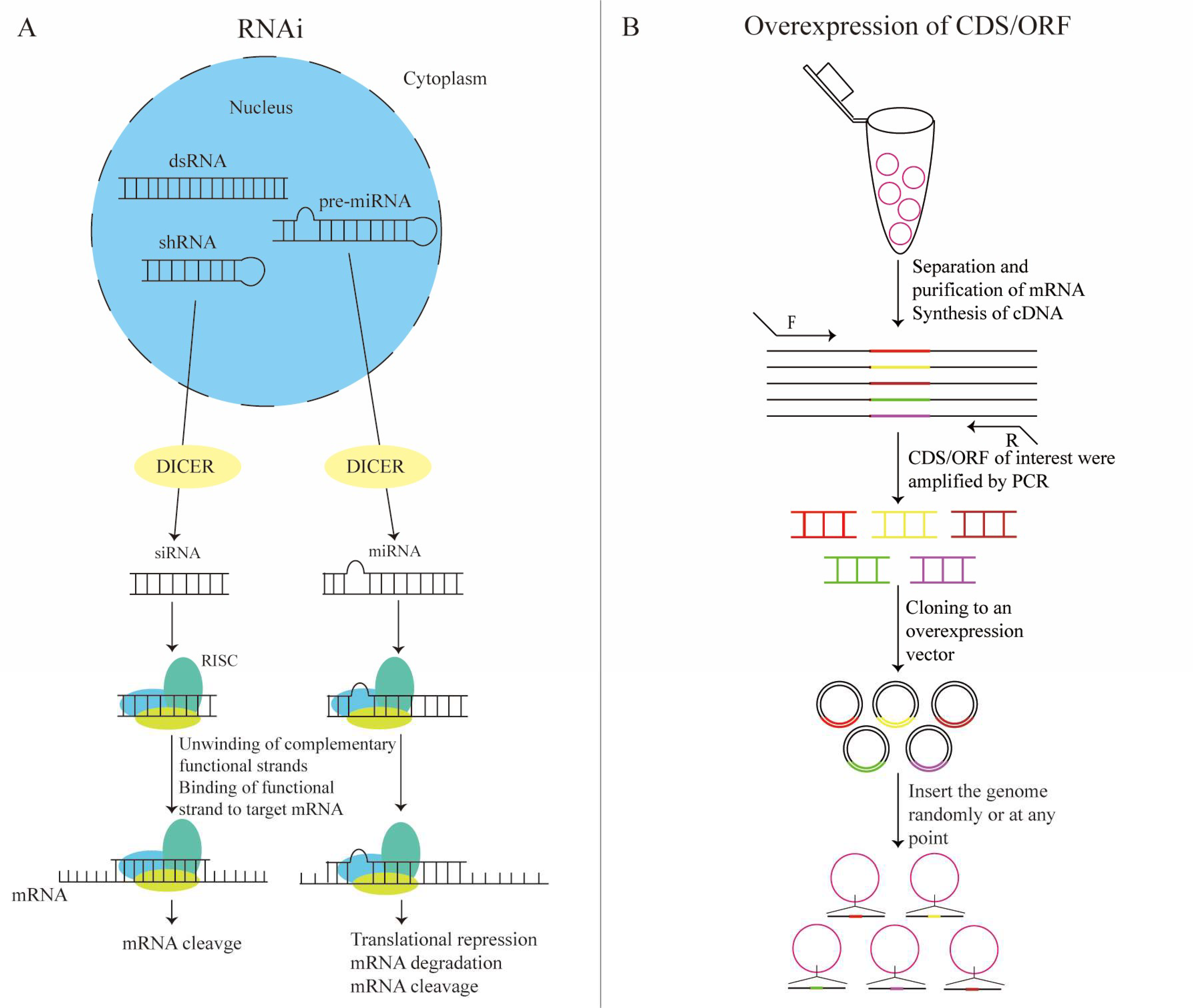
2.1. RNAi Screening
2.2. Overexpression of cDNA/ORF
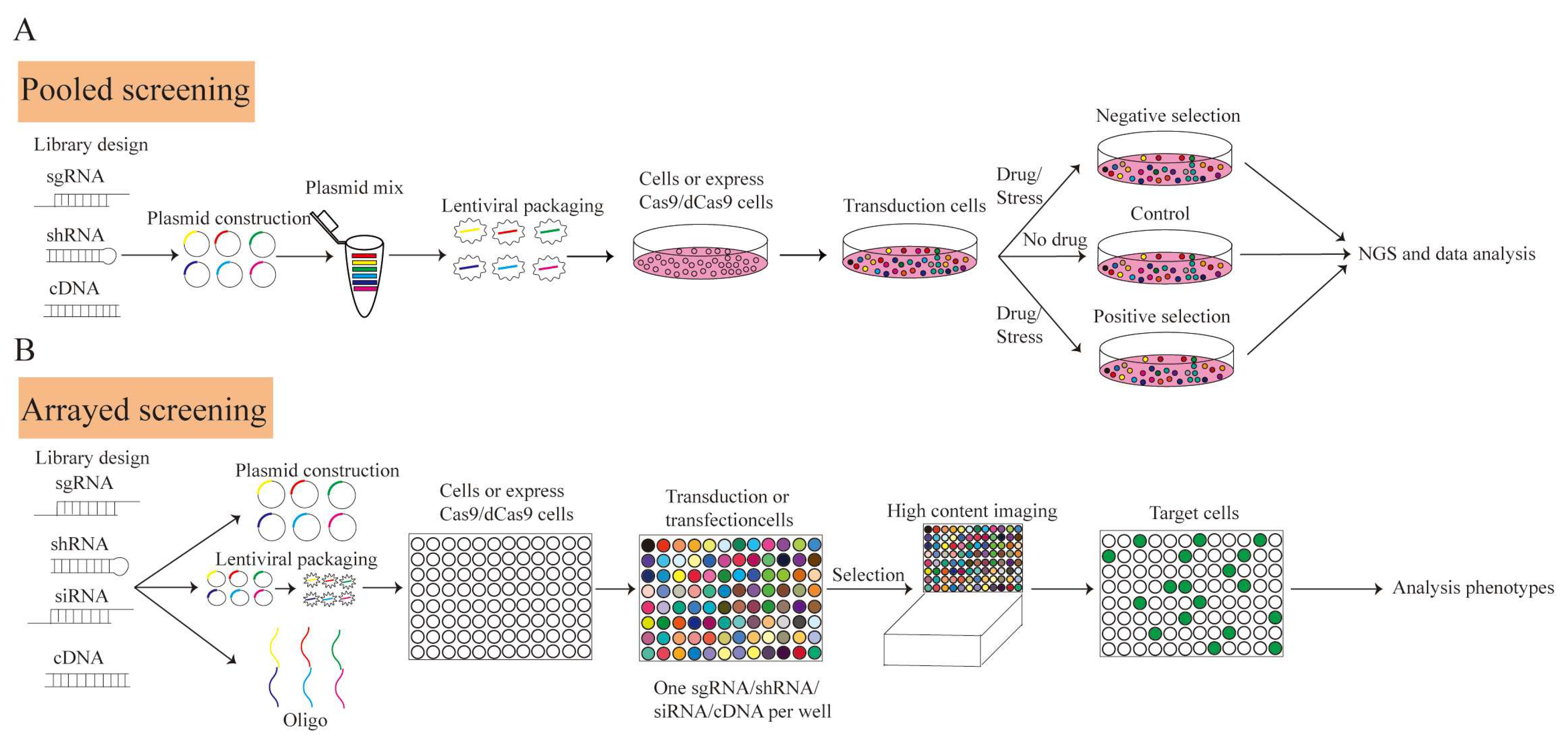
3. CRISPR Screening
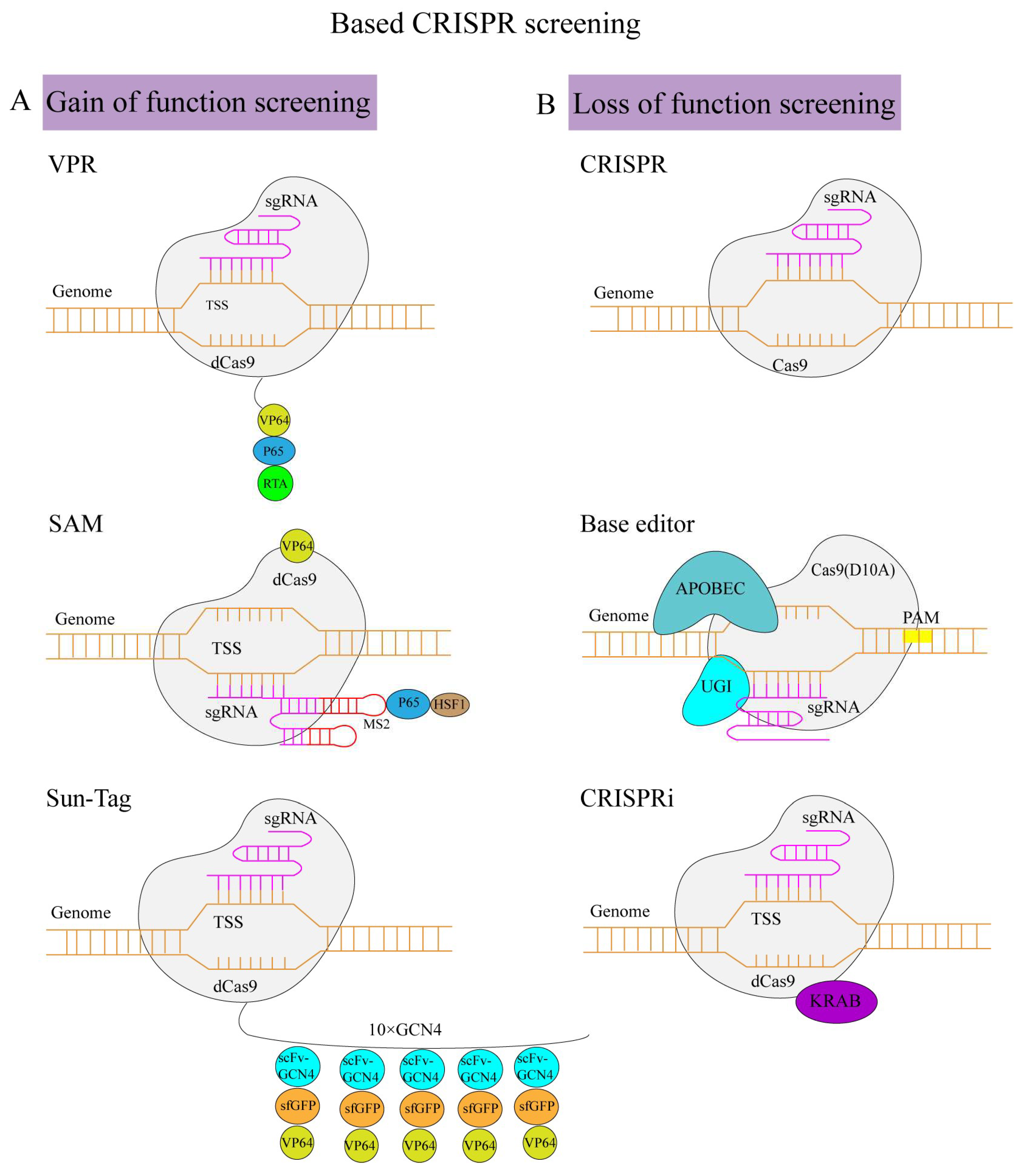
3.1. CRISPR Activation Screening
3.2. CRISPR Knock-Out Screening
3.3. CRISPR Interference Screening
3.4. Base Editing Screening
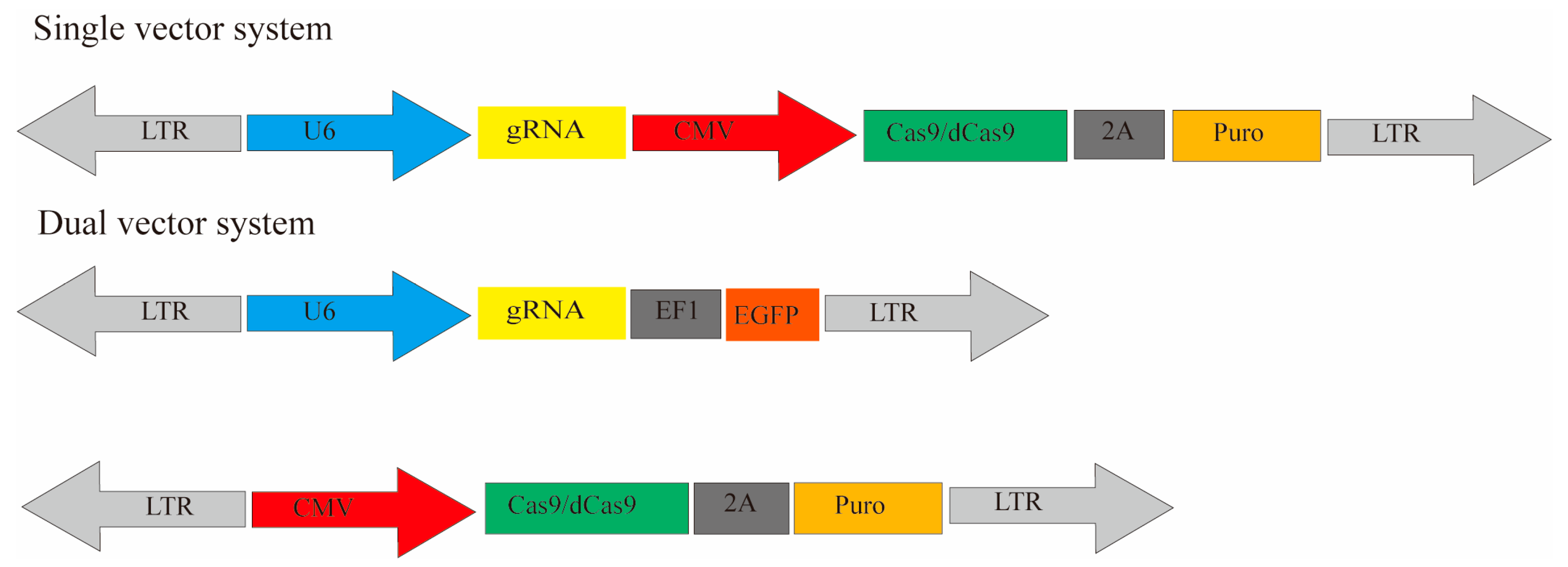
4. Screening Delivery Systems
5. Bioinformatics Analysis
6. Joint Applications with Other Technologies
7. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Chen, C.; Wang, Z.; Qin, Y. CRISPR/Cas9 system: Recent applications in immuno-oncology and cancer immunotherapy. Exp. Hematol. Oncol. 2023, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, M.; Maddalo, D. Applications of CRISPR-Cas9 for advancing precision medicine in oncology: From target discovery to disease modeling. Front. Genet. 2023, 14, 1273994. [Google Scholar] [CrossRef] [PubMed]
- Meyers, S.; Demeyer, S.; Cools, J. CRISPR screening in hematology research: From bulk to single-cell level. J. Hematol. Oncol. 2023, 16, 107. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Uno, N.; Ding, X.; Avery, L.; Banach, D.; Liu, C. Bioinspired CRISPR-Mediated Cascade Reaction Biosensor for Molecular Detection of HIV Using a Glucose Meter. ACS Nano 2023, 17, 3966–3975. [Google Scholar] [CrossRef] [PubMed]
- Nouri, R.; Jiang, Y.; Politza, A.J.; Liu, T.; Greene, W.H.; Zhu, Y.; Nunez, J.J.; Lian, X.; Guan, W. STAMP-Based Digital CRISPR-Cas13a for Amplification-Free Quantification of HIV-1 Plasma Viral Loads. ACS Nano 2023, 17, 10701–10712. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Su, S.; Zhu, Y.; Cheng, X.; Cheng, C.; Chen, L.; Lei, A.; Zhang, L.; Xu, Y.; Ye, D.; et al. Metabolic Reprogramming via ACOD1 depletion enhances function of human induced pluripotent stem cell-derived CAR-macrophages in solid tumors. Nat. Commun. 2023, 14, 5778. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chao, J.; Wang, C.; Sun, G.; Roeth, D.; Liu, W.; Chen, X.; Li, L.; Tian, E.; Feng, L.; et al. Astrocytic response mediated by the CLU risk allele inhibits OPC proliferation and myelination in a human iPSC model. Cell Rep. 2023, 42, 112841. [Google Scholar] [CrossRef]
- Samelson, A.J.; Ariqat, N.; McKetney, J.; Rohanitazangi, G.; Bravo, C.P.; Goodness, D.; Tian, R.; Grosjean, P.; Abskharon, R.; Eisenberg, D.; et al. CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Ebrahimi, N.; Manavi, M.S.; Nazari, A.; Momayezi, A.; Faghihkhorasani, F.; Rasool Riyadh Abdulwahid, A.H.; Rezaei-Tazangi, F.; Kavei, M.; Rezaei, R.; Mobarak, H.; et al. Nano-scale delivery systems for siRNA delivery in cancer therapy: New era of gene therapy empowered by nanotechnology. Environ. Res. 2023, 239 Pt 2, 117263. [Google Scholar] [CrossRef]
- Biswal, P.; Lalruatfela, A.; Behera, S.K.; Biswal, S.; Mallick, B. miR-203a-A multifaceted regulator modulating cancer hallmarks and therapy response. IUBMB Life 2023. [CrossRef]
- Jiang, L.; Qi, Y.; Yang, L.; Miao, Y.; Ren, W.; Liu, H.; Huang, Y.; Huang, S.; Chen, S.; Shi, Y.; et al. Remodeling the tumor immune microenvironment via siRNA therapy for precision cancer treatment. Asian J. Pharm. Sci. 2023, 18, 100852. [Google Scholar] [CrossRef] [PubMed]
- Umer, Z.; Akhtar, J.; Khan, M.H.F.; Shaheen, N.; Haseeb, M.A.; Mazhar, K.; Mithani, A.; Anwar, S.; Tariq, M. Genome-wide RNAi screen in Drosophila reveals Enok as a novel trithorax group regulator. Epigenetics Chromatin 2019, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Neault, N.; O’Reilly, S.; Baig, A.T.; Plaza-Diaz, J.; Azimi, M.; Farooq, F.; Baird, S.D.; MacKenzie, A. High-throughput kinome-RNAi screen identifies protein kinase R activator (PACT) as a novel genetic modifier of CUG foci integrity in myotonic dystrophy type 1 (DM1). PLoS ONE 2021, 16, e0256276. [Google Scholar] [CrossRef] [PubMed]
- Houseman, M.; Huang, M.Y.; Huber, M.; Staiger, M.; Zhang, L.; Hoffmann, A.; Lippuner, C.; Stüber, F. Flow cytometry-based high-throughput RNAi screening for miRNAs regulating MHC class II HLA-DR surface expression. Eur. J. Immunol. 2022, 52, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Žemaitis, K.; Subramaniam, A.; Galeev, R.; Prosz, A.; Jassinskaja, M.; Hansson, J.; Larsson, J. RNAi Screen Identifies MTA1 as an Epigenetic Modifier of Differentiation Commitment in Human HSPCs. Exp. Hematol. 2022, 115, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Booker, M.; Samsonova, A.A.; Kwon, Y.; Flockhart, I.; Mohr, S.E.; Perrimon, N. False negative rates in Drosophila cell-based RNAi screens: A case study. BMC Genom. 2011, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. RNA interference in human cells is restricted to the cytoplasm. RNA 2002, 8, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Su, K.C.; Tsang, M.J.; Emans, N.; Cheeseman, I.M. CRISPR/Cas9-based gene targeting using synthetic guide RNAs enables robust cell biological analyses. Mol. Biol. Cell 2018, 29, 2370–2377. [Google Scholar] [CrossRef]
