Efficacy of Novel Quaternary Ammonium and Phosphonium Salts Differing in Cation Type and Alkyl Chain Length against Antibiotic-Resistant Staphylococcus aureus
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemical Studies
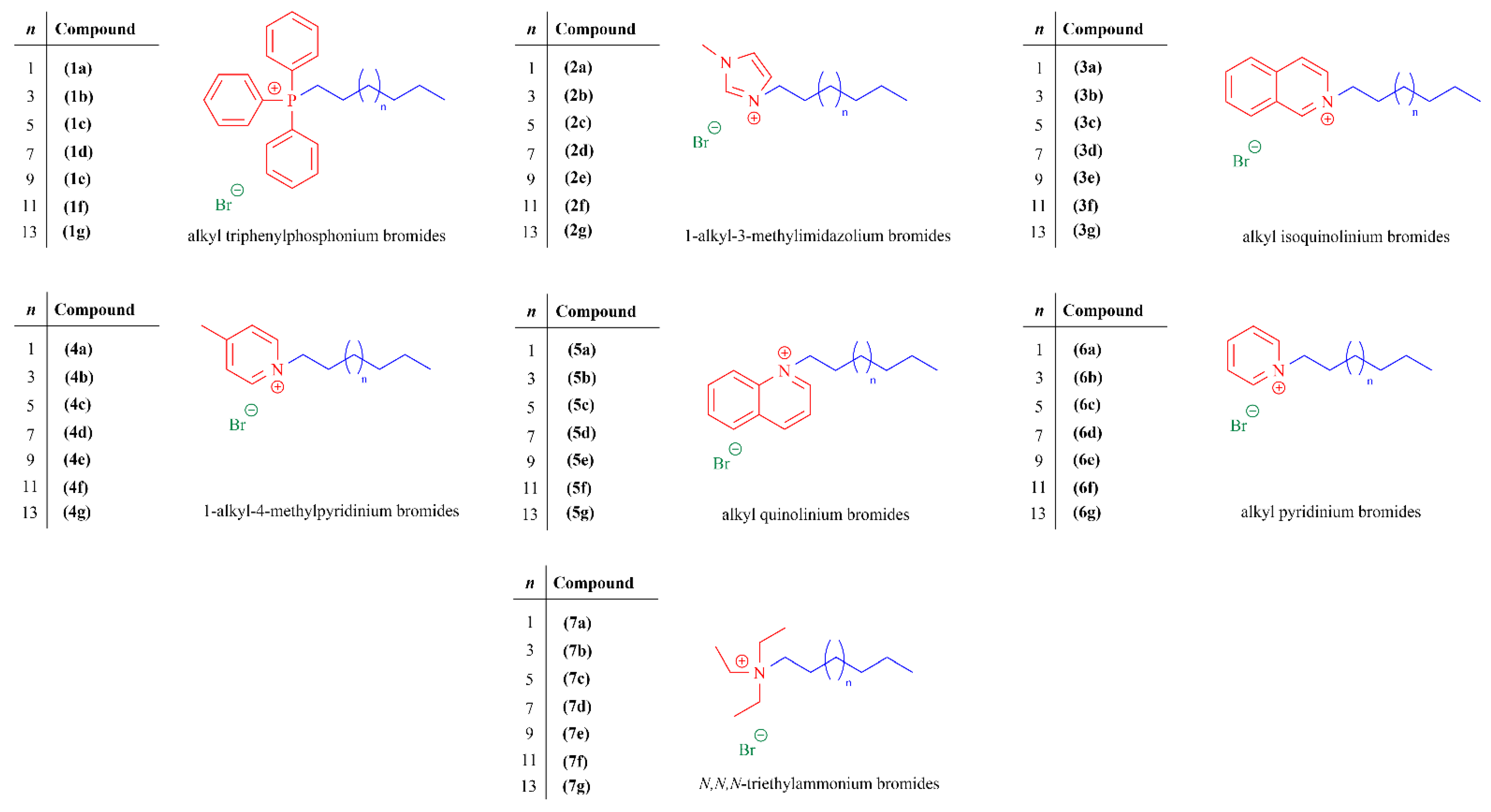
2.1.1. Synthesis of Quaternary Ammonium and Phosphonium Salts
2.1.2. Determination of the Chromatographic Hydrophobicity Index
2.2. Microbiological Studies
2.2.1. Determination of Inhibitory/Bactericidal Activities of Quaternary Ammonium and Phosphonium Salts against an Antibiotic-Susceptible S. aureus Strain: Structure–Activity Relationship Analyses
2.2.2. Determination of Inhibitory/Bactericidal Activities of the Most Promising Quaternary Ammonium and Phosphonium Salts against Antibiotic-Resistant S. aureus Strains
2.2.3. Quaternary Ammonium and Phosphonium Salts Activity against Planktonic Bacteria: Dose–Response
2.3. Cytotoxicity Cell-Based Studies
Evaluation of the Cytotoxicity Profile in Human Hepatocellular Carcinoma Cells
3. Materials and Methods
3.1. Chemical Studies
3.1.1. Reagents and Apparatus
3.1.2. Chemical Synthesis and Structural Characterization
Synthesis of Triphenylphosphonium, Methylimidazolium, Isoquinolinium, Methylpyridinium, Quinolinium, and Pyridinium Salts
Synthesis of Triethylammonium Salts
3.1.3. Evaluation of the Chromatographic Hydrophobicity Index
3.2. Microbiological Studies
3.2.1. Preparation of the Quaternary Ammonium and Phosphonium Salts
3.2.2. Bacterial Strains and Culture Conditions
3.2.3. Evaluation of Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
3.2.4. Antimicrobial Susceptibility Test: Dose–Response
3.3. Cytotoxicity Cell-Based Studies
3.3.1. Reagents and Solvents
3.3.2. Preparation of the Quaternary Ammonium and Phosphonium Salts
