Targeting Farnesoid X Receptor in Tumor and the Tumor Microenvironment: Implication for Therapy
Abstract
1. Introduction
2. Bile Acid Synthesis
3. Bile Acid Receptors
4. Farnesoid X Receptor (FXR) Structure
5. Post-Translational Modifications of FXR
5.1. Sumoylation
5.2. Acetylation
5.3. Methylation
5.4. Phosphorylation
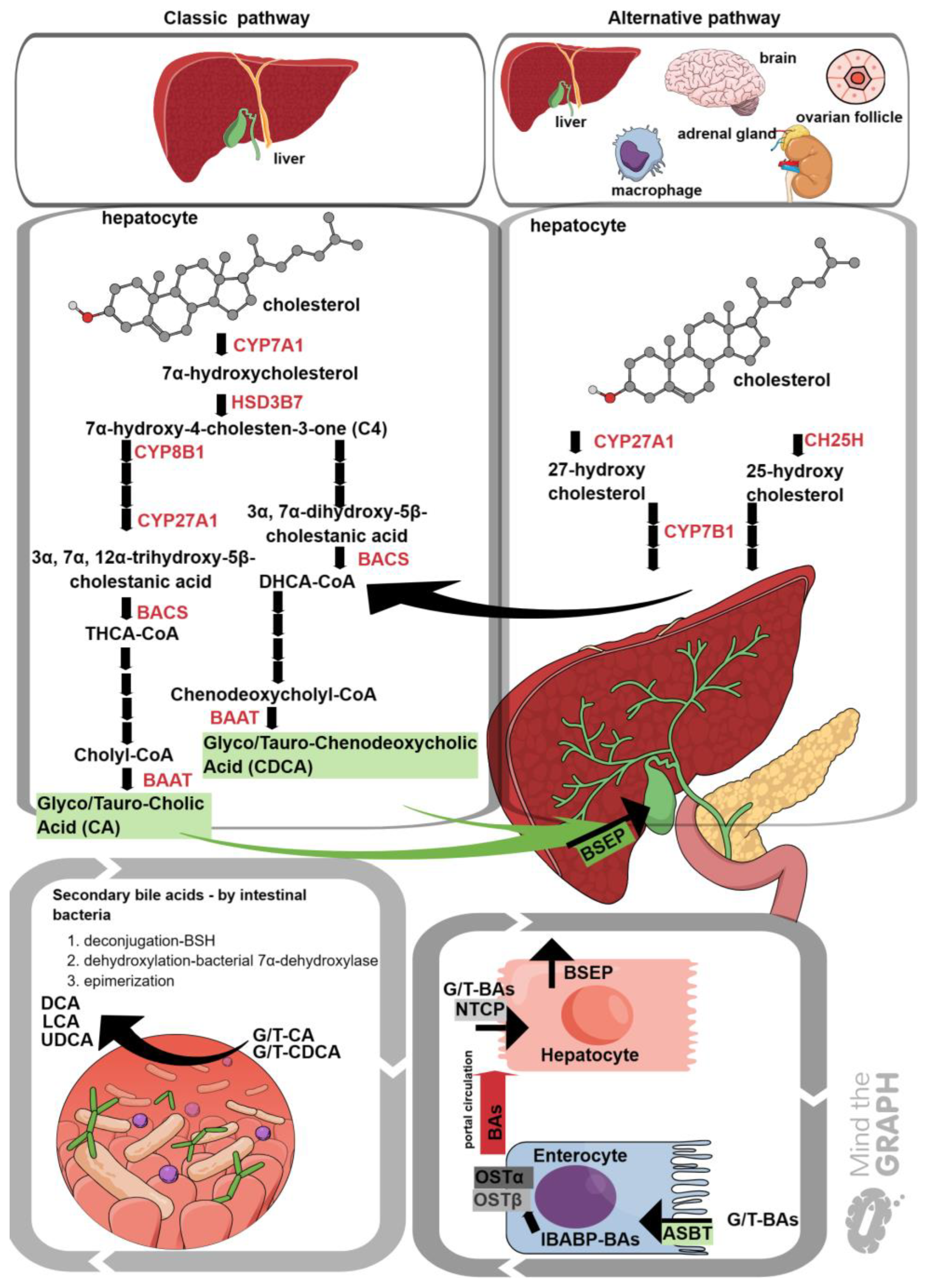
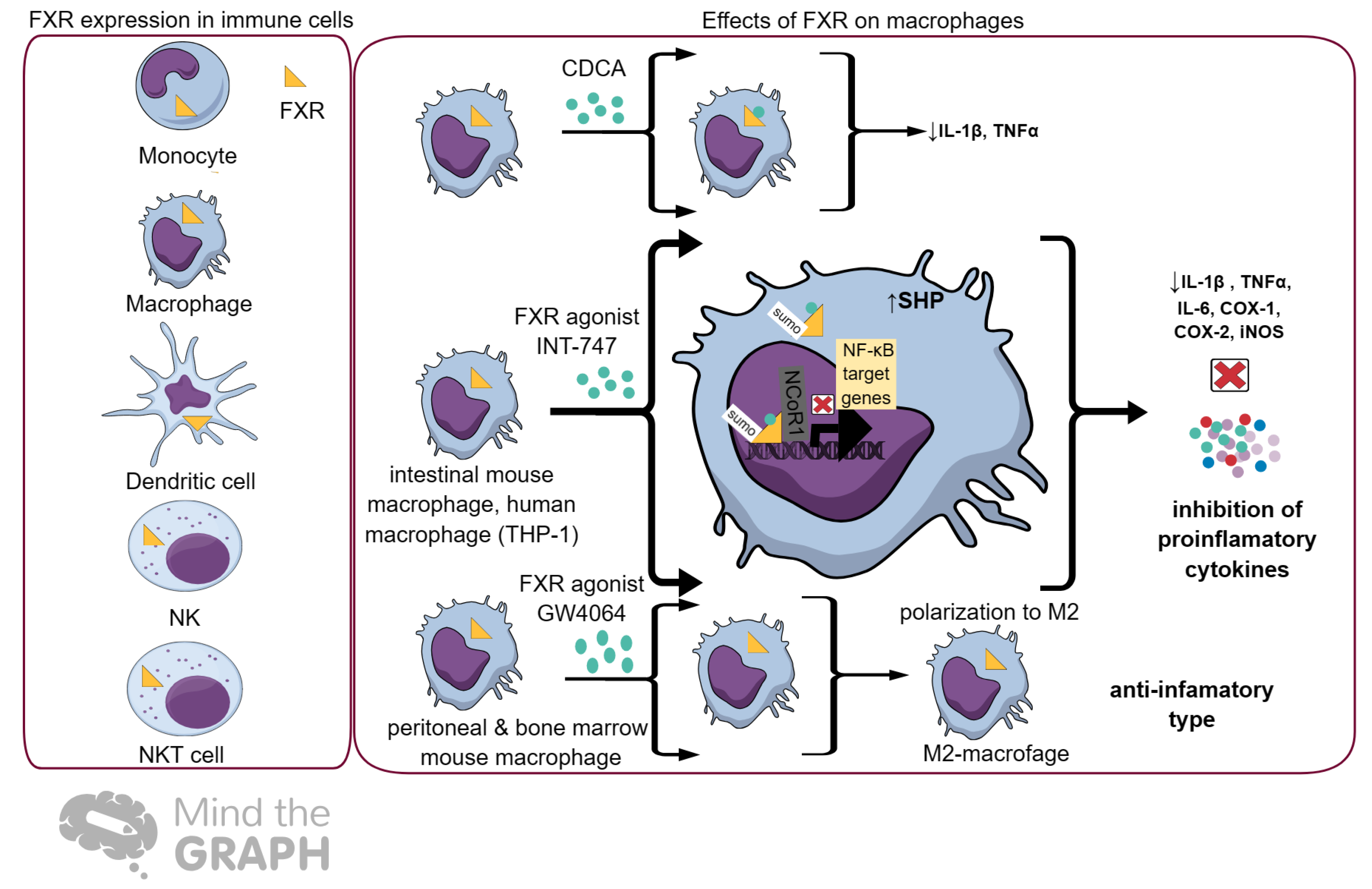
6. FXR and Immunity
7. Farnesoid X Receptor (FXR) in Cancer
7.1. Hepatocellular Carcinoma (HCC)
7.2. Biliary Tract Cancer (BTC)
7.3. Colorectal Carcinoma (CRC)
7.4. Esophageal and Gastric Cancer
7.5. Pancreatic Cancer
7.6. Breast and Cervical Cancer
7.7. Renal Cell Carcinoma (RCC) and Bladder Carcinoma
7.8. Prostate Cancer
7.9. In Other Tumors
8. FXR in Tumor Microenvironment and Implication for Anti-PD1/PD-L1 Immunotherapy
8.1. Lung TME
8.2. Liver TME
8.3. Colon TME
8.4. Breast TME
8.5. Kidney TME
9. Influence of Other Bile Acid Receptors on the TME and the Implication for Immunotherapy
9.1. TGR5
9.2. S1PR2
9.3. CHRM2 and CHRM3
9.4. VDR
10. The Potential Role of FXR Agonists and Antagonists in Cancer Therapy
10.1. Single-Drug Therapy
 represents activation, while
represents activation, while  represents inhibition.
represents inhibition.
 represents activation, while
represents activation, while  represents inhibition.