- Grimm, D. The dose can make the poison: Lessons learned from adverse in vivo toxicities caused by RNAi overexpression. Silence 2011, 2, 8. [Google Scholar] [CrossRef]
- Frecot, D.I.; Froehlich, T.; Rothbauer, U. 30 years of nanobodies—An ongoing success story of small binders in biological research. J. Cell Sci. 2023, 136, jcs261395. [Google Scholar] [CrossRef]
- Mita, M. Relaxin-like Gonad-Stimulating Peptides in Asteroidea. Biomolecules 2023, 13, 781. [Google Scholar] [CrossRef]
- Verwilt, J.; Mestdagh, P.; Vandesompele, J. Artifacts and biases of the reverse transcription reaction in RNA sequencing. RNA 2023, 29, 889–897. [Google Scholar] [CrossRef]
- Zhang, P.; Kratz, A.S.; Salama, M.; Elabd, S.; Heinrich, T.; Wittbrodt, J.; Blattner, C.; Davidson, G. Expression screening using a Medaka cDNA library identifies evolutionarily conserved regulators of the p53/Mdm2 pathway. BMC Biotechnol. 2015, 15, 92. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Dong, C.; Liu, T.; Shi, Y.; Wang, H.; Tao, Z.; Liang, Y.; Lian, J. Improved Functional Expression of Cytochrome P450s in Saccharomyces cerevisiae Through Screening a cDNA Library From Arabidopsis thaliana. Front. Bioeng. Biotechnol. 2021, 9, 764851. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Miyamoto, T.; Higuchi, S.; Ono, M.; Kobara, H.; Asaka, R.; Ando, H.; Suzuki, A.; Shiozawa, T. cDNA expression library screening revealed novel functional genes involved in clear cell carcinogenesis of the ovary in vitro. J. Obstet. Gynaecol. J. Inst. Obstet. Gynaecol. 2021, 41, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.H.M.; Khoshakhlagh, P.; Rojo Arias, J.E.; Pasquini, G.; Wang, K.; Swiersy, A.; Shipman, S.L.; Appleton, E.; Kiaee, K.; Kohman, R.E.; et al. A comprehensive library of human transcription factors for cell fate engineering. Nat. Biotechnol. 2021, 39, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Legut, M.; Gajic, Z.; Guarino, M.; Daniloski, Z.; Rahman, J.A.; Xue, X.; Lu, C.; Lu, L.; Mimitou, E.P.; Hao, S.; et al. A genome-scale screen for synthetic drivers of T cell proliferation. Nature 2022, 603, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef]
- Bhaya, D.; Davison, M.; Barrangou, R. CRISPR-Cas Systems in Bacteria and Archaea: Versatile Small RNAs for Adaptive Defense and Regulation. Annu. Rev. Genet. 2011, 45, 273–297. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Lou, Z.; Minter-Dykhouse, K.; Franco, S.; Gostissa, M.; Rivera, M.A.; Celeste, A.; Manis, J.P.; van Deursen, J.; Nussenzweig, A.; Paull, T.T.; et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 2006, 21, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R.; Ma, Y.; Pannicke, U.; Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003, 4, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Kanafi, M.M.; Tavallaei, M. Overview of advances in CRISPR/deadCas9 technology and its applications in human diseases. Gene 2022, 830, 146518. [Google Scholar] [CrossRef] [PubMed]
- Coukos, R.; Yao, D.; Sanchez, M.I.; Strand, E.T.; Olive, M.E.; Udeshi, N.D.; Weissman, J.S.; Carr, S.A.; Bassik, M.C.; Ting, A.Y. An engineered transcriptional reporter of protein localization identifies regulators of mitochondrial and ER membrane protein trafficking in high-throughput CRISPRi screens. eLife 2021, 10, e69142. [Google Scholar] [CrossRef] [PubMed]
- De Bakker, V.; Liu, X.; Bravo, A.M.; Veening, J.W. CRISPRi-seq for genome-wide fitness quantification in bacteria. Nat. Protoc. 2022, 17, 252–281. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.-A.; Frick, L.; Scheidmann, M.C.; Liu, T.; Trevisan, C.; Dhingra, A.; Spinelli, A.; Wu, Y.; Yao, L.; Vena, D.L.; et al. Robust and Versatile Arrayed Libraries for Human Genome-Wide CRISPR Activation, Deletion and Silencing. bioRxiv 2023. [Google Scholar] [CrossRef]