3.3.3. Cell Culture and Conditions
3.3.4. Evaluation of the Cytotoxicity
3.3.5. Data Analysis and Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nikfarjam, N.; Ghomi, M.; Agarwal, T.; Hassanpour, M.; Sharifi, E.; Khorsandi, D.; Ali Khan, M.; Rossi, F.; Rossetti, A.; Nazarzadeh Zare, E.; et al. Antimicrobial Ionic Liquid-Based Materials for Biomedical Applications. Adv. Funct. Mater. 2021, 31, 2104148. [Google Scholar] [CrossRef]
- Hassan, R.; Asghar, M.A.; Iqbal, M.; Qaisar, A.; Habib, U.; Ahmad, B. A Comparative Evaluation of Antibacterial Activities of Imidazolium-, Pyridinium-, and Phosphonium-Based Ionic Liquids Containing Octyl Side Chains. Heliyon 2022, 8, e09533. [Google Scholar] [CrossRef] [PubMed]
- Duman, A.N.; Ozturk, I.; Tunçel, A.; Ocakoglu, K.; Colak, S.G.; Hoşgör-Limoncu, M.; Yurt, F. Synthesis of New Water-Soluble Ionic Liquids and Their Antibacterial Profile against Gram-Positive and Gram-Negative Bacteria. Heliyon 2019, 5, e02607. [Google Scholar] [CrossRef] [PubMed]
- Brunel, F.; Lautard, C.; Garzino, F.; Raimundo, J.-M.; Bolla, J.-M.; Camplo, M. Phosphonium-Ammonium-Based Di-Cationic Ionic Liquids as Antibacterial over the ESKAPE Group. Bioorganic Med. Chem. Lett. 2020, 30, 127389. [Google Scholar] [CrossRef] [PubMed]
- Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Chavarria, D.; Borges, A.; Benfeito, S.; Sequeira, L.; Ribeiro, M.; Oliveira, C.; Borges, F.; Simões, M.; Cagide, F. Phytochemicals and Quaternary Phosphonium Ionic Liquids: Connecting the Dots to Develop a New Class of Antimicrobial Agents. J. Adv. Res. 2023, 54, 251–269. [Google Scholar] [CrossRef] [PubMed]
- 2021 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis; World Health Organization: Geneva, Switzerland, 2021; ISBN 9789240047655.