represents inhibition.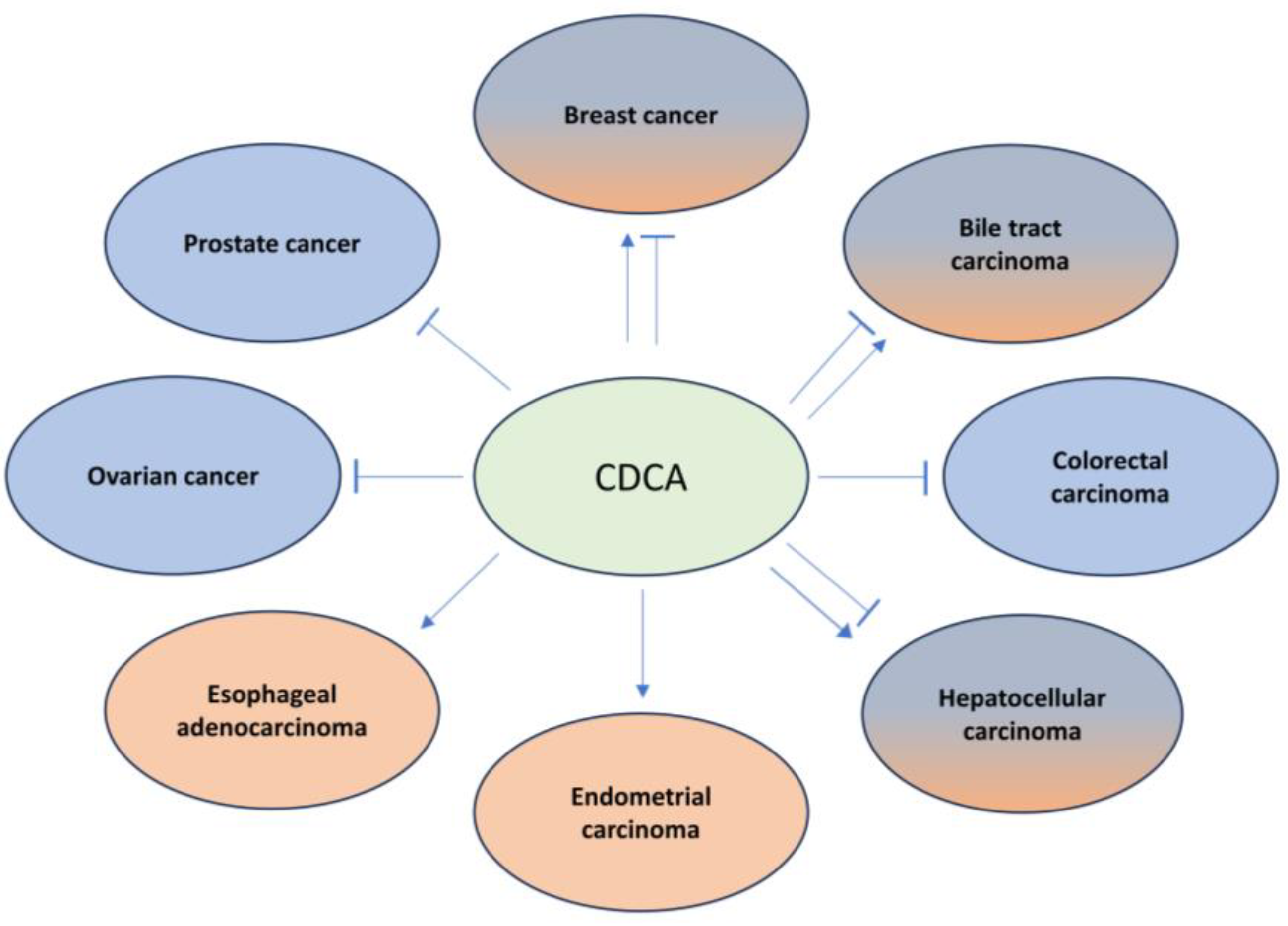
 represents activation, while
represents activation, while  represents inhibition.
represents inhibition.
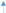 represents activation, while
represents activation, while  represents inhibition.
represents inhibition.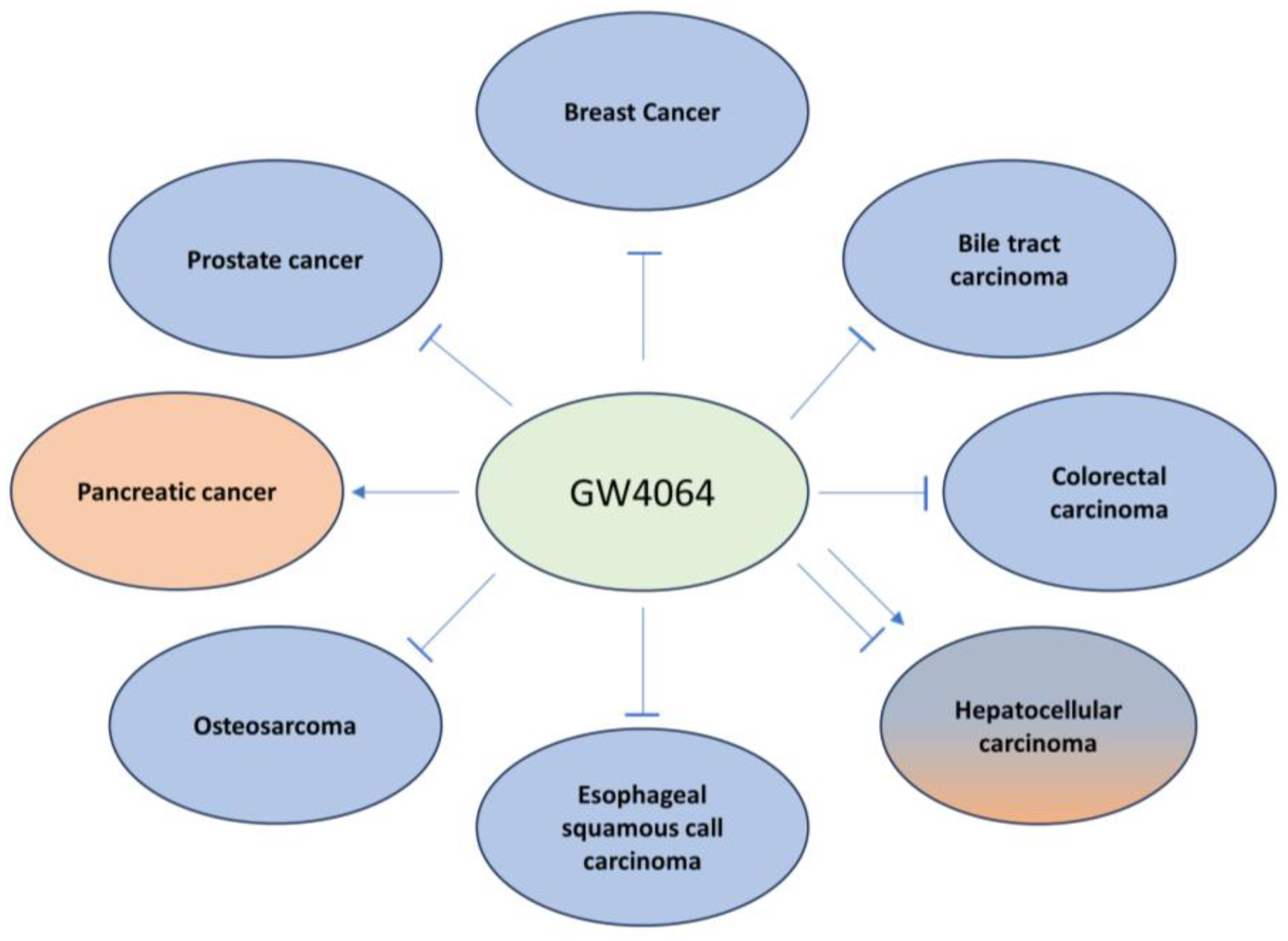
10.2. Combination Therapy
11. Challenges and Future Perspectives
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease Collaborative Network. Global Burden of Disease Study 2016 (GBD 2016) Covariates 1980–2016; Institute for Health Metrics and Evaluation (IHME): Seattle, WA, USA, 2017. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Vedsted, P.; Emery, J. Improving cancer outcomes: Better access to diagnostics in primary care could be critical. Br. J. Gen. Pract. 2011, 61, 317–318. [Google Scholar] [CrossRef]
- Režen, T.; Rozman, D.; Kovács, T.; Kovács, P.; Sipos, A.; Bai, P.; Mikó, E. The role of bile acids in carcinogenesis. Cell. Mol. Life Sci. 2022, 79, 243. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Houten, S.M.; Mataki, C.; Christoffolete, M.A.; Kim, B.W.; Sato, H.; Messaddeq, N.; Harney, J.W.; Ezaki, O.; Kodama, T.; et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006, 439, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef]
- Forman, B.M.; Goode, E.; Chen, J.; Oro, A.E.; Bradley, D.J.; Perlmann, T.; Noonan, D.J.; Burka, L.T.; McMorris, T.; Lamph, W.W.; et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 1995, 81, 687–693. [Google Scholar] [CrossRef]
- Makishima, M.; Okamoto, A.Y.; Repa, J.J.; Tu, H.; Learned, R.M.; Luk, A.; Hull, M.V.; Lustig, K.D.; Mangelsdorf, D.J.; Shan, B. Identification of a nuclear receptor for bile acids. Science 1999, 284, 1362–1365. [Google Scholar] [CrossRef]
- Parks, D.J.; Blanchard, S.G.; Bledsoe, R.K.; Chandra, G.; Consler, T.G.; Kliewer, S.A.; Stimmel, J.B.; Willson, T.M.; Zavacki, A.M.; Moore, D.D.; et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science 1999, 284, 1365–1368. [Google Scholar] [CrossRef]
- Girisa, S.; Henamayee, S.; Parama, D.; Rana, V.; Dutta, U.; Kunnumakkara, A.B. Targeting Farnesoid X receptor (FXR) for developing novel therapeutics against cancer. Mol. Biomed. 2021, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, W.T.; Luo, Q.Q.; Wang, B.; Chen, X.; Yan, X.J.; Qiu, H.Y.; Chen, S.L. Bile acids induce visceral hypersensitivity via mucosal mast cell-to-nociceptor signaling that involves the farnesoid X receptor/nerve growth factor/transient receptor potential vanilloid 1 axis. FASEB J. 2019, 33, 2435–2450. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Zampella, A.; Ricci, P.; Distrutti, E.; Biagioli, M. Immunomodulatory functions of FXR. Mol. Cell. Endocrinol. 2022, 551, 111650. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.F. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 1999, 159, 2647–2658. [Google Scholar] [CrossRef]
- Staels, B.; Fonseca, V.A. Bile acids and metabolic regulation: Mechanisms and clinical responses to bile acid sequestration. Diabetes Care 2009, 32 (Suppl. S2), S237–S245. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.C.P.; Ma, J.; Loiola, R.A.; Chen, X.; Jia, W. Bile acid-activated receptors in innate and adaptive immunity: Targeted drugs and biological agents. Eur. J. Immunol. 2023, 53, 2250299. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Diabetes and Digestive and Kidney Diseases. LiverTox: Clinical and Research Information on Drug-Induced Liver Injury; National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2012. [Google Scholar]
- Baloni, P.; Funk, C.C.; Yan, J.; Yurkovich, J.T.; Kueider-Paisley, A.; Nho, K.; Heinken, A.; Jia, W.; Mahmoudiandehkordi, S.; Louie, G.; et al. Metabolic Network Analysis Reveals Altered Bile Acid Synthesis and Metabolism in Alzheimer’s Disease. Cell Rep. Med. 2020, 1, 100138. [Google Scholar] [CrossRef]
- Zurkinden, L.; Sviridov, D.; Vogt, B.; Escher, G. Downregulation of Cyp7a1 by Cholic Acid and Chenodeoxycholic Acid in Cyp27a1/ApoE Double Knockout Mice: Differential Cardiovascular Outcome. Front. Endocrinol. 2020, 11, 586980. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Up to date on cholesterol 7 alpha-hydroxylase (CYP7A1) in bile acid synthesis. Liver Res. 2020, 4, 47–63. [Google Scholar] [CrossRef]
- Honda, A.; Yoshida, T.; Xu, G.; Matsuzaki, Y.; Fukushima, S.; Tanaka, N.; Doy, M.; Shefer, S.; Salen, G. Significance of plasma 7alpha-hydroxy-4-cholesten-3-one and 27-hydroxycholesterol concentrations as markers for hepatic bile acid synthesis in cholesterol-fed rabbits. Metabolism 2004, 53, 42–48. [Google Scholar] [CrossRef]
- Honda, A.; Yamashita, K.; Numazawa, M.; Ikegami, T.; Doy, M.; Matsuzaki, Y.; Miyazaki, H. Highly sensitive quantification of 7alpha-hydroxy-4-cholesten-3-one in human serum by LC-ESI-MS/MS. J. Lipid Res. 2007, 48, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu. Rev. Nutr. 2019, 39, 175–200. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.; Macias, R.I.; Briz, O.; Banales, J.M.; Monte, M.J. Bile Acids in Physiology, Pathology and Pharmacology. Curr. Drug Metab. 2015, 17, 4–29. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Bile acid metabolism and signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, T.; Honda, A. Reciprocal interactions between bile acids and gut microbiota in human liver diseases. Hepatol. Res. 2018, 48, 15–27. [Google Scholar] [CrossRef]
- Smith, L.P.; Nierstenhoefer, M.; Yoo, S.W.; Penzias, A.S.; Tobiasch, E.; Usheva, A. The bile acid synthesis pathway is present and functional in the human ovary. PLoS ONE 2009, 4, e7333. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G554–G573. [Google Scholar] [CrossRef]
- Wise, J.L.; Cummings, B.P. The 7-α-dehydroxylation pathway: An integral component of gut bacterial bile acid metabolism and potential therapeutic target. Front. Microbiol. 2023, 13, 1093420. [Google Scholar] [CrossRef]
- Xiang, D.; Yang, J.; Liu, L.; Yu, H.; Gong, X.; Liu, D. The regulation of tissue-specific farnesoid X receptor on genes and diseases involved in bile acid homeostasis. Biomed. Pharmacother. 2023, 168, 115606. [Google Scholar] [CrossRef]
- Fuchs, C.D.; Trauner, M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 432–450. [Google Scholar] [CrossRef] [PubMed]
- Long, X.Q.; Liu, M.Z.; Liu, Z.H.; Xia, L.Z.; Lu, S.P.; Xu, X.P.; Wu, M.H. Bile acids and their receptors: Potential therapeutic targets in inflammatory bowel disease. World J. Gastroenterol. 2023, 29, 4252–4270. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Pellicciari, R.; Pruzanski, M.; Auwerx, J.; Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 2008, 7, 678–693. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, P.; Cariou, B.; Lien, F.; Kuipers, F.; Staels, B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 2009, 89, 147–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Chen, W.D.; Yu, D.; Forman, B.M.; Huang, W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology 2011, 54, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- McMahan, R.H.; Wang, X.X.; Cheng, L.L.; Krisko, T.; Smith, M.; El Kasmi, K.; Pruzanski, M.; Adorini, L.; Golden-Mason, L.; Levi, M.; et al. Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J. Biol. Chem. 2013, 288, 11761–11770. [Google Scholar] [CrossRef] [PubMed]
- Ihunnah, C.A.; Jiang, M.; Xie, W. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim. Biophys. Acta 2011, 1812, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Wallace, K.; Cowie, D.E.; Konstantinou, D.K.; Hill, S.J.; Tjelle, T.E.; Axon, A.; Koruth, M.; White, S.A.; Carlsen, H.; Mann, D.A.; et al. The PXR is a drug target for chronic inflammatory liver disease. J. Steroid Biochem. Mol. Biol. 2010, 120, 137–148. [Google Scholar] [CrossRef]