- Chong, Z.S.; Wright, G.J.; Sharma, S. Investigating Cellular Recognition Using CRISPR/Cas9 Genetic Screening. Trends Cell Biol. 2020, 30, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.C.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.R.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Chavez, A.; Tuttle, M.; Pruitt, B.W.; Ewen-Campen, B.; Chari, R.; Ter-Ovanesyan, D.; Haque, S.J.; Cecchi, R.J.; Kowal, E.J.K.; Buchthal, J.; et al. Comparison of Cas9 activators in multiple species. Nat. Methods 2016, 13, 563–567. [Google Scholar] [CrossRef]
- Alda-Catalinas, C.; Bredikhin, D.; Hernando-Herraez, I.; Santos, F.; Kubinyecz, O.; Eckersley-Maslin, M.A.; Stegle, O.; Reik, W. A Single-Cell Transcriptomics CRISPR-Activation Screen Identifies Epigenetic Regulators of the Zygotic Genome Activation Program. Cell Syst. 2020, 11, 25–41.e9. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, C.; Daley, T.P.; Wang, F.; Cao, W.S.; Bhate, S.; Lin, X.; Still, C., 2nd; Liu, H.; Zhao, D.; et al. CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell 2018, 23, 758–771.e8. [Google Scholar] [CrossRef]
- Yang, J.; Rajan, S.S.; Friedrich, M.J.; Lan, G.; Zou, X.; Ponstingl, H.; Garyfallos, D.A.; Liu, P.; Bradley, A.; Metzakopian, E. Genome-Scale CRISPRa Screen Identifies Novel Factors for Cellular Reprogramming. Stem Cell Rep. 2019, 12, 757–771. [Google Scholar] [CrossRef]
- Schmidt, R.; Steinhart, Z.; Layeghi, M.; Freimer, J.W.; Bueno, R.; Nguyen, V.Q.; Blaeschke, F.; Ye, C.J.; Marson, A. CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science 2022, 375, eabj4008. [Google Scholar] [CrossRef]
- Liu, S.; Striebel, J.; Pasquini, G.; Ng, A.H.M.; Khoshakhlagh, P.; Church, G.M.; Busskamp, V. Neuronal Cell-type Engineering by Transcriptional Activation. Front. Genome Ed. 2021, 3, 715697. [Google Scholar] [CrossRef]
- Hazan, J.; Bester, A.C. CRISPR-Based Approaches for the High-Throughput Characterization of Long Non-Coding RNAs. Non-Coding RNA 2021, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, J.S.L.; Tilgner, K.; Ong, S.H.; Koike-Yusa, H.; Yusa, K. Genome-wide CRISPR-KO Screen Uncovers mTORC1-Mediated Gsk3 Regulation in Naive Pluripotency Maintenance and Dissolution. Cell Rep. 2018, 24, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Shen, H.; Huang, W.; He, S.; Chen, J.; Zhang, D.; Shen, Y.; Sun, Y. Genome-scale CRISPR-Cas9 knockout screening in hepatocellular carcinoma with lenvatinib resistance. Cell Death Discov. 2021, 7, 359. [Google Scholar] [CrossRef] [PubMed]
- Loo, C.S.; Gatchalian, J.; Liang, Y.; Leblanc, M.; Xie, M.; Ho, J.; Venkatraghavan, B.; Hargreaves, D.C.; Zheng, Y. A Genome-wide CRISPR Screen Reveals a Role for the Non-canonical Nucleosome-Remodeling BAF Complex in Foxp3 Expression and Regulatory T Cell Function. Immunity 2020, 53, 143–157.e8. [Google Scholar] [CrossRef] [PubMed]
- Sutra Del Galy, A.; Menegatti, S.; Fuentealba, J.; Lucibello, F.; Perrin, L.; Helft, J.; Darbois, A.; Saitakis, M.; Tosello, J.; Rookhuizen, D.; et al. In vivo genome-wide CRISPR screens identify SOCS1 as intrinsic checkpoint of CD4(+) T(H)1 cell response. Sci. Immunol. 2021, 6, eabe8219. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Evers, B.; Jastrzebski, K.; Heijmans, J.P.; Grernrum, W.; Beijersbergen, R.L.; Bernards, R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016, 34, 631–633. [Google Scholar] [CrossRef]
- Kampmann, M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 2020, 16, 465–480. [Google Scholar] [CrossRef]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018, 24, 927–930. [Google Scholar] [CrossRef]
- Yuen, G.; Khan, F.J.; Gao, S.; Stommel, J.M.; Batchelor, E.; Wu, X.; Luo, J. CRISPR/Cas9-mediated gene knockout is insensitive to target copy number but is dependent on guide RNA potency and Cas9/sgRNA threshold expression level. Nucleic Acids Res. 2017, 45, 12039–12053. [Google Scholar] [CrossRef] [PubMed]
- Sanson, K.R.; Hanna, R.E.; Hegde, M.; Donovan, K.F.; Strand, C.; Sullender, M.E.; Vaimberg, E.W.; Goodale, A.; Root, D.E.; Piccioni, F.; et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 2018, 9, 5416. [Google Scholar] [CrossRef]
- Groner, A.C.; Meylan, S.; Ciuffi, A.; Zangger, N.; Ambrosini, G.; Dénervaud, N.; Bucher, P.; Trono, D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010, 6, e1000869. [Google Scholar] [CrossRef] [PubMed]
- La Russa, M.F.; Qi, L.S. The New State of the Art: Cas9 for Gene Activation and Repression. Mol. Cell. Biol. 2015, 35, 3800–3809. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Gachechiladze, M.A.; Ludwig, C.H.; Laurie, M.T.; Hong, J.Y.; Nathaniel, D.; Prabhu, A.V.; Fernandopulle, M.S.; Patel, R.; Abshari, M.; et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 2019, 104, 239–255.e12. [Google Scholar] [CrossRef] [PubMed]