- Cortes, E.; Mora, J.; Márquez, E. Modelling the Anti-Methicillin-Resistant Staphylococcus aureus (MRSA) Activity of Cannabinoids: A QSAR and Docking Study. Crystals 2020, 10, 692. [Google Scholar] [CrossRef]
- Tălăpan, D.; Sandu, A.-M.; Rafila, A. Antimicrobial Resistance of Staphylococcus aureus Isolated between 2017 and 2022 from Infections at a Tertiary Care Hospital in Romania. Antibiotics 2023, 12, 974. [Google Scholar] [CrossRef]
- Jain, M.; Stitt, G.; Son, L.; Enioutina, E.Y. Probiotics and Their Bioproducts: A Promising Approach for Targeting Methicillin-Resistant Staphylococcus aureus and Vancomycin-Resistant Enterococcus. Microorganisms 2023, 11, 2393. [Google Scholar] [CrossRef]
- Dague, A.L.; Valeeva, L.R.; McCann, N.M.; Sharipova, M.R.; Valentovic, M.A.; Bogomolnaya, L.M.; Shakirov, E.V. Identification and Analysis of Antimicrobial Activities from a Model Moss Ceratodon Purpureus. Metabolites 2023, 13, 350. [Google Scholar] [CrossRef]
- Ermolaev, V.V.; Arkhipova, D.M.; Miluykov, V.A.; Lyubina, A.P.; Amerhanova, S.K.; Kulik, N.V.; Voloshina, A.D.; Ananikov, V.P. Sterically Hindered Quaternary Phosphonium Salts (QPSs): Antimicrobial Activity and Hemolytic and Cytotoxic Properties. Int. J. Mol. Sci. 2021, 23, 86. [Google Scholar] [CrossRef]
- Xie, X.; Cong, W.; Zhao, F.; Li, H.; Xin, W.; Hou, G.; Wang, C. Synthesis, Physiochemical Property and Antimicrobial Activity of Novel Quaternary Ammonium Salts. J. Enzym. Inhib. Med. Chem. 2018, 33, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Xiao, H.; Zhang, Y. Antimicrobial Polymeric Materials with Quaternary Ammonium and Phosphonium Salts. Int. J. Mol. Sci. 2015, 16, 3626–3655. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.V.; Zeldi, M.I.; Pugachev, M.V.; Sapozhnikov, S.V.; Shtyrlin, N.V.; Kuznetsova, S.V.; Evtygin, V.E.; Bogachev, M.I.; Kayumov, A.R.; Shtyrlin, Y.G. Antibacterial Effects of Quaternary Bis-Phosphonium and Ammonium Salts of Pyridoxine on Staphylococcus aureus Cells: A Single Base Hitting Two Distinct Targets? World J. Microbiol. Biotechnol. 2016, 32, 5. [Google Scholar] [CrossRef] [PubMed]
- Kula, N.; Lamch, Ł.; Futoma-Kołoch, B.; Wilk, K.A.; Obłąk, E. The Effectiveness of Newly Synthesized Quaternary Ammonium Salts Differing in Chain Length and Type of Counterion against Priority Human Pathogens. Sci. Rep. 2022, 12, 21799. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.M. Quaternary Ammonium Disinfectants and Antiseptics: Tolerance, Resistance and Potential Impact on Antibiotic Resistance. Antimicrob. Resist. Infect. Control 2023, 12, 32. [Google Scholar] [CrossRef]
- Vereshchagin, A.N.; Frolov, N.A.; Egorova, K.S.; Seitkalieva, M.M.; Ananikov, V.P. Quaternary Ammonium Compounds (QACs) and Ionic Liquids (ILs) as Biocides: From Simple Antiseptics to Tunable Antimicrobials. Int. J. Mol. Sci. 2021, 22, 6793. [Google Scholar] [CrossRef]