- Ivanov, I.I.; McKenzie, B.S.; Zhou, L.; Tadokoro, C.E.; Lepelley, A.; Lafaille, J.J.; Cua, D.J.; Littman, D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 2006, 126, 1121–1133. [Google Scholar] [CrossRef]
- Chiang, J.Y. Sphingosine-1-phosphate receptor 2: A novel bile acid receptor and regulator of hepatic lipid metabolism? Hepatology 2015, 61, 1118–1120. [Google Scholar] [CrossRef]
- Wang, Y.M.; Ong, S.S.; Chai, S.C.; Chen, T. Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin. Drug Metab. Toxicol. 2012, 8, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Schaap, F.G.; Trauner, M.; Jansen, P.L. Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.Y.; Chen, S.M.; Pan, C.X.; Li, Y. FXR: Structures, biology, and drug development for NASH and fibrosis diseases. Acta Pharmacol. Sin. 2022, 43, 1120–1132. [Google Scholar] [CrossRef] [PubMed]
- Davignon, J.; Ganz, P. Role of endothelial dysfunction in atherosclerosis. Circulation 2004, 109, III27–III32. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.; Mathie, A.; Peters, J. Guide to Receptors and Channels (GRAC), 5th edition. Br. J. Pharmacol. 2011, 164, S1–S2. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.T.; Collins, J.L.; Pearce, K.H. The nuclear receptor superfamily and drug discovery. ChemMedChem 2006, 1, 504–523. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Chen, W.D.; Moore, D.D.; Huang, W. FXR: A metabolic regulator and cell protector. Cell Res. 2008, 18, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zou, H.; Zhang, T.; Zhang, J. Nuclear Receptors-Mediated Endocrine Disrupting Effects of Non-Phthalate Plasticizers: A Review. J. Agric. Food Chem. 2023. ahead of print. [Google Scholar] [CrossRef]
- Merk, D.; Sreeramulu, S.; Kudlinzki, D.; Saxena, K.; Linhard, V.; Gande, S.L.; Hiller, F.; Lamers, C.; Nilsson, E.; Aagaard, A.; et al. Molecular tuning of farnesoid X receptor partial agonism. Nat. Commun. 2019, 10, 2915. [Google Scholar] [CrossRef]
- Krattinger, R.; Boström, A.; Schiöth, H.B.; Thasler, W.E.; Mwinyi, J.; Kullak-Ublick, G.A. microRNA-192 suppresses the expression of the farnesoid X receptor. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G1044–G1051. [Google Scholar] [CrossRef]
- Vaquero, J.; Monte, M.J.; Dominguez, M.; Muntané, J.; Marin, J.J.G. Differential activation of the human farnesoid X receptor depends on the pattern of expressed isoforms and the bile acid pool composition. Biochem. Pharmacol. 2013, 86, 926–939. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.B.; Masson, H.L.P.; Bäumler, A.J. Bile acids as modulators of gut microbiota composition and function. Gut Microbes 2023, 15, 2172671. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, J.; Briz, O.; Herraez, E.; Muntané, J.; Marin, J.J. Activation of the nuclear receptor FXR enhances hepatocyte chemoprotection and liver tumor chemoresistance against genotoxic compounds. Biochim. Biophys. Acta 2013, 1833, 2212–2219. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.Q.; Luo, Y.; Backos, D.S.; Xu, S.; Balasubramaniyan, N.; Reigan, P.; Suchy, F.J. Identification of functionally relevant lysine residues that modulate human farnesoid X receptor activation. Mol. Pharmacol. 2013, 83, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Tao, W.; Zhai, K.; Fang, X.; Huang, Z.; Yu, J.S.; Sloan, A.E.; Rich, J.N.; Zhou, W.; Bao, S. Protein sumoylation with SUMO1 promoted by Pin1 in glioma stem cells augments glioblastoma malignancy. Neuro Oncol. 2020, 22, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniyan, N.; Luo, Y.; Sun, A.-Q.; Suchy, F.J. SUMOylation of the Farnesoid X Receptor (FXR) Regulates the Expression of FXR Target Genes*. J. Biol. Chem. 2013, 288, 13850–13862. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Nawaz, Z. 35—Molecular Biology of Estrogen Receptor Action. In Hormones, Brain and Behavior, 2nd ed.; Pfaff, D.W., Arnold, A.P., Etgen, A.M., Fahrbach, S.E., Rubin, R.T., Eds.; Academic Press: San Diego, CA, USA, 2009; pp. 1187–1220. [Google Scholar]
- Pascual, G.; Fong, A.L.; Ogawa, S.; Gamliel, A.; Li, A.C.; Perissi, V.; Rose, D.W.; Willson, T.M.; Rosenfeld, M.G.; Glass, C.K. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 2005, 437, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Vavassori, P.; Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 2009, 183, 6251–6261. [Google Scholar] [CrossRef]
- Kemper, J.K.; Xiao, Z.; Ponugoti, B.; Miao, J.; Fang, S.; Kanamaluru, D.; Tsang, S.; Wu, S.Y.; Chiang, C.M.; Veenstra, T.D. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009, 10, 392–404. [Google Scholar] [CrossRef]
- Fang, S.; Tsang, S.; Jones, R.; Ponugoti, B.; Yoon, H.; Wu, S.Y.; Chiang, C.M.; Willson, T.M.; Kemper, J.K. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J. Biol. Chem. 2008, 283, 35086–35095. [Google Scholar] [CrossRef]
- Kim, D.-H.; Xiao, Z.; Kwon, S.; Sun, X.; Ryerson, D.; Tkac, D.; Ma, P.; Wu, S.-Y.; Chiang, C.-M.; Zhou, E.; et al. A dysregulated acetyl/SUMO switch of FXR promotes hepatic inflammation in obesity. EMBO J. 2015, 34, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zang, C.; Rosenfeld, J.A.; Schones, D.E.; Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Peng, W.; Zhang, M.Q.; et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008, 40, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Zhou, V.W.; Goren, A.; Bernstein, B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011, 12, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniyan, N.; Ananthanarayanan, M.; Suchy, F.J. Direct methylation of FXR by Set7/9, a lysine methyltransferase, regulates the expression of FXR target genes. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G937–G947. [Google Scholar] [CrossRef] [PubMed]
- Ananthanarayanan, M.; Li, Y.; Surapureddi, S.; Balasubramaniyan, N.; Ahn, J.; Goldstein, J.A.; Suchy, F.J. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G771–G781. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gineste, R.; Sirvent, A.; Paumelle, R.; Helleboid, S.; Aquilina, A.; Darteil, R.; Hum, D.W.; Fruchart, J.C.; Staels, B. Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol. Endocrinol. 2008, 22, 2433–2447. [Google Scholar] [CrossRef] [PubMed]
- Schote, A.B.; Turner, J.D.; Schiltz, J.; Muller, C.P. Nuclear receptors in human immune cells: Expression and correlations. Mol. Immunol. 2007, 44, 1436–1445. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Bile acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr. Rev. 2002, 23, 443–463. [Google Scholar] [CrossRef]
- Beigneux, A.P.; Moser, A.H.; Shigenaga, J.K.; Grunfeld, C.; Feingold, K.R. The acute phase response is associated with retinoid X receptor repression in rodent liver. J. Biol. Chem. 2000, 275, 16390–16399. [Google Scholar] [CrossRef]