- Genga, R.M.J.; Kernfeld, E.M.; Parsi, K.M.; Parsons, T.J.; Ziller, M.J.; Maehr, R. Single-Cell RNA-Sequencing-Based CRISPRi Screening Resolves Molecular Drivers of Early Human Endoderm Development. Cell Rep. 2019, 27, 708–718.e10. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Ni, K.; Liu, M.; Li, Y.; Wang, L.; Wang, Y.; Liu, Y.; Yu, Z.; Qi, Y.; Lu, Z.; et al. Direct-seq: Programmed gRNA scaffold for streamlined scRNA-seq in CRISPR screen. Genome Biol. 2020, 21, 136. [Google Scholar] [CrossRef]
- Haswell, J.R.; Mattioli, K.; Gerhardinger, C.; Maass, P.G.; Foster, D.J.; Peinado, P.; Wang, X.; Medina, P.P.; Rinn, J.L.; Slack, F.J. Genome-wide CRISPR interference screen identifies long non-coding RNA loci required for differentiation and pluripotency. PLoS ONE 2021, 16, e0252848. [Google Scholar] [CrossRef]
- Cress, B.F.; Leitz, Q.D.; Kim, D.C.; Amore, T.D.; Suzuki, J.Y.; Linhardt, R.J.; Koffas, M.A. CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Factories 2017, 16, 10. [Google Scholar] [CrossRef]
- Mandegar, M.A.; Huebsch, N.; Frolov, E.B.; Shin, E.; Truong, A.; Olvera, M.P.; Chan, A.H.; Miyaoka, Y.; Holmes, K.; Spencer, C.I.; et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 2016, 18, 541–553. [Google Scholar] [CrossRef]
- Rosenbluh, J.; Xu, H.; Harrington, W.; Gill, S.; Wang, X.; Vazquez, F.; Root, D.E.; Tsherniak, A.; Hahn, W.C. Complementary information derived from CRISPR Cas9 mediated gene deletion and suppression. Nat. Commun. 2017, 8, 15403. [Google Scholar] [CrossRef]
- Cui, L.; Vigouroux, A.; Rousset, F.; Varet, H.; Khanna, V.; Bikard, D. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 2018, 9, 1912. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420. [Google Scholar] [CrossRef] [PubMed]
- Ranzoni, A.M.; Tangherloni, A.; Berest, I.; Riva, S.G.; Myers, B.; Strzelecka, P.M.; Xu, J.; Panada, E.; Mohorianu, I.; Zaugg, J.B.; et al. Integrative Single-Cell RNA-Seq and ATAC-Seq Analysis of Human Developmental Hematopoiesis. Cell Stem Cell 2021, 28, 472–487.e7. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.E.; Hegde, M.; Fagre, C.R.; DeWeirdt, P.C.; Sangree, A.K.; Szegletes, Z.; Griffith, A.; Feeley, M.N.; Sanson, K.R.; Baidi, Y.; et al. Massively parallel assessment of human variants with base editor screens. Cell 2021, 184, 1064–1080.e20. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, G.; Wu, J.; Liang, J.; Wang, X. Identification of pathogenic variants in cancer genes using base editing screens with editing efficiency correction. Genome Biol. 2021, 22, 80. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Liu, F.; Ren, Z.; Chen, W.; Chen, Y.; Liu, T.; Ma, Y.; Cao, N.; Wang, J. Parallel functional assessment of m(6)A sites in human endodermal differentiation with base editor screens. Nat. Commun. 2022, 13, 478. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Gao, X.D.; Podracky, C.J.; Nelson, A.T.; Koblan, L.W.; Raguram, A.; Levy, J.M.; Mercer, J.A.M.; Liu, D.R. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 2022, 40, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Liu, Z.; Liu, Y.; Ma, H.; Xu, Y.; Bao, Y.; Zhu, S.; Cao, Z.; Wu, Z.; Zhou, Z.; et al. Genome-wide interrogation of gene functions through base editor screens empowered by barcoded sgRNAs. Nat. Biotechnol. 2021, 39, 1403–1413. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Sakata, R.C.; Ishiguro, S.; Mori, H.; Tanaka, M.; Tatsuno, K.; Ueda, H.; Yamamoto, S.; Seki, M.; Masuyama, N.; Nishida, K.; et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020, 38, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Luft, C.; Ketteler, R. Electroporation Knows No Boundaries: The Use of Electrostimulation for siRNA Delivery in Cells and Tissues. J. Biomol. Screen. 2015, 20, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Xue, W.; Zhao, Y.; Ning, G.; Wang, J. CRISPR-based modular assembly of a UAS-cDNA/ORF plasmid library for more than 5500 Drosophila genes conserved in humans. Genome Res. 2020, 30, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Konermann, S.; Gootenberg, J.S.; Abudayyeh, O.O.; Platt, R.J.; Brigham, M.D.; Sanjana, N.E.; Zhang, F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017, 12, 828–863. [Google Scholar] [CrossRef] [PubMed]