- Tischer, M.; Pradel, G.; Ohlsen, K.; Holzgrabe, U. Quaternary Ammonium Salts and Their Antimicrobial Potential: Targets or Nonspecific Interactions? ChemMedChem 2012, 7, 22–31. [Google Scholar] [CrossRef]
- Kwaśniewska, D.; Chen, Y.-L.; Wieczorek, D. Biological Activity of Quaternary Ammonium Salts and Their Derivatives. Pathogens 2020, 9, 459. [Google Scholar] [CrossRef]
- Sikora, K.; Jędrzejczak, J.; Bauer, M.; Neubauer, D.; Jaśkiewicz, M.; Szaryńska, M. Quaternary Ammonium Salts of Cationic Lipopeptides with Lysine Residues—Synthesis, Antimicrobial, Hemolytic and Cytotoxic Activities. Probiotics Antimicrob. Proteins 2023, 15, 1465–1483. [Google Scholar] [CrossRef]
- Pugachev, M.V.; Shtyrlin, N.V.; Sysoeva, L.P.; Nikitina, E.V.; Abdullin, T.I.; Iksanova, A.G.; Ilaeva, A.A.; Musin, R.Z.; Berdnikov, E.A.; Shtyrlin, Y.G. Synthesis and Antibacterial Activity of Novel Phosphonium Salts on the Basis of Pyridoxine. Bioorg. Med. Chem. 2013, 21, 4388–4395. [Google Scholar] [CrossRef]
- Noroozi-Shad, N.; Gholizadeh, M.; Sabet-Sarvestani, H. Quaternary Phosphonium Salts in the Synthetic Chemistry: Recent Progress, Development, and Future Perspectives. J. Mol. Struct. 2022, 1257, 132628. [Google Scholar] [CrossRef]
- Pugachev, M.V.; Shtyrlin, N.V.; Sapozhnikov, S.V.; Sysoeva, L.P.; Iksanova, A.G.; Nikitina, E.V.; Musin, R.Z.; Lodochnikova, O.A.; Berdnikov, E.A.; Shtyrlin, Y.G. Bis-Phosphonium Salts of Pyridoxine: The Relationship between Structure and Antibacterial Activity. Bioorg. Med. Chem. 2013, 21, 7330–7342. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhou, H.; Gai, F.; Chi, X.; Zhao, Y.; Zhang, F.; Zhao (Kent), Z. Synthesis of Quaternary Phosphonium N-Chloramine Biocides for Antimicrobial Applications. RSC Adv. 2017, 7, 13244–13249. [Google Scholar] [CrossRef]
- Khasiyatullina, N.R.; Mironov, V.F.; Voloshina, A.D.; Sapunova, A.S. Synthesis and Antimicrobial Properties of Novel Phosphonium Salts Bearing 1,4-Dihydroxyaryl Fragment. Chem. Biodivers. 2019, 16, e1900039. [Google Scholar] [CrossRef]
- Akshay Ravindra, P.; Karpagam, S. Synthesis and Biological Activity of Azine Heterocycle Functionalized Quaternary Phosphonium Salts. IOP Conf. Ser. Mater. Sci. Eng. 2017, 263, 022017. [Google Scholar] [CrossRef]
- Florio, W.; Becherini, S.; D’Andrea, F.; Lupetti, A.; Chiappe, C.; Guazzelli, L. Comparative Evaluation of Antimicrobial Activity of Different Types of Ionic Liquids. Mater. Sci. Eng. C 2019, 104, 109907. [Google Scholar] [CrossRef]
- Pendleton, J.N.; Gilmore, B.F. The Antimicrobial Potential of Ionic Liquids: A Source of Chemical Diversity for Infection and Biofilm Control. Int. J. Antimicrob. Agents 2015, 46, 131–139. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, J.; Han, J.; Gao, L.; Liu, H.; Lu, Z.; Zhao, H.; Bie, X. Insights into the Antimicrobial Activity and Cytotoxicity of Engineered α-Helical Peptide Amphiphiles. J. Med. Chem. 2016, 59, 10946–10962. [Google Scholar] [CrossRef]