- Fiorucci, S.; Baldoni, M.; Ricci, P.; Zampella, A.; Distrutti, E.; Biagioli, M. Bile acid-activated receptors and the regulation of macrophages function in metabolic disorders. Curr. Opin. Pharmacol. 2020, 53, 45–54. [Google Scholar] [CrossRef]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile Acids Activated Receptors Regulate Innate Immunity. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Jaroonwitchawan, T.; Arimochi, H.; Sasaki, Y.; Ishifune, C.; Kondo, H.; Otsuka, K.; Tsukumo, S.I.; Yasutomo, K. Stimulation of the farnesoid X receptor promotes M2 macrophage polarization. Front. Immunol. 2023, 14, 1065790. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.; Banota, T.; Guo, G.L.; Smith, L.C.; Meshanni, J.A.; Lee, J.; Kong, B.; Abramova, E.V.; Goedken, M.; Gow, A.J.; et al. Farnesoid X receptor regulates lung macrophage activation and injury following nitrogen mustard exposure. Toxicol. Appl. Pharmacol. 2022, 454, 116208. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.; Marchildon, F.; Michaels, A.J.; Takemoto, N.; van der Veeken, J.; Schizas, M.; Pritykin, Y.; Leslie, C.S.; Intlekofer, A.M.; Cohen, P.; et al. FXR mediates T cell-intrinsic responses to reduced feeding during infection. Proc. Natl. Acad. Sci. USA 2020, 117, 33446–33454. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Lu, Y.; Li, X.Y. Farnesoid X receptor: A master regulator of hepatic triglyceride and glucose homeostasis. Acta Pharmacol. Sin. 2015, 36, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huang, X.; Yi, T.; Yen, Y.; Moore, D.D.; Huang, W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007, 67, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Morimura, K.; Shah, Y.; Yang, Q.; Ward, J.M.; Gonzalez, F.J. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 2007, 28, 940–946. [Google Scholar] [CrossRef]
- Deuschle, U.; Schüler, J.; Schulz, A.; Schlüter, T.; Kinzel, O.; Abel, U.; Kremoser, C. FXR controls the tumor suppressor NDRG2 and FXR agonists reduce liver tumor growth and metastasis in an orthotopic mouse xenograft model. PLoS ONE 2012, 7, e43044. [Google Scholar] [CrossRef]
- Nie, X.; Liu, H.; Wei, X.; Li, L.; Lan, L.; Fan, L.; Ma, H.; Liu, L.; Zhou, Y.; Hou, R.; et al. miRNA-382-5p Suppresses the Expression of Farnesoid X Receptor to Promote Progression of Liver Cancer. Cancer Manag. Res. 2021, 13, 8025–8035. [Google Scholar] [CrossRef]
- Nomoto, M.; Miyata, M.; Yin, S.; Kurata, Y.; Shimada, M.; Yoshinari, K.; Gonzalez, F.J.; Suzuki, K.; Shibasaki, S.; Kurosawa, T.; et al. Bile acid-induced elevated oxidative stress in the absence of farnesoid X receptor. Biol. Pharm. Bull. 2009, 32, 172–178. [Google Scholar] [CrossRef]
- Zheng, P.; Zeng, B.; Zhou, C.; Liu, M.; Fang, Z.; Xu, X.; Zeng, L.; Chen, J.; Fan, S.; Du, X.; et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 2016, 21, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.F.; Zhao, W.Y.; Huang, W.D. FXR and liver carcinogenesis. Acta Pharmacol. Sin. 2015, 36, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Kainuma, M.; Takada, I.; Makishima, M.; Sano, K. Farnesoid X Receptor Activation Enhances Transforming Growth Factor β-Induced Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2018, 19, 1898. [Google Scholar] [CrossRef] [PubMed]
- Asano, Y.; Hiramoto, T.; Nishino, R.; Aiba, Y.; Kimura, T.; Yoshihara, K.; Koga, Y.; Sudo, N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1288–G1295. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.; Thomas, A.; Edwards, G.; Jaseja, R.; Guo, G.L.; Apte, U. Increased activation of the Wnt/β-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J. Pharmacol. Exp. Ther. 2011, 338, 12–21. [Google Scholar] [CrossRef]
- Guo, Y.; Peng, Q.; Hao, L.; Ji, J.; Zhang, Z.; Xue, Y.; Liu, Y.; Gao, Y.; Li, C.; Shi, X. Dihydroartemisinin promoted FXR expression independent of YAP1 in hepatocellular carcinoma. FASEB J. 2022, 36, e22361. [Google Scholar] [CrossRef]
- Lozano, E.; Sanchez-Vicente, L.; Monte, M.J.; Herraez, E.; Briz, O.; Banales, J.M.; Marin, J.J.; Macias, R.I. Cocarcinogenic effects of intrahepatic bile acid accumulation in cholangiocarcinoma development. Mol. Cancer Res. 2014, 12, 91–100. [Google Scholar] [CrossRef]
- Lv, B.; Ma, L.; Tang, W.; Huang, P.; Yang, B.; Wang, L.; Chen, S.; Gao, Q.; Zhang, S.; Xia, J. FXR Acts as a Metastasis Suppressor in Intrahepatic Cholangiocarcinoma by Inhibiting IL-6-Induced Epithelial-Mesenchymal Transition. Cell. Physiol. Biochem. 2018, 48, 158–172. [Google Scholar] [CrossRef]
- Dai, J.; Wang, H.; Shi, Y.; Dong, Y.; Zhang, Y.; Wang, J. Impact of bile acids on the growth of human cholangiocarcinoma via FXR. J. Hematol. Oncol. 2011, 4, 41. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef]
- Zhong, X.Y.; Yu, J.H.; Zhang, W.G.; Wang, Z.D.; Dong, Q.; Tai, S.; Cui, Y.F.; Li, H. MicroRNA-421 functions as an oncogenic miRNA in biliary tract cancer through down-regulating farnesoid X receptor expression. Gene 2012, 493, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhan, M.; Liu, Q.; Wang, J. Glycochenodeoxycholate promotes the metastasis of gallbladder cancer cells by inducing epithelial to mesenchymal transition via activation of SOCS3/JAK2/STAT3 signaling pathway. J. Cell. Physiol. 2020, 235, 1615–1623. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yin, X.; Li, G.; Yi, J.; Wang, J. Expressions of farnesoid X receptor and myeloid cell leukemia sequence 1 protein are associated with poor prognosis in patients with gallbladder cancer. Chin. Med. J. 2014, 127, 2637–2642. [Google Scholar] [PubMed]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Edwards, P.A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef]
- Modica, S.; Murzilli, S.; Salvatore, L.; Schmidt, D.R.; Moschetta, A. Nuclear Bile Acid Receptor FXR Protects against Intestinal Tumorigenesis. Cancer Res. 2008, 68, 9589–9594. [Google Scholar] [CrossRef]
- Lax, S.; Schauer, G.; Prein, K.; Kapitan, M.; Silbert, D.; Berghold, A.; Berger, A.; Trauner, M. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int. J. Cancer 2012, 130, 2232–2239. [Google Scholar] [CrossRef]
- Maran, R.R.; Thomas, A.; Roth, M.; Sheng, Z.; Esterly, N.; Pinson, D.; Gao, X.; Zhang, Y.; Ganapathy, V.; Gonzalez, F.J.; et al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharmacol. Exp. Ther. 2009, 328, 469–477. [Google Scholar] [CrossRef]
- Smith, D.L.; Keshavan, P.; Avissar, U.; Ahmed, K.; Zucker, S.D. Sodium taurocholate inhibits intestinal adenoma formation in APCMin/+ mice, potentially through activation of the farnesoid X receptor. Carcinogenesis 2010, 31, 1100–1109. [Google Scholar] [CrossRef]
- Peng, Z.; Raufman, J.P.; Xie, G. Src-mediated cross-talk between farnesoid X and epidermal growth factor receptors inhibits human intestinal cell proliferation and tumorigenesis. PLoS ONE 2012, 7, e48461. [Google Scholar] [CrossRef]
- Fu, T.; Coulter, S.; Yoshihara, E.; Oh, T.G.; Fang, S.; Cayabyab, F.; Zhu, Q.; Zhang, T.; Leblanc, M.; Liu, S.; et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 2019, 176, 1098–1112.e1018. [Google Scholar] [CrossRef]
- Yu, J.; Yang, K.; Zheng, J.; Zhao, P.; Xia, J.; Sun, X.; Zhao, W. Correction: Activation of FXR and inhibition of EZH2 synergistically inhibit colorectal cancer through cooperatively accelerating FXR nuclear location and upregulating CDX2 expression. Cell Death Dis. 2023, 14, 101. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.M.; Zhan, L.; Maru, D.; Shureiqi, I.; Pickering, C.R.; Kiriakova, G.; Izzo, J.; He, N.; Wei, C.; Baladandayuthapani, V.; et al. FXR silencing in human colon cancer by DNA methylation and KRAS signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G48–G58. [Google Scholar] [CrossRef] [PubMed]
- De Gottardi, A.; Touri, F.; Maurer, C.A.; Perez, A.; Maurhofer, O.; Ventre, G.; Bentzen, C.L.; Niesor, E.J.; Dufour, J.F. The bile acid nuclear receptor FXR and the bile acid binding protein IBABP are differently expressed in colon cancer. Dig. Dis. Sci. 2004, 49, 982–989. [Google Scholar] [CrossRef]