- Segel, M.; Lash, B.; Song, J.; Ladha, A.; Liu, C.C.; Jin, X.; Mekhedov, S.L.; Macrae, R.K.; Koonin, E.V.; Zhang, F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021, 373, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.C.; Kim, H.S.; Chen, Y.; Li, Y.; LaMere, M.W.; Chen, C.; Wang, H.; Gong, J.; Palumbo, C.D.; Ashton, J.M.; et al. Scaffold-mediated CRISPR-Cas9 delivery system for acute myeloid leukemia therapy. Sci. Adv. 2021, 7, eabg3217. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.S.; Lee, J.S.; Jeong, J.; Kim, H.; Byun, J.; Kim, S.A.; Lee, H.J.; Chung, H.S.; Lee, J.B.; Ahn, D.R. Poly-sgRNA/siRNA ribonucleoprotein nanoparticles for targeted gene disruption. J. Control. Release 2017, 250, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Mout, R.; Ray, M.; Tonga, G.Y.; Lee, Y.W.; Tay, T.; Sasaki, K.; Rotello, V.M. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano 2017, 11, 2452–2458. [Google Scholar] [CrossRef]
- Mout, R.; Rotello, V.M. A General Method for Intracellular Protein Delivery through ‘E-tag’ Protein Engineering and Arginine Functionalized Gold Nanoparticles. Bio-Protocol 2017, 7, e2661. [Google Scholar] [CrossRef]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. Engl. 2015, 54, 12029–12033. [Google Scholar] [CrossRef]
- Zhou, W.H.; Cui, H.D.; Ying, L.M.; Yu, X.F. Enhanced Cytosolic Delivery and Release of CRISPR/Cas9 by Black Phosphorus Nanosheets for Genome Editing. Angew. Chem. Int. Ed. 2018, 57, 10268–10272. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Zhou, X.; Cheng, M.; Xing, D. Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing. Nanoscale 2018, 10, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Alsaiari, S.K.; Patil, S.; Alyami, M.; Alamoudi, K.O.; Aleisa, F.A.; Merzaban, J.S.; Li, M.; Khashab, N.M. Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework. J. Am. Chem. Soc. 2018, 140, 143–146. [Google Scholar] [CrossRef]
- Chou, L.Y.; Ming, K.; Chan, W.C. Strategies for the intracellular delivery of nanoparticles. Chem. Soc. Rev. 2011, 40, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 2014, 43, 744–764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, H.; Bao, G. Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano 2015, 9, 8655–8671. [Google Scholar] [CrossRef] [PubMed]
- O’Keeffe Ahern, J.; Lara-Saez, I.; Zhou, D.; Murillas, R.; Bonafont, J.; Mencia, A.; Garcia, M.; Manzanares, D.; Lynch, J.; Foley, R.; et al. Non-viral delivery of CRISPR-Cas9 complexes for targeted gene editing via a polymer delivery system. Gene Ther. 2022, 29, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Avci-Adali, M.; Santos, H.A. Current trends in delivery of non-viral nucleic acid-based therapeutics for improved efficacy. Adv. Drug Deliv. Rev. 2022, 185, 114297. [Google Scholar] [CrossRef]
- Liu, N.; Zhu, L.; Sun, H.; Zhou, Z.; Dong, J.; Sun, M. Crosslinked Protein Delivery Strategy with Precise Activity Regulation Properties for Cancer Therapy and Gene Editing. Adv. Healthc. Mater. 2022, 11, e2102329. [Google Scholar] [CrossRef]
- Johnson, L.T.; Zhang, D.; Zhou, K.J.; Lee, S.M.; Liu, S.; Dilliard, S.A.; Farbiak, L.; Chatterjee, S.; Lin, Y.H.; Siegwart, D.J. Lipid Nanoparticle (LNP) Chemistry Can Endow Unique In Vivo RNA Delivery Fates within the Liver That Alter Therapeutic Outcomes in a Cancer Model. Mol. Pharm. 2022, 19, 3973–3986. [Google Scholar] [CrossRef]
- Dubey, A.K.; Mostafavi, E. Biomaterials-mediated CRISPR/Cas9 delivery: Recent challenges and opportunities in gene therapy. Front. Chem. 2023, 11, 1259435. [Google Scholar] [CrossRef] [PubMed]
- Haldrup, J.; Andersen, S.; Labial, A.R.L.; Wolff, J.H.; Frandsen, F.P.; Skov, T.W.; Rovsing, A.B.; Nielsen, I.; Jakobsen, T.S.; Askou, A.L.; et al. Engineered lentivirus-derived nanoparticles (LVNPs) for delivery of CRISPR/Cas ribonucleoprotein complexes supporting base editing, prime editing and in vivo gene modification. Nucleic Acids Res. 2023, 51, 10059–10074. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.B.; Stokes, L.D.; Kelly, I.; Henderson, K.M.; Vallecillo-Viejo, I.C.; Colazo, J.M.; Wong, B.V.; Yu, F.; d’Arcy, R.; Struthers, M.N.; et al. Nonviral Delivery of CRISPR-Cas9 Using Protein-Agnostic, High-Loading Porous Silicon and Polymer Nanoparticles. ACS Nano 2023, 17, 16412–16431. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, J.; Xu, C.; Wang, J.; Han, W.; Yang, J.; Wu, S.; An, J.; Liu, J.; Zhang, Z.; et al. Biomimetic Mineralized CRISPR/Cas RNA Nanoparticles for Efficient Tumor-Specific Multiplex Gene Editing. ACS Nano 2023, 17, 15025–15043. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Manan, R.S.; Liang, S.Q.; Gordon, A.; Jiang, A.; Varley, A.; Gao, G.; Langer, R.; Xue, W.; Anderson, D. Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing. Nat. Biotechnol. 