- Demberelnyamba, D.; Kim, K.-S.; Choi, S.; Park, S.-Y.; Lee, H.; Kim, C.-J.; Yoo, I.-D. Synthesis and Antimicrobial Properties of Imidazolium and Pyrrolidinonium Salts. Bioorg. Med. Chem. 2004, 12, 853–857. [Google Scholar] [CrossRef]
- Gilmore, B.F.; Andrews, G.P.; Borberly, G.; Earle, M.J.; Gilea, M.A.; Gorman, S.P.; Lowry, A.F.; McLaughlin, M.; Seddon, K.R. Enhanced Antimicrobial Activities of 1-Alkyl-3-Methyl Imidazolium Ionic Liquids Based on Silver or Copper Containing Anions. New J. Chem. 2013, 37, 873. [Google Scholar] [CrossRef]
- Hodyna, D.; Kovalishyn, V.; Rogalsky, S.; Blagodatnyi, V.; Petko, K.; Metelytsia, L. Antibacterial Activity of Imidazolium-Based Ionic Liquids Investigated by QSAR Modeling and Experimental Studies. Chem. Biol. Drug Des. 2016, 88, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.J.L.P.; Arco, S.D. Solvent-Free Sonochemical Synthesis and Antifungal Activity of 1-Alkyl-3-Methylimidazolium Bromide [RMIM]Br Ionic Liquids. J. Chin. Chem. Soc. 2014, 61, 935–939. [Google Scholar] [CrossRef]
- Metelytsia, L.O.; Hodyna, D.M.; Semenyuta, I.V.; Kovalishyn, V.V.; Rogalsky, S.P.; Derevianko, K.Y.; Brovarets, V.S.; Tetko, I.V. Theoretical and Experimental Studies of Phosphonium Ionic Liquids as Potential Antibacterials of MDR Acinetobacter baumannii. Antibiotics 2022, 11, 491. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.R.P.; Paredes, X.; Cristino, A.F.; Santos, F.J.V.; Queirós, C.S.G.P. Ionic Liquids—A Review of Their Toxicity to Living Organisms. Int. J. Mol. Sci. 2021, 22, 5612. [Google Scholar] [CrossRef] [PubMed]
- Mester, P.; Wagner, M.; Rossmanith, P. Antimicrobial Effects of Short Chained Imidazolium-Based Ionic Liquids—Influence of Anion Chaotropicity. Ecotoxicol. Environ. Saf. 2015, 111, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Valko, K.; Nunhuck, S.; Bevan, C.; Abraham, M.H.; Reynolds, D.P. Fast Gradient HPLC Method to Determine Compounds Binding to Human Serum Albumin. Relationships with Octanol/Water and Immobilized Artificial Membrane Lipophilicity. J. Pharm. Sci. 2003, 92, 2236–2248. [Google Scholar] [CrossRef] [PubMed]
- Rokitskaya, T.I.; Kotova, E.A.; Luzhkov, V.B.; Kirsanov, R.S.; Aleksandrova, E.V.; Korshunova, G.A.; Tashlitsky, V.N.; Antonenko, Y.N. Lipophilic Ion Aromaticity Is Not Important for Permeability across Lipid Membranes. Biochim. Biophys. Acta (BBA)-Biomembr. 2021, 1863, 183483. [Google Scholar] [CrossRef]
- Valkó, K.; Bevan, C.; Reynolds, D. Chromatographic Hydrophobicity Index by Fast-Gradient RP-HPLC: A High-Throughput Alternative to Log P/Log D. Anal. Chem. 1997, 69, 2022–2029. [Google Scholar] [CrossRef]