- Modica, S.; Cariello, M.; Morgano, A.; Gross, I.; Vegliante, M.C.; Murzilli, S.; Salvatore, L.; Freund, J.N.; Sabbà, C.; Moschetta, A. Transcriptional regulation of the intestinal nuclear bile acid farnesoid X receptor (FXR) by the caudal-related homeobox 2 (CDX2). J. Biol. Chem. 2014, 289, 28421–28432. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 2009, 15, 3329–3340. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Li, H.; Yang, Z.; Hoque, A.; Xu, X. Inhibition of farnesoid X receptor controls esophageal cancer cell growth in vitro and in nude mouse xenografts. Cancer 2013, 119, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Zhang, H.; Yao, D.; Zhang, X.; Chen, W.D.; Wang, Y.D. Activation of FXR Suppresses Esophageal Squamous Cell Carcinoma Through Antagonizing ERK1/2 Signaling Pathway. Cancer Manag. Res. 2021, 13, 5907–5918. [Google Scholar] [CrossRef]
- De Gottardi, A.; Dumonceau, J.-M.; Bruttin, F.; Vonlaufen, A.; Morard, I.; Spahr, L.; Rubbia-Brandt, L.; Frossard, J.-L.; Dinjens, W.N.M.; Rabinovitch, P.S.; et al. Expression of the bile acid receptor FXR in Barrett’s esophagus and enhancement of apoptosis by guggulsterone in vitro. Mol. Cancer 2006, 5, 48. [Google Scholar] [CrossRef]
- Correa, P.; Piazuelo, M.B. The gastric precancerous cascade. J. Dig. Dis. 2012, 13, 2–9. [Google Scholar] [CrossRef]
- Zhou, H.; Ni, Z.; Li, T.; Su, L.; Zhang, L.; Liu, N.; Shi, Y. Activation of FXR promotes intestinal metaplasia of gastric cells via SHP-dependent upregulation of the expression of CDX2. Oncol. Lett. 2018, 15, 7617–7624. [Google Scholar] [CrossRef]
- Wang, N.; Wu, S.; Zhao, J.; Chen, M.; Zeng, J.; Lu, G.; Wang, J.; Zhang, J.; Liu, J.; Shi, Y. Bile acids increase intestinal marker expression via the FXR/SNAI2/miR-1 axis in the stomach. Cell Oncol. 2021, 44, 1119–1131. [Google Scholar] [CrossRef] [PubMed]
- Lian, F.; Xing, X.; Yuan, G.; Schäfer, C.; Rauser, S.; Walch, A.; Röcken, C.; Ebeling, M.; Wright, M.B.; Schmid, R.M.; et al. Farnesoid X receptor protects human and murine gastric epithelial cells against inflammation-induced damage. Biochem. J. 2011, 438, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Tran, Q.T.; Tran, V.H.; Sendler, M.; Doller, J.; Wiese, M.; Bolsmann, R.; Wilden, A.; Glaubitz, J.; Modenbach, J.M.; Thiel, F.G.; et al. Role of Bile Acids and Bile Salts in Acute Pancreatitis: From the Experimental to Clinical Studies. Pancreas 2021, 50, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.Y.; Chen, Y.C. Role of bile acids in carcinogenesis of pancreatic cancer: An old topic with new perspective. World J. Gastroenterol. 2016, 22, 7463–7477. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wu, L.L.; Han, T.; Zhuo, M.; Lei, W.; Cui, J.J.; Jiao, F.; Wang, L.W. Correlated high expression of FXR and Sp1 in cancer cells confers a poor prognosis for pancreatic cancer: A study based on TCGA and tissue microarray. Oncotarget 2017, 8, 33265–33275. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Xie, K.X.; Yang, Z.L.; Yuan, L.W. Expression of FXR and HRG and their clinicopathological significance in benign and malignant pancreatic lesions. Int. J. Clin. Exp. Pathol. 2019, 12, 2111–2120. [Google Scholar]
- Castellsagué, X.; Muñoz, N.; Pitisuttithum, P.; Ferris, D.; Monsonego, J.; Ault, K.; Luna, J.; Myers, E.; Mallary, S.; Bautista, O.M.; et al. End-of-study safety, immunogenicity, and efficacy of quadrivalent HPV (types 6, 11, 16, 18) recombinant vaccine in adult women 24–45 years of age. Br. J. Cancer 2011, 105, 28–37. [Google Scholar] [CrossRef]
- Giaginis, C.; Koutsounas, I.; Alexandrou, P.; Zizi-Serbetzoglou, A.; Patsouris, E.; Kouraklis, G.; Theocharis, S. Elevated Farnesoid X Receptor (FXR) and Retinoid X Receptors (RXRs) expression is associated with less tumor aggressiveness and favourable prognosis in patients with pancreatic adenocarcinoma. Neoplasma 2015, 62, 332–341. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, K.T.; Lee, J.K.; Lee, K.H.; Jang, K.T.; Heo, J.S.; Choi, S.H.; Kim, Y.; Rhee, J.C. Farnesoid X receptor, overexpressed in pancreatic cancer with lymph node metastasis promotes cell migration and invasion. Br. J. Cancer 2011, 104, 1027–1037. [Google Scholar] [CrossRef]
- Joshi, S.; Cruz, E.; Rachagani, S.; Guha, S.; Brand, R.E.; Ponnusamy, M.P.; Kumar, S.; Batra, S.K. Bile acids-mediated overexpression of MUC4 via FAK-dependent c-Jun activation in pancreatic cancer. Mol. Oncol. 2016, 10, 1063–1077. [Google Scholar] [CrossRef]
- Swales, K.E.; Korbonits, M.R.; Carpenter, R.; Walsh, D.T.; Warner, T.D.; Bishop-Bailey, D. The Farnesoid X Receptor Is Expressed in Breast Cancer and Regulates Apoptosis and Aromatase Expression. Cancer Res. 2006, 66, 10120–10126. [Google Scholar] [CrossRef] [PubMed]
- Journe, F.; Durbecq, V.; Chaboteaux, C.; Rouas, G.; Laurent, G.; Nonclercq, D.; Sotiriou, C.; Body, J.J.; Larsimont, D. Association between farnesoid X receptor expression and cell proliferation in estrogen receptor-positive luminal-like breast cancer from postmenopausal patients. Breast Cancer Res. Treat. 2009, 115, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Absil, L.; Journé, F.; Larsimont, D.; Body, J.J.; Tafforeau, L.; Nonclercq, D. Farnesoid X receptor as marker of osteotropism of breast cancers through its role in the osteomimetism of tumor cells. BMC Cancer 2020, 20, 640. [Google Scholar] [CrossRef] [PubMed]
- Alasmael, N.; Mohan, R.; Meira, L.B.; Swales, K.E.; Plant, N.J. Activation of the Farnesoid X-receptor in breast cancer cell lines results in cytotoxicity but not increased migration potential. Cancer Lett. 2016, 370, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Catalano, S.; Panza, S.; Vizza, D.; Barone, I.; Bonofiglio, D.; Gelsomino, L.; Rizza, P.; Fuqua, S.A.W.; Andò, S. Farnesoid X receptor inhibits tamoxifen-resistant MCF-7 breast cancer cell growth through downregulation of HER2 expression. Oncogene 2011, 30, 4129–4140. [Google Scholar] [CrossRef]
- Barone, I.; Vircillo, V.; Giordano, C.; Gelsomino, L.; Győrffy, B.; Tarallo, R.; Rinaldi, A.; Bruno, G.; Caruso, A.; Romeo, F.; et al. Activation of Farnesoid X Receptor impairs the tumor-promoting function of breast cancer-associated fibroblasts. Cancer Lett. 2018, 437, 89–99. [Google Scholar] [CrossRef]
- Huang, X.; Wang, B.; Chen, R.; Zhong, S.; Gao, F.; Zhang, Y.; Niu, Y.; Li, C.; Shi, G. The Nuclear Farnesoid X Receptor Reduces p53 Ubiquitination and Inhibits Cervical Cancer Cell Proliferation. Front. Cell Dev. Biol. 2021, 9, 583146. [Google Scholar] [CrossRef]
- Huang, X.; Wang, B.; Shen, H.; Huang, D.; Shi, G. Farnesoid X receptor functions in cervical cancer via the p14ARF-mouse double minute 2-p53 pathway. Mol. Biol. Rep. 2022, 49, 3617–3625. [Google Scholar] [CrossRef]
- Herman-Edelstein, M.; Weinstein, T.; Levi, M. Bile acid receptors and the kidney. Curr. Opin. Nephrol. Hypertens. 2018, 27, 56–62. [Google Scholar] [CrossRef]
- Fujino, T.; Sakamaki, R.; Ito, H.; Furusato, Y.; Sakamoto, N.; Oshima, T.; Hayakawa, M. Farnesoid X receptor regulates the growth of renal adenocarcinoma cells without affecting that of a normal renal cell-derived cell line. J. Toxicol. Sci. 2017, 42, 259–265. [Google Scholar] [CrossRef]
- Fujino, T.; Sugizaki, K.; Kato, R.; Beppu, M.; Murakami, S.; Lee, H.; Oshima, T.; Hayakawa, M. Farnesoid X receptor and liver X receptors regulate Oct3/4 expression by multiple feedback regulating system in normal renal-derived cells and renal adenocarcinoma cells. J. Toxicol. Sci. 2020, 45, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Hou, Y.; Hu, M.; Hu, J.; Liu, X. Clinical significance and oncogenic function of NR1H4 in clear cell renal cell carcinoma. BMC Cancer 2022, 22, 995. [Google Scholar] [CrossRef] [PubMed]