2023, 41, 1410–1415. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Toyonaga, S.; Anderson, D.G. In Vivo RNA Delivery to Hematopoietic Stem and Progenitor Cells via Targeted Lipid Nanoparticles. Nano Lett. 2023, 23, 2938–2944. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, X.; Xie, R.; Wang, Y.; Xu, X.; Burger, J.; Gong, S. Guanidinium-Rich Lipopeptide-Based Nanoparticle Enables Efficient Gene Editing in Skeletal Muscles. ACS Appl. Mater. Interfaces 2023, 15, 10464–10476. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.Y.; Carlaw, T.; Thomson, T.; Birkenshaw, A.; Basha, G.; Kurek, D.; Huang, C.; Kulkarni, J.; Zhang, L.H.; Ross, C.J.D. A luciferase reporter mouse model to optimize in vivo gene editing validated by lipid nanoparticle delivery of adenine base editors. Mol. Ther. 2023, 31, 1159–1166. [Google Scholar] [CrossRef]
- Onuma, H.; Sato, Y.; Harashima, H. Lipid nanoparticle-based ribonucleoprotein delivery for in vivo genome editing. J. Control. Release 2023, 355, 406–416. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Hardcastle, T.J.; Kelly, K.A. baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinform. 2010, 11, 422. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.A.; Qin, H.; Ramalho-Santos, M.; Song, J.S. HiTSelect: A comprehensive tool for high-complexity-pooled screen analysis. Nucleic Acids Res. 2015, 43, e16. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, H.; Xiao, T.; Cong, L.; Love, M.I.; Zhang, F.; Irizarry, R.A.; Liu, J.S.; Brown, M.; Liu, X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014, 15, 554. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Koster, J.; Xu, H.; Chen, C.H.; Xiao, T.; Liu, J.S.; Brown, M.; Liu, X.S. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol. 2015, 16, 281. [Google Scholar] [CrossRef]
- Yu, J.; Silva, J.; Califano, A. ScreenBEAM: A novel meta-analysis algorithm for functional genomics screens via Bayesian hierarchical modeling. Bioinformatics 2016, 32, 260–267. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Wang, B.; Wang, M.; Zhang, W.; Xiao, T.; Chen, C.H.; Wu, A.; Wu, F.; Traugh, N.; Wang, X.; Li, Z.; et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat. Protoc. 2019, 14, 756–780. [Google Scholar] [CrossRef]
- Hart, T.; Moffat, J. BAGEL: A computational framework for identifying essential genes from pooled library screens. BMC Bioinform. 2016, 17, 164. [Google Scholar] [CrossRef]
- Papalexi, E.; Mimitou, E.P.; Butler, A.W.; Foster, S.; Bracken, B.; Mauck, W.M., 3rd; Wessels, H.H.; Hao, Y.; Yeung, B.Z.; Smibert, P.; et al. Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nat. Genet. 2021, 53, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Dixit, A.; Parnas, O.; Li, B.; Chen, J.; Fulco, C.P.; Jerby-Arnon, L.; Marjanovic, N.D.; Dionne, D.; Burks, T.; Raychowdhury, R.; et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016, 167, 1853–1866.e17. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhu, Y.; Yu, H.; Cheng, X.; Chen, S.; Chu, Y.; Huang, H.; Zhang, J.; Li, W. scMAGeCK links genotypes with multiple phenotypes in single-cell CRISPR screens. Genome Biol. 2020, 21, 19. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Falcon, S.; Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007, 23, 257–258. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Przybyla, L.; Gilbert, L.A. A new era in functional genomics screens. Nat. Rev. Genet. 2022, 23, 89–103. [Google Scholar] [CrossRef]
- Datlinger, P.; Rendeiro, A.F.; Schmidl, C.; Krausgruber, T.; Traxler, P.; Klughammer, J.; Schuster, L.C.; Kuchler, A.; Alpar, D.; Bock, C. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017, 14, 297–301. [Google Scholar] [CrossRef]
- Xie, S.; Duan, J.; Li, B.; Zhou, P.; Hon, G.C. Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Mol. Cell 2017, 66, 285–299.e5. [Google Scholar] [CrossRef]
- Jaitin, D.A.; Weiner, A.; Yofe, I.; Lara-Astiaso, D.; Keren-Shaul, H.; David, E.; Salame, T.M.; Tanay, A.; van Oudenaarden, A.; Amit, I. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 2016, 167, 1883–1896.e15. [Google Scholar] [CrossRef]
- Replogle, J.M.; Norman, T.M.; Xu, A.; Hussmann, J.A.; Chen, J.; Cogan, J.Z.; Meer, E.J.; Terry, J.M.; Riordan, D.P.; Srinivas, N.; et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 2020, 38, 954–961. [Google Scholar] [CrossRef]
- Replogle, J.M.; Saunders, R.A.; Pogson, A.N.; Hussmann, J.A.; Lenail, A.; Guna, A.; Mascibroda, L.; Wagner, E.J.; Adelman, K.; Lithwick-Yanai, G.; et al. Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell 2022, 185, 2559–2575.e28. [Google Scholar] [CrossRef]