- Chavarria, D.; Benfeito, S.; Soares, P.; Lima, C.; Garrido, J.; Serrão, P.; Soares-da-Silva, P.; Remião, F.; Oliveira, P.J.; Borges, F. Boosting Caffeic Acid Performance as Antioxidant and Monoamine Oxidase B/Catechol-O-Methyltransferase Inhibitor. Eur. J. Med. Chem. 2022, 243, 114740. [Google Scholar] [CrossRef]
- Jain, P.; Kumar, A. Concentration-Dependent Apparent Partition Coefficients of Ionic Liquids Possessing Ethyl- and Bi-Sulphate Anions. Phys. Chem. Chem. Phys. 2016, 18, 1105–1113. [Google Scholar] [CrossRef]
- Abreu, A.C.; Serra, S.C.; Borges, A.; Saavedra, M.J.; Salgado, A.J.; Simões, M. Evaluation of the Best Method to Assess Antibiotic Potentiation by Phytochemicals against Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 2014, 79, 125–134. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abreu, A.C.; Serra, S.C.; Borges, A.; Saavedra, M.J.; Mcbain, A.J.; Salgado, A.J.; Simões, M. Combinatorial Activity of Flavonoids with Antibiotics Against Drug-Resistant Staphylococcus aureus. Microb. Drug Resist. 2015, 21, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.; Borges, A.; Saavedra, M.J.; Borges, F.; Simões, M. Screening of Natural Molecules as Adjuvants to Topical Antibiotics to Treat Staphylococcus aureus from Diabetic Foot Ulcer Infections. Antibiotics 2022, 11, 620. [Google Scholar] [CrossRef] [PubMed]
- Pinto, T.C.A.; Banerjee, A.; Nazarov, P.A. Triphenyl phosphonium-based substances are alternatives to common antibiotics. Bull. Russ. State Med. Univ. 2018, 1, 16–21. [Google Scholar] [CrossRef]
- Strobykina, I.Y.; Voloshina, A.D.; Andreeva, O.V.; Sapunova, A.S.; Lyubina, A.P.; Amerhanova, S.K.; Belenok, M.G.; Saifina, L.F.; Semenov, V.E.; Kataev, V.E. Synthesis, Antimicrobial Activity and Cytotoxicity of Triphenylphosphonium (TPP) Conjugates of 1,2,3-Triazolyl Nucleoside Analogues. Bioorg.Chem. 2021, 116, 105328. [Google Scholar] [CrossRef] [PubMed]
- Ventura, S.P.M.; Gonçalves, A.M.M.; Sintra, T.; Pereira, J.L.; Gonçalves, F.; Coutinho, J.A.P. Designing Ionic Liquids: The Chemical Structure Role in the Toxicity. Ecotoxicology 2013, 22, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Petkovic, M.; Seddon, K.R.; Rebelo, L.P.N.; Silva Pereira, C. Ionic Liquids: A Pathway to Environmental Acceptability. Chem. Soc. Rev. 2011, 40, 1383–1403. [Google Scholar] [CrossRef]
- de Almeida, C.G.; Garbois, G.D.; Amaral, L.M.; Diniz, C.C.; Le Hyaric, M. Relationship between Structure and Antibacterial Activity of Lipophilic N-Acyldiamines. Biomed. Pharmacother. 2010, 64, 287–290. [Google Scholar] [CrossRef]
- Brown, P.; Abdulle, O.; Boakes, S.; Divall, N.; Duperchy, E.; Ganeshwaran, S.; Lester, R.; Moss, S.; Rivers, D.; Simonovic, M.; et al. Influence of Lipophilicity on the Antibacterial Activity of Polymyxin Derivatives and on Their Ability to Act as Potentiators of Rifampicin. ACS Infect. Dis. 2021, 7, 894–905. [Google Scholar] [CrossRef]