- Strauss, P.; Rivedal, M.; Scherer, A.; Eikrem, Ø.; Nakken, S.; Beisland, C.; Bostad, L.; Flatberg, A.; Skandalou, E.; Beisvåg, V.; et al. A multiomics disease progression signature of low-risk ccRCC. Sci. Rep. 2022, 12, 13503. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.R.; Wang, H.H.; Chang, H.H.; Tsai, Y.L.; Tsai, W.C.; Lee, C.R.; Changchien, C.Y.; Cheng, Y.C.; Wu, S.T.; Chen, Y. Enhancement of Farnesoid X Receptor Inhibits Migration, Adhesion and Angiogenesis through Proteasome Degradation and VEGF Reduction in Bladder Cancers. Int. J. Mol. Sci. 2022, 23, 5259. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.R.; Tsai, Y.L.; Tsai, W.C.; Chen, T.M.; Chang, H.H.; Changchien, C.Y.; Wu, S.T.; Wang, H.H.; Chen, Y.; Lin, Y.H. Farnesoid X Receptor Overexpression Decreases the Migration, Invasion and Angiogenesis of Human Bladder Cancers via AMPK Activation and Cholesterol Biosynthesis Inhibition. Cancers 2022, 14, 4398. [Google Scholar] [CrossRef] [PubMed]
- Massie, C.E.; Lynch, A.; Ramos-Montoya, A.; Boren, J.; Stark, R.; Fazli, L.; Warren, A.; Scott, H.; Madhu, B.; Sharma, N.; et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011, 30, 2719–2733. [Google Scholar] [CrossRef] [PubMed]
- Kaeding, J.; Bouchaert, E.; Bélanger, J.; Caron, P.; Chouinard, S.; Verreault, M.; Larouche, O.; Pelletier, G.; Staels, B.; Bélanger, A.; et al. Activators of the farnesoid X receptor negatively regulate androgen glucuronidation in human prostate cancer LNCAP cells. Biochem. J. 2008, 410, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tong, S.J.; Wang, X.; Qu, L.X. Farnesoid X receptor inhibits LNcaP cell proliferation via the upregulation of PTEN. Exp. Ther. Med. 2014, 8, 1209–1212. [Google Scholar] [CrossRef]
- Liu, N.; Zhao, J.; Wang, J.; Teng, H.; Fu, Y.; Yuan, H. Farnesoid X receptor ligand CDCA suppresses human prostate cancer cells growth by inhibiting lipid metabolism via targeting sterol response element binding protein 1. Am. J. Transl. Res. 2016, 8, 5118–5124. [Google Scholar]
- Id Boufker, H.; Lagneaux, L.; Fayyad-Kazan, H.; Badran, B.; Najar, M.; Wiedig, M.; Ghanem, G.; Laurent, G.; Body, J.J.; Journé, F. Role of farnesoid X receptor (FXR) in the process of differentiation of bone marrow stromal cells into osteoblasts. Bone 2011, 49, 1219–1231. [Google Scholar] [CrossRef]
- Wu, B.; Xing, C.; Tao, J. Upregulation of microRNA-23b-3p induced by farnesoid X receptor regulates the proliferation and apoptosis of osteosarcoma cells. J. Orthop. Surg. Res. 2019, 14, 398. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Chen, B.; Liu, X.; Xue, S.; Qin, H.; Jiang, H. Farnesoid X receptor, a novel proto-oncogene in non-small cell lung cancer, promotes tumor growth via directly transactivating CCND1. Sci. Rep. 2017, 7, 591. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.N.; Chen, J.R.; Chen, J.L. Role of Farnesoid X Receptor in the Pathogenesis of Respiratory Diseases. Can. Respir. J. 2020, 2020, 9137251. [Google Scholar] [CrossRef] [PubMed]
- Shaik, F.B.; Prasad, D.V.; Narala, V.R. Role of farnesoid X receptor in inflammation and resolution. Inflamm. Res. 2015, 64, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.M.; Gayer, C.P. The Pathophysiology of Farnesoid X Receptor (FXR) in the GI Tract: Inflammation, Barrier Function and Innate Immunity. Cells 2021, 10, 3206. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi Rad, H.; Monkman, J.; Warkiani, M.E.; Ladwa, R.; O’Byrne, K.; Rezaei, N.; Kulasinghe, A. Understanding the tumor microenvironment for effective immunotherapy. Med. Res. Rev. 2021, 41, 1474–1498. [Google Scholar] [CrossRef] [PubMed]
- Petitprez, F.; Meylan, M.; de Reyniès, A.; Sautès-Fridman, C.; Fridman, W.H. The Tumor Microenvironment in the Response to Immune Checkpoint Blockade Therapies. Front. Immunol. 2020, 11, 784. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer 2019, 18, 10. [Google Scholar] [CrossRef]
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef]
- Lee, J.B.; Kim, H.R.; Ha, S.J. Immune Checkpoint Inhibitors in 10 Years: Contribution of Basic Research and Clinical Application in Cancer Immunotherapy. Immune Netw. 2022, 22, e2. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Daud, A.I.; Wolchok, J.D.; Robert, C.; Hwu, W.J.; Weber, J.S.; Ribas, A.; Hodi, F.S.; Joshua, A.M.; Kefford, R.; Hersey, P.; et al. Programmed Death-Ligand 1 Expression and Response to the Anti-Programmed Death 1 Antibody Pembrolizumab in Melanoma. J. Clin. Oncol. 2016, 34, 4102–4109. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Du, Y.; Xue, C.; Wu, P.; Du, N.; Zhu, G.; Xu, H.; Zhu, Z. Efficacy and safety of anti-PD-1/PD-L1 therapy in the treatment of advanced colorectal cancer: A meta-analysis. BMC Gastroenterol. 2022, 22, 431. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Li, L.; Sun, D.; Liu, X.; Xia, Z.; Xue, S.; Chen, B.; Qin, H.; Ai, J.; Jiang, H. Farnesoid X Receptor Constructs an Immunosuppressive Microenvironment and Sensitizes FXR(high)PD-L1(low) NSCLC to Anti-PD-1 Immunotherapy. Cancer Immunol. Res. 2019, 7, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Gui, Y.; Wei, Y.; Shang, B.; Sun, J.; Ma, S.; You, W.; Jiang, S. Z-guggulsterone induces PD-L1 upregulation partly mediated by FXR, Akt and Erk1/2 signaling pathways in non-small cell lung cancer. Int. Immunopharmacol. 2021, 93, 107395. [Google Scholar] [CrossRef]
- Mencarelli, A.; Renga, B.; Migliorati, M.; Cipriani, S.; Distrutti, E.; Santucci, L.; Fiorucci, S. The bile acid sensor farnesoid X receptor is a modulator of liver immunity in a rodent model of acute hepatitis. J. Immunol. 2009, 183, 6657–6666. [Google Scholar] [CrossRef]
- Gong, Y.; Li, K.; Qin, Y.; Zeng, K.; Liu, J.; Huang, S.; Chen, Y.; Yu, H.; Liu, W.; Ye, L.; et al. Norcholic Acid Promotes Tumor Progression and Immune Escape by Regulating Farnesoid X Receptor in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 711448. [Google Scholar] [CrossRef]
- Ji, G.; Ma, L.; Yao, H.; Ma, S.; Si, X.; Wang, Y.; Bao, X.; Ma, L.; Chen, F.; Ma, C.; et al. Precise delivery of obeticholic acid via nanoapproach for triggering natural killer T cell-mediated liver cancer immunotherapy. Acta Pharm. Sin. B 2020, 10, 2171–2182. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, D.; Li, X.; Cao, Y.; Yi, C.; Wiredu Ocansey, D.K.; Zhou, Y.; Mao, F. Farnesoid-X receptor as a therapeutic target for inflammatory bowel disease and colorectal cancer. Front. Pharmacol. 2022, 13, 1016836. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.S.; Kumar, V.; Al-Abbasi, F.A.; Kamal, M.A.; Anwar, F. Risk of colorectal cancer in inflammatory bowel diseases. Semin. Cancer Biol. 2020, 64, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Dong, X. Abstract 3442: FXR mediates macrophage intrinsic responses to suppress colon cancer progression. Cancer Res. 2023, 83, 3442. [Google Scholar] [CrossRef]
- Lu, L.; Jiang, Y.X.; Liu, X.X.; Jin, J.M.; Gu, W.J.; Luan, X.; Guan, Y.Y.; Zhang, L.J. FXR agonist GW4064 enhances anti-PD-L1 immunotherapy in colorectal cancer. Oncoimmunology 2023, 12, 2217024. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Thorell, A.; Claudel, T.; Jha, P.; Koefeler, H.; Lackner, C.; Hoesel, B.; Fauler, G.; Stojakovic, T.; Einarsson, C.; et al. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J. Hepatol. 2015, 62, 1398–1404. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Lu, C.; Song, Z.; Qiao, C.; Wang, J.; Chen, J.; Zhang, C.; Zeng, X.; Ma, Z.; Chen, T.; et al. Ursodeoxycholic acid reduces antitumor immunosuppression by inducing CHIP-mediated TGF-β degradation. Nat. Commun. 2022, 13, 3419. [Google Scholar] [CrossRef]