- Feldman, D.; Singh, A.; Schmid-Burgk, J.L.; Carlson, R.J.; Mezger, A.; Garrity, A.J.; Zhang, F.; Blainey, P.C. Optical Pooled Screens in Human Cells. Cell 2019, 179, 787–799.e17. [Google Scholar] [CrossRef] [PubMed]
- Binan, L.; Danquah, S.; Valakh, V.; Simonton, B.; Bezney, J.; Nehme, R.; Cleary, B.; Farhi, S.L. Simultaneous CRISPR screening and spatial transcriptomics reveals intracellular, intercellular, and functional transcriptional circuits. bioRxiv 2023. [Google Scholar] [CrossRef]
- Cuella-Martin, R.; Hayward, S.B.; Fan, X.; Chen, X.; Huang, J.W.; Taglialatela, A.; Leuzzi, G.; Zhao, J.; Rabadan, R.; Lu, C.; et al. Functional interrogation of DNA damage response variants with base editing screens. Cell 2021, 184, 1081–1097.e19. [Google Scholar] [CrossRef] [PubMed]
- Alemany, A.; Florescu, M.; Baron, C.S.; Peterson-Maduro, J.; van Oudenaarden, A. Whole-organism clone tracing using single-cell sequencing. Nature 2018, 556, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Raj, B.; Wagner, D.E.; McKenna, A.; Pandey, S.; Klein, A.M.; Shendure, J.; Gagnon, J.A.; Schier, A.F. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 2018, 36, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.M.; Smith, Z.D.; Grosswendt, S.; Kretzmer, H.; Norman, T.M.; Adamson, B.; Jost, M.; Quinn, J.J.; Yang, D.; Jones, M.G.; et al. Molecular recording of mammalian embryogenesis. Nature 2019, 570, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bowling, S.; McGeary, S.E.; Yu, Q.; Lemke, B.; Alcedo, K.; Jia, Y.; Liu, X.; Ferreira, M.; Klein, A.M.; et al. A mouse model with high clonal barcode diversity for joint lineage, transcriptomic, and epigenomic profiling in single cells. Cell 2023, 186, 5183–5199.e22. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Liu, H.; You, Z.; Wang, L.; Li, Y.; Zhang, X.; Ji, X.; He, H.; Yuan, T.; Zheng, W.; et al. Comprehensive spatiotemporal mapping of single-cell lineages in developing mouse brain by CRISPR-based barcoding. Nat. Methods 2023, 20, 1244–1255. [Google Scholar] [CrossRef]
- Frieda, K.L.; Linton, J.M.; Hormoz, S.; Choi, J.; Chow, K.K.; Singer, Z.S.; Budde, M.W.; Elowitz, M.B.; Cai, L. Synthetic recording and in situ readout of lineage information in single cells. Nature 2017, 541, 107–111. [Google Scholar] [CrossRef]
- Liscovitch-Brauer, N.; Montalbano, A.; Deng, J.; Mendez-Mancilla, A.; Wessels, H.H.; Moss, N.G.; Kung, C.Y.; Sookdeo, A.; Guo, X.; Geller, E.; et al. Profiling the genetic determinants of chromatin accessibility with scalable single-cell CRISPR screens. Nat. Biotechnol. 2021, 39, 1270–1277. [Google Scholar] [CrossRef]
- Rubin, A.J.; Parker, K.R.; Satpathy, A.T.; Qi, Y.; Wu, B.; Ong, A.J.; Mumbach, M.R.; Ji, A.L.; Kim, D.S.; Cho, S.W.; et al. Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 2019, 176, 361–376.e17. [Google Scholar] [CrossRef]
- Pierce, S.E.; Granja, J.M.; Greenleaf, W.J. High-throughput single-cell chromatin accessibility CRISPR screens enable unbiased identification of regulatory networks in cancer. Nat. Commun. 2021, 12, 2969. [Google Scholar] [CrossRef]
- Frangieh, C.J.; Melms, J.C.; Thakore, P.I.; Geiger-Schuller, K.R.; Ho, P.; Luoma, A.M.; Cleary, B.; Jerby-Arnon, L.; Malu, S.; Cuoco, M.S.; et al. Multimodal pooled Perturb-CITE-seq screens in patient models define mechanisms of cancer immune evasion. Nat. Genet. 2021, 53, 332–341. [Google Scholar] [CrossRef]
- Dhainaut, M.; Rose, S.A.; Akturk, G.; Wroblewska, A.; Nielsen, S.R.; Park, E.S.; Buckup, M.; Roudko, V.; Pia, L.; Sweeney, R.; et al. Spatial CRISPR genomics identifies regulators of the tumor microenvironment. Cell 2022, 185, 1223–1239.e20. [Google Scholar] [CrossRef] [PubMed]
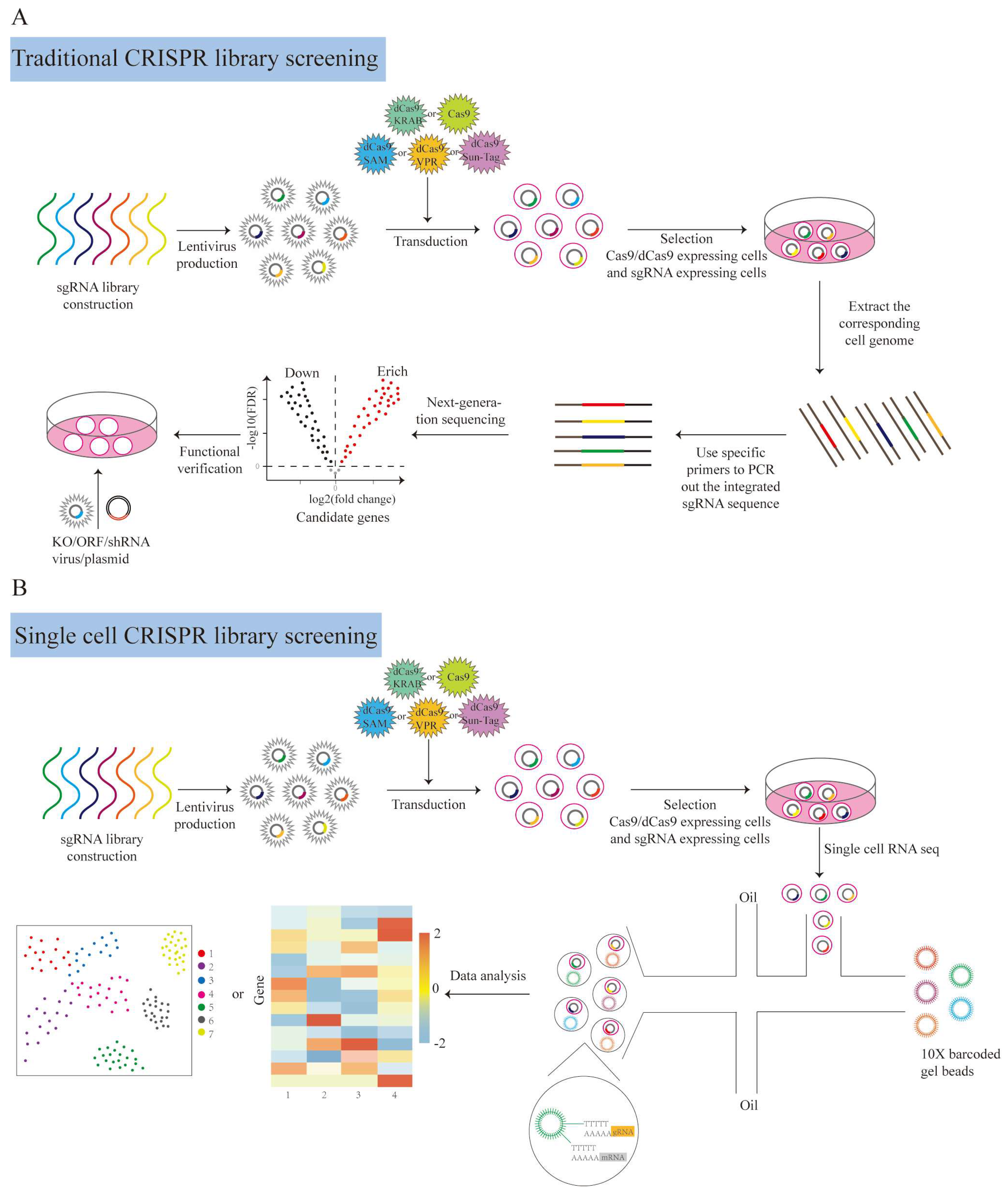
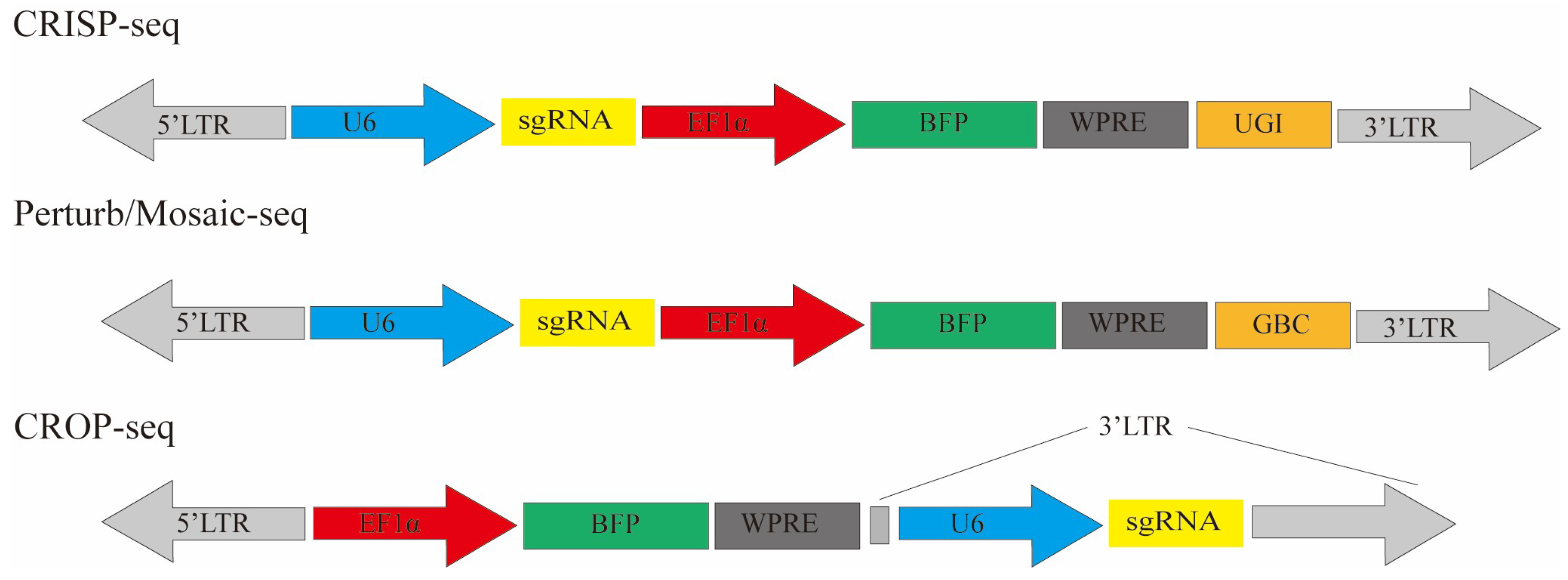
| Type | Size | Source | Dicer Enzyme Processing Method | Target Site | Mechanism of Action | Cellular Delivery Method |
|---|---|---|---|---|---|---|
| shRNA | 50–100 base pairs | Local complementary paired double-stranded RNA formed by a hairpin structure | Double-stranded RNA cleavage on both strands | Arbitrary position of mRNA | mRNA degradation (transcriptional level regulation) | Requires vector, such as lentiviral transduction shRNA library |
| siRNA | 20–25 base pairs | Artificially synthesized linear double-stranded RNA with fully complementary base pairing in the duplex region | Double-stranded RNA cleavage on both strands RNA | Arbitrary position of mRNA | mRNA degradation (transcriptional level regulation) | Transfection of dsRNA or siRNA |