- Valls, A.; Andreu, J.J.; Falomir, E.; Luis, S.V.; Atrián-Blasco, E.; Mitchell, S.G.; Altava, B. Imidazole and Imidazolium Antibacterial Drugs Derived from Amino Acids. Pharmaceuticals 2020, 13, 482. [Google Scholar] [CrossRef]
- Preiss, U.; Jungnickel, C.; Thöming, J.; Krossing, I.; Łuczak, J.; Diedenhofen, M.; Klamt, A. Predicting the Critical Micelle Concentrations of Aqueous Solutions of Ionic Liquids and Other Ionic Surfactants. Chem.-Eur. J. 2009, 15, 8880–8885. [Google Scholar] [CrossRef] [PubMed]
- Kiran Kumar Reddy, G.; Nancharaiah, Y.V. Alkylimidazolium Ionic Liquids for Biofilm Control: Experimental Studies on Controlling Multispecies Biofilms in Natural Waters. J. Mol. Liq. 2021, 336, 116859. [Google Scholar] [CrossRef]
- Busetti, A.; Crawford, D.E.; Earle, M.J.; Gilea, M.A.; Gilmore, B.F.; Gorman, S.P.; Laverty, G.; Lowry, A.F.; McLaughlin, M.; Seddon, K.R. Antimicrobial and Antibiofilm Activities of 1-Alkylquinolinium Bromide Ionic Liquids. Green Chem. 2010, 12, 420–442. [Google Scholar] [CrossRef]
- Salajkova, S.; Benkova, M.; Marek, J.; Sleha, R.; Prchal, L.; Malinak, D.; Dolezal, R.; Sepčić, K.; Gunde-Cimerman, N.; Kuca, K.; et al. Wide-Antimicrobial Spectrum of Picolinium Salts. Molecules 2020, 25, 2254. [Google Scholar] [CrossRef] [PubMed]
- Carson, L.; Chau, P.K.W.; Earle, M.J.; Gilea, M.A.; Gilmore, B.F.; Gorman, S.P.; McCann, M.T.; Seddon, K.R. Antibiofilm Activities of 1-Alkyl-3-Methylimidazolium Chloride Ionic Liquids. Green Chem. 2009, 11, 492–497. [Google Scholar] [CrossRef]
- Garcia, M.T.; Ribosa, I.; Perez, L.; Manresa, A.; Comelles, F. Self-Assembly and Antimicrobial Activity of Long-Chain Amide-Functionalized Ionic Liquids in Aqueous Solution. Colloids Surf. B Biointerfaces 2014, 123, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Pernak, J.; Sobaszkiewicz, K.; Mirska, I. Anti-Microbial Activities of Ionic Liquids. Green Chem. 2003, 5, 52–56. [Google Scholar] [CrossRef]
- Garcia, M.T.; Ribosa, I.; Perez, L.; Manresa, A.; Comelles, F. Micellization and Antimicrobial Properties of Surface-Active Ionic Liquids Containing Cleavable Carbonate Linkages. Langmuir 2017, 33, 6511–6520. [Google Scholar] [CrossRef]
- Łuczak, J.; Jungnickel, C.; Łącka, I.; Stolte, S.; Hupka, J. Antimicrobial and Surface Activity of 1-Alkyl-3-Methylimidazolium Derivatives. Green Chem. 2010, 12, 593. [Google Scholar] [CrossRef]
- Forero-Doria, O.; Araya-Maturana, R.; Barrientos-Retamal, A.; Morales-Quintana, L.; Guzmán, L. N-Alkylimidazolium Salts Functionalized with p-Coumaric and Cinnamic Acid: A Study of Their Antimicrobial and Antibiofilm Effects. Molecules 2019, 24, 3484. [Google Scholar] [CrossRef]
- Gilmore, B.F. Antimicrobial Ionic Liquids. In Ionic Liquids: Applications and Perspectives; Kokorin, A., Ed.; IntechOpen: London, UK, 2011; pp. 587–604. [Google Scholar]