- Tang, W.; Putluri, V.; Ambati, C.R.; Dorsey, T.H.; Putluri, N.; Ambs, S. Liver- and Microbiome-derived Bile Acids Accumulate in Human Breast Tumors and Inhibit Growth and Improve Patient Survival. Clin. Cancer Res. 2019, 25, 5972–5983. [Google Scholar] [CrossRef]
- Wu, R.; Yu, I.; Tokumaru, Y.; Asaoka, M.; Oshi, M.; Yan, L.; Okuda, S.; Ishikawa, T.; Takabe, K. Elevated bile acid metabolism and microbiome are associated with suppressed cell proliferation and better survival in breast cancer. Am. J. Cancer Res. 2022, 12, 5271–5285. [Google Scholar]
- Giordano, C.; Barone, I.; Vircillo, V.; Panza, S.; Malivindi, R.; Gelsomino, L.; Pellegrino, M.; Rago, V.; Mauro, L.; Lanzino, M.; et al. Activated FXR Inhibits Leptin Signaling and Counteracts Tumor-promoting Activities of Cancer-Associated Fibroblasts in Breast Malignancy. Sci. Rep. 2016, 6, 21782. [Google Scholar] [CrossRef]
- Kawamata, Y.; Fujii, R.; Hosoya, M.; Harada, M.; Yoshida, H.; Miwa, M.; Fukusumi, S.; Habata, Y.; Itoh, T.; Shintani, Y.; et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003, 278, 9435–9440. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Nakamura, T.; Itadani, H.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Duan, G.; Wei, D.; Zhao, C.; Ma, Y. The Bile Acid Membrane Receptor TGR5 in Cancer: Friend or Foe? Molecules 2022, 27, 5292. [Google Scholar] [CrossRef]
- Guan, Z.; Luo, L.; Liu, S.; Guan, Z.; Zhang, Q.; Wu, Z.; Tao, K. The role of TGR5 as an onco-immunological biomarker in tumor staging and prognosis by encompassing the tumor microenvironment. Front. Oncol. 2022, 12, 953091. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, H.; Liu, X.; Xue, S.; Chen, D.; Zou, J.; Jiang, H. TGR5 deficiency activates antitumor immunity in non-small cell lung cancer via restraining M2 macrophage polarization. Acta Pharm. Sin. B 2022, 12, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Studer, E.; Zhou, X.; Zhao, R.; Wang, Y.; Takabe, K.; Nagahashi, M.; Pandak, W.M.; Dent, P.; Spiegel, S.; Shi, R.; et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology 2012, 55, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhao, R.; Zhou, X.; Liang, X.; Campbell, D.J.; Zhang, X.; Zhang, L.; Shi, R.; Wang, G.; Pandak, W.M.; et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology 2014, 60, 908–918. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, X.; Qiang, X.; Luo, L.; Hylemon, P.B.; Jiang, Z.; Zhang, L.; Zhou, H. Taurocholate Induces Cyclooxygenase-2 Expression via the Sphingosine 1-phosphate Receptor 2 in a Human Cholangiocarcinoma Cell Line. J. Biol. Chem. 2015, 290, 30988–31002. [Google Scholar] [CrossRef]
- Nagahashi, M.; Yuza, K.; Hirose, Y.; Nakajima, M.; Ramanathan, R.; Hait, N.C.; Hylemon, P.B.; Zhou, H.; Takabe, K.; Wakai, T. The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J. Lipid Res. 2016, 57, 1636–1643. [Google Scholar] [CrossRef]
- Bryan, A.M.; Del Poeta, M. Sphingosine-1-phosphate receptors and innate immunity. Cell Microbiol. 2018, 20, e12836. [Google Scholar] [CrossRef]
- Grigorova, I.L.; Schwab, S.R.; Phan, T.G.; Pham, T.H.M.; Okada, T.; Cyster, J.G. Cortical sinus probing, S1P1-dependent entry and flow-based capture of egressing T cells. Nat. Immunol. 2009, 10, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, H.; Lu, J.; Lv, Q.; Yun, B.; Ge, Z.; Yan, L. Down-regulation of S1PR2 is correlated with poor prognosis and immune infiltrates in cervical squamous cell carcinoma and endocervical adenocarcinoma. Int. J. Immunopathol. Pharmacol. 2023, 37, 3946320231178131. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, Y.I.; Campos, L.E.; Castro, M.G.; Aladhami, A.; Oskeritzian, C.A.; Alvarez, S.E. Sphingosine-1 Phosphate: A New Modulator of Immune Plasticity in the Tumor Microenvironment. Front. Oncol. 2016, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Gao, M.; Lin, D.; Du, G.; Cai, Y. Prognostic and Immunological Roles of MMP-9 in Pan-Cancer. Biomed. Res. Int. 2022, 2022, 2592962. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.; Aoki, H.; Wu, R.; Aoki, M.; Hylemon, P.; Zhou, H.; Takabe, K. Conjugated Bile Acids Accelerate Progression of Pancreatic Cancer Metastasis via S1PR2 Signaling in Cholestasis. Ann. Surg. Oncol. 2023, 30, 1630–1641. [Google Scholar] [CrossRef] [PubMed]
- Schledwitz, A.; Sundel, M.H.; Alizadeh, M.; Hu, S.; Xie, G.; Raufman, J.P. Differential Actions of Muscarinic Receptor Subtypes in Gastric, Pancreatic, and Colon Cancer. Int. J. Mol. Sci. 2021, 22, 13153. [Google Scholar] [CrossRef] [PubMed]
- Calaf, G.M.; Crispin, L.A.; Muñoz, J.P.; Aguayo, F.; Bleak, T.C. Muscarinic Receptors Associated with Cancer. Cancers 2022, 14, 2322. [Google Scholar] [CrossRef]
- Arora, J.; Wang, J.; Weaver, V.; Zhang, Y.; Cantorna, M.T. Novel insight into the role of the vitamin D receptor in the development and function of the immune system. J. Steroid Biochem. Mol. Biol. 2022, 219, 106084. [Google Scholar] [CrossRef]
- Pols, T.W.H.; Puchner, T.; Korkmaz, H.I.; Vos, M.; Soeters, M.R.; de Vries, C.J.M. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the Vitamin D receptor. PLoS ONE 2017, 12, e0176715. [Google Scholar] [CrossRef]
- Li, P.; Zhu, X.; Cao, G.; Wu, R.; Li, K.; Yuan, W.; Chen, B.; Sun, G.; Xia, X.; Zhang, H.; et al. 1α,25(OH)(2)D(3) reverses exhaustion and enhances antitumor immunity of human cytotoxic T cells. J. Immunother. Cancer 2022, 10, e003477. [Google Scholar] [CrossRef]
- Khazan, N.; Quarato, E.R.; Singh, N.A.; Snyder, C.W.A.; Moore, T.; Miller, J.P.; Yasui, M.; Teramoto, Y.; Goto, T.; Reshi, S.; et al. Vitamin D Receptor Antagonist MeTC7 Inhibits PD-L1. Cancers 2023, 15, 3432. [Google Scholar] [CrossRef] [PubMed]
- Fukase, K.; Ohtsuka, H.; Onogawa, T.; Oshio, H.; Ii, T.; Mutoh, M.; Katayose, Y.; Rikiyama, T.; Oikawa, M.; Motoi, F.; et al. Bile acids repress E-cadherin through the induction of Snail and increase cancer invasiveness in human hepatobiliary carcinoma. Cancer Sci. 2008, 99, 1785–1792. [Google Scholar] [CrossRef] [PubMed]
- Di Matteo, S.; Nevi, L.; Costantini, D.; Overi, D.; Carpino, G.; Safarikia, S.; Giulitti, F.; Napoletano, C.; Manzi, E.; De Rose, A.M.; et al. The FXR agonist obeticholic acid inhibits the cancerogenic potential of human cholangiocarcinoma. PLoS ONE 2019, 14, e0210077. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zeng, Y.; Wang, X.; Ma, X.; Li, Q.; Li, N.; Su, H.; Huang, W. FXR blocks the growth of liver cancer cells through inhibiting mTOR-s6K pathway. Biochem. Biophys. Res. Commun. 2016, 474, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Erice, O.; Labiano, I.; Arbelaiz, A.; Santos-Laso, A.; Munoz-Garrido, P.; Jimenez-Agüero, R.; Olaizola, P.; Caro-Maldonado, A.; Martín-Martín, N.; Carracedo, A.; et al. Differential effects of FXR or TGR5 activation in cholangiocarcinoma progression. Biochim. Biophys. Acta Mol. Basis. Dis. 2018, 1864, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Chen, J.; Drachenberg, C.B.; Raufman, J.P.; Xie, G. Farnesoid X receptor represses matrix metalloproteinase 7 expression, revealing this regulatory axis as a promising therapeutic target in colon cancer. J. Biol. Chem. 2019, 294, 8529–8542. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.J.; Jia, X.L.; Li, M.; Yang, N.; Li, Y.P.; Zhang, X.; Gao, N.; Dang, S.S. Guggulsterone induces apoptosis of human hepatocellular carcinoma cells through intrinsic mitochondrial pathway. World J. Gastroenterol. 2015, 21, 13277–13287. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Hu, Y.; Liu, H.X.; Wan, Y.J. MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J. Biol. Chem. 2015, 290, 6507–6515. [Google Scholar] [CrossRef]