| miRNA | 21–23 nucleotides | Local duplex formed by a hairpin structure, often with partial complementarity | Stem-loop removal | Target gene 3′-UTR region | mRNA degradation (transcriptional level regulation) or inhibition of mRNA translation (translational level regulation) | Transfection of miRNA |
| Functional Genomics Screening Technique | Element | Toxicity | Off-Target Effect | Gain/Loss-of-Function Type | Reversibility | sgRNA Target Region | Adaptability |
|---|---|---|---|---|---|---|---|
| RNAi | shRNA or siRNA | Non-toxicity | High: false positives and false negatives | Knock-down | Reversible | RNA | Mature RNA in the cytoplasm |
| CRISPRi | dCas9/sgRNA/transcriptional repressor | Non-toxicity | Low | Knock-down | Reversible | 50 bp upstream and 300 bp downstream of the TSS of genomic DNA | Coding RNA and lncRNA, i.e., the entire genome |
| CRISPRko | Cas9/sgRNA | DSB toxicity | Low | Knock-out | Irreversible | Arbitrary target site of genomic DNA | Coding RNA and lncRNA, i.e., the entire genome |
| Base editor | dCas9/sgRNA/APOBEC1 | Non-toxicity | Low | Base change | Irreversible | Arbitrary target site of genomic DNA | Genomic DNA |
| cDNA overexpression | cDNA library plasmid | Non-toxicity | - | Overexpression | Irreversible | Arbitrary target site of genomic DNA | Genomic DNA |
| CRISPRa | Cas9/sgRNA/transcriptional repressor | Non-toxicity | Low | Activation | Reversible | 50–500 bp upstream of the TSS of genomic DNA | Coding RNA and lncRNA, i.e., the entire genome |
| System | Transcriptional Activation Element | Molecular Tether |
|---|---|---|
| dCas9-VPR | VP64, p65, Rta | - |
| dCas9-Sun Tag | scFv-GCN4, sfGFP, VP64 | Multimeric GCN4 |
| dCas9-SAM | MCP, p65, HSF1 | MS2 Hairpin |
| Vector Type | Genome Type | Immunogenicity | Integration into Host Genome | Range of Infected Cells | Uniformity of Copy Number Post-Transfection/Transduction | Security and Reasons | Persistence of Exogenous Gene Expression | Packaging System |
|---|---|---|---|---|---|---|---|---|
| Plasmid vector | dsDNA | None | Non-integrating | Limited; difficult to transfect neurons and other cells | Non-uniform | Secure | Transient expression | Plasmid |
| Lentivirus vector | RNA | Low | Random integration | Broad; nearly all mammalian cells | Uniform | Secure 1. Viral packaging requires helper plasmid. 2. 5′ LTR inactivation. | Stable | One transfer plasmid, one membrane protein expression plasmid, two packaging plasmids |
| Adenoviral vector | dsDNA | High | Non-integrating | Relatively widespread; however, difficult to transduce endothelial cells, neurons, etc. | Uniform | Secure 1. Can transduce target cells only. unable to replicate. 2. Viral packaging requires helper plasmid. | Transient expression | Transfer plasmid containing adenoviral genome sequence with the deletion of E1/E3 genes |
| AAV vector | ssDNA | Extremely low | Non-integrating | Relatively widespread; different serotypes correspond to different types of cells | Uniform | Safest, with replication defects | Transient expression | Transfer plasmid, Rep and Cap expression plasmids, and one helper plasmid |
| Transposon vector | dsDNA | None | Integrating | Limited; difficult to transfect neurons and other cells | Non-uniform | Secure | Stable | Helper plasmid encoding transposase, transposon plasmid |
| Retroviral vector | RNA | Low | Random integration | Limited; cannot infect non-dividing cells | Uniform | Secure | Stable | Transfer plasmid, membrane protein expression plasmid, packaging plasmid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Lei, Y.; Ren, T.; Yao, M. The Current Situation and Development Prospect of Whole-Genome Screening. Int. J. Mol. Sci. 2024, 25, 658. https://doi.org/10.3390/ijms25010658
Yang C, Lei Y, Ren T, Yao M. The Current Situation and Development Prospect of Whole-Genome Screening. International Journal of Molecular Sciences. 2024; 25(1):658. https://doi.org/10.3390/ijms25010658
Chicago/Turabian StyleYang, Caiting, Yu Lei, Tinglin Ren, and Mingze Yao. 2024. "The Current Situation and Development Prospect of Whole-Genome Screening" International Journal of Molecular Sciences 25, no. 1: 658. https://doi.org/10.3390/ijms25010658
APA StyleYang, C., Lei, Y., Ren, T., & Yao, M. (2024). The Current Situation and Development Prospect of Whole-Genome Screening. International Journal of Molecular Sciences, 25(1), 658. https://doi.org/10.3390/ijms25010658