- Feder-Kubis, J.; Wnętrzak, A.; Suchodolski, J.; Tomasz Mitkowski, P.; Krasowska, A. Imidazolium Room-Temperature Ionic Liquids with Alkoxymethyl Substituent: A Quest for Improved Microbiological Selectivity. Chem. Eng. J. 2022, 442, 136062. [Google Scholar] [CrossRef]
- Gibbons, S. A Novel Inhibitor of Multidrug Efflux Pumps in Staphylococcus aureus. J. Antimicrob. Chemother. 2003, 51, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.; Udo, E.E. The Effect of Reserpine, a Modulator of Multidrug Efflux Pumps, on the in Vitro Activity of Tetracycline against Clinical Isolates of Methicillin Resistant Staphylococcus aureus (MRSA) Possessing the Tet(K) Determinant. Phytother. Res. 2000, 14, 139–140. [Google Scholar] [CrossRef]
- Zimmermann, S.; Klinger-Strobel, M.; Bohnert, J.A.; Wendler, S.; Rödel, J.; Pletz, M.W.; Löffler, B.; Tuchscherr, L. Clinically Approved Drugs Inhibit the Staphylococcus aureus Multidrug NorA Efflux Pump and Reduce Biofilm Formation. Front. Microbiol. 2019, 10, 2762. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Suss, P.H.; Telles, J.P.; Dantas, L.R.; Borges, N.H.; Ribeiro, V.S.T. Antimicrobial Treatment of Staphylococcus aureus Biofilms. Antibiotics 2023, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Russell, A.D.; McDonnell, G. Concentration: A Major Factor in Studying Biocidal Action. J. Hosp. Infect. 2000, 44, 1–3. [Google Scholar] [CrossRef]
- Rita Pereira, A.; Gomes, I.B.; Simões, M. Choline-Based Ionic Liquids for Planktonic and Biofilm Growth Control of Bacillus cereus and Pseudomonas fluorescens. J. Mol. Liq. 2022, 346, 117077. [Google Scholar] [CrossRef]
- Fernandes, S.; Gomes, I.B.; Simões, M. Antimicrobial Activity of Glycolic Acid and Glyoxal against Bacillus cereus and Pseudomonas fluorescens. Food Res. Int. 2020, 136, 109346. [Google Scholar] [CrossRef]
- Maillard, J.-Y. Bacterial Target Sites for Biocide Action. J. Appl. Microbiol. 2002, 92, 16S–27S. [Google Scholar] [CrossRef]
- Hugo, W.B.; Denyer, S.P. Preservatives in the Food, Pharmaceutical and Environmental Industries. In The Concentration Exponent of Disinfectants and Preservatives (Biocides); Board, R.G., Allwood, M.C., Banks, J.G., Eds.; Blackwell Scientific Publications: Nottingham, UK, 1987; pp. 281–291. ISBN 9780632017270. [Google Scholar]
- Xuan, J.; Chen, S.; Ning, B.; Tolleson, W.H.; Guo, L. Development of HepG2-Derived Cells Expressing Cytochrome P450s for Assessing Metabolism-Associated Drug-Induced Liver Toxicity. Chem. Biol. Interact. 2016, 255, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Donato, M.T.; Tolosa, L.; Gómez-Lechón, M.J. Culture and Functional Characterization of Human Hepatoma HepG2 Cells. Methods Mol. Biol. 2015, 1250, 77–93. [Google Scholar] [PubMed]
- Martínez-Sena, T.; Moro, E.; Moreno-Torres, M.; Quintás, G.; Hengstler, J.; Castell, J.V. Metabolomics-Based Strategy to Assess Drug Hepatotoxicity and Uncover the Mechanisms of Hepatotoxicity Involved. Arch. Toxicol. 