- Horowitz, N.S.; Hua, J.; Powell, M.A.; Gibb, R.K.; Mutch, D.G.; Herzog, T.J. Novel cytotoxic agents from an unexpected source: Bile acids and ovarian tumor apoptosis. Gynecol. Oncol. 2007, 107, 344–349. [Google Scholar] [CrossRef]
- Casaburi, I.; Avena, P.; Lanzino, M.; Sisci, D.; Giordano, F.; Maris, P.; Catalano, S.; Morelli, C.; Andò, S. Chenodeoxycholic acid through a TGR5-dependent CREB signaling activation enhances cyclin D1 expression and promotes human endometrial cancer cell proliferation. Cell Cycle 2012, 11, 2699–2710. [Google Scholar] [CrossRef]
- Sharma, R.; Quilty, F.; Gilmer, J.F.; Long, A.; Byrne, A.M. Unconjugated secondary bile acids activate the unfolded protein response and induce golgi fragmentation via a src-kinase-dependant mechanism. Oncotarget 2017, 8, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Chen, X.; Wang, C.; Li, W.; Li, J. Effects and mechanism of the bile acid (farnesoid X) receptor on the Wnt/β-catenin signaling pathway in colon cancer. Oncol. Lett. 2020, 20, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Choi, H.I.; Park, J.S.; Kim, C.S.; Bae, E.H.; Ma, S.K.; Kim, S.W. Farnesoid X receptor protects against cisplatin-induced acute kidney injury by regulating the transcription of ferroptosis-related genes. Redox Biol. 2022, 54, 102382. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zheng, J.; Mu, M.; Chen, Z.; Xu, Z.; Zhao, C.; Yang, K.; Qin, X.; Sun, X.; Yu, J. GW4064 enhances the chemosensitivity of colorectal cancer to oxaliplatin by inducing pyroptosis. Biochem. Biophys. Res. Commun. 2021, 548, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yang, K.; Zheng, J.; Zhao, W.; Sun, X. Synergistic tumor inhibition of colon cancer cells by nitazoxanide and obeticholic acid, a farnesoid X receptor ligand. Cancer Gene Ther. 2021, 28, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhan, M.; Li, Q.; Chen, W.; Chu, H.; Huang, Q.; Hou, Z.; Man, M.; Wang, J. FXR agonists enhance the sensitivity of biliary tract cancer cells to cisplatin via SHP dependent inhibition of Bcl-xL expression. Oncotarget 2016, 7, 34617–34629. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Shirakami, Y.; Shimizu, M.; Kubota, M.; Sakai, H.; Yasuda, Y.; Kochi, T.; Tsurumi, H.; Moriwaki, H. Synergistic growth inhibition of human hepatocellular carcinoma cells by acyclic retinoid and GW4064, a farnesoid X receptor ligand. Cancer Lett. 2012, 323, 215–222. [Google Scholar] [CrossRef]
- Lee, S.; Cho, Y.Y.; Cho, E.J.; Yu, S.J.; Lee, J.H.; Yoon, J.H.; Kim, Y.J. Synergistic effect of ursodeoxycholic acid on the antitumor activity of sorafenib in hepatocellular carcinoma cells via modulation of STAT3 and ERK. Int. J. Mol. Med. 2018, 42, 2551–2559. [Google Scholar] [CrossRef]
- Lee, J.; Hong, E.M.; Kim, J.H.; Kim, J.H.; Jung, J.H.; Park, S.W.; Koh, D.H. Ursodeoxycholic acid inhibits epithelial-mesenchymal transition, suppressing invasiveness of bile duct cancer cells: An in vitro study. Oncol. Lett. 2022, 24, 448. [Google Scholar] [CrossRef]
- Alberts, D.S.; Martínez, M.E.; Hess, L.M.; Einspahr, J.G.; Green, S.B.; Bhattacharyya, A.K.; Guillen, J.; Krutzsch, M.; Batta, A.K.; Salen, G.; et al. Phase III trial of ursodeoxycholic acid to prevent colorectal adenoma recurrence. J. Natl. Cancer Inst. 2005, 97, 846–853. [Google Scholar] [CrossRef]
- Zhang, T.; Feng, S.; Li, J.; Wu, Z.; Deng, Q.; Yang, W.; Li, J.; Pan, G. Farnesoid X receptor (FXR) agonists induce hepatocellular apoptosis and impair hepatic functions via FXR/SHP pathway. Arch. Toxicol. 2022, 96, 1829–1843. [Google Scholar] [CrossRef] [PubMed]
- Panzitt, K.; Zollner, G.; Marschall, H.U.; Wagner, M. Recent advances on FXR-targeting therapeutics. Mol. Cell Endocrinol. 2022, 552, 111678. [Google Scholar] [CrossRef] [PubMed]
- Jose, S.; Devi, S.S.; Sajeev, A.; Girisa, S.; Alqahtani, M.S.; Abbas, M.; Alshammari, A.; Sethi, G.; Kunnumakkara, A.B. Repurposing FDA-approved drugs as FXR agonists: A structure based in silico pharmacological study. Biosci. Rep. 2023, 43, BSR20212791. [Google Scholar] [CrossRef] [PubMed]
- Baghdasaryan, A.; Claudel, T.; Gumhold, J.; Silbert, D.; Adorini, L.; Roda, A.; Vecchiotti, S.; Gonzalez, F.J.; Schoonjans, K.; Strazzabosco, M.; et al. Dual farnesoid X receptor/TGR5 agonist INT-767 reduces liver injury in the Mdr2−/− (Abcb4−/−) mouse cholangiopathy model by promoting biliary HCO−3 output. Hepatology 2011, 54, 1303–1312. [Google Scholar] [CrossRef]
| Compound | Cancer Type | Influence on Immunotherapy | Molecular Mechanism | Reference |
|---|---|---|---|---|
| GW4064 | HCC | sensitizes anti-PD1/PD-L1 therapy | [163] | |
| obeticholic acid | HCC | increases the secretion of CXCL16, | [164] | |
| IFN-γ, and NKT cell populations | ||||
| GW4064 | CRC | sensitizes anti-PD1/PD-L1 therapy | increases CD8+ T cells and activates | [168] |
| FXR and MAPK pathways | ||||
| GW4064 | breast cancer | decreases CAF migration | [129] | |
| UDCA | CRC | sensitizes anti-PD1/PD-L1 therapy | increases CD8+ T-cell responses, | [170] |
| decreases Treg cells | ||||
| guggulsterone | LLC | upregulates PD-L1 expression | inhibits FXR and activates AKT and | [161] |
| MAPK pathways |
| Compound | Anti-Cancer Drug | Cancer Type | Mechanism | Reference |
|---|---|---|---|---|
| Agonist | ||||
| GW4064 | oxaliplatin | CRC | enhances the chemosensitivity of cells to oxaliplatin by induction | [207] |
| of BAX/caspase-3/GSDME-mediated pyroptosis in vitro | ||||
| GW4064 | NTZ | CRC | synergisticly inhibits tumor growth both in vitro and in vivo by | [208] |
| upregulating SHP and downregulating β-catenin | ||||
| GW4064 | cisplatin | BTC | enhances chemosensitivity by upregulating SHP and | [209] |
| downregulating STAT3 phosphorylation in vitro and in vivo | ||||
| OCA | gemcitabine/cisplatin | BTC | enhances the anti-proliferative and pro-apoptotic effects of | [196] |
| chemotherapeutics in vitro and in vivo | ||||
| GW4064 | tamoxifen | Breast Ca. | inhibits tamoxifen-resistant breast cancer cell growth in vitro | [128] |
| PX20350 | sorafenib | HCC | reduces HCC metastasis in the lymph nodes in vivo | [81] |
| GW4064 | doxorubicin, mitomycin C | HCC | enhances tumor chemoresistance against genotoxic drugs | [55] |
| GW4064 | ACR | HCC | synergistically inhibits the HCC growth by inducing apoptosis in vitro | [210] |
| Antagonist | ||||
| UDCA | sorafenib | HCC | inhibits proliferation and induces apoptosis through ROS-dependent | [211] |
| activation of ERK and dephosphorylation of STAT3 in vitro | ||||
| UDCA | gefitinib | BTC | suppresses tumor invasiveness by inhibition of EMT in vitro | [212] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nenkov, M.; Shi, Y.; Ma, Y.; Gaßler, N.; Chen, Y. Targeting Farnesoid X Receptor in Tumor and the Tumor Microenvironment: Implication for Therapy. Int. J. Mol. Sci. 2024, 25, 6. https://doi.org/10.3390/ijms25010006
Nenkov M, Shi Y, Ma Y, Gaßler N, Chen Y. Targeting Farnesoid X Receptor in Tumor and the Tumor Microenvironment: Implication for Therapy. International Journal of Molecular Sciences. 2024; 25(1):6. https://doi.org/10.3390/ijms25010006
Chicago/Turabian StyleNenkov, Miljana, Yihui Shi, Yunxia Ma, Nikolaus Gaßler, and Yuan Chen. 2024. "Targeting Farnesoid X Receptor in Tumor and the Tumor Microenvironment: Implication for Therapy" International Journal of Molecular Sciences 25, no. 1: 6. https://doi.org/10.3390/ijms25010006
APA StyleNenkov, M., Shi, Y., Ma, Y., Gaßler, N., & Chen, Y. (2024). Targeting Farnesoid X Receptor in Tumor and the Tumor Microenvironment: Implication for Therapy. International Journal of Molecular Sciences, 25(1), 6. https://doi.org/10.3390/ijms25010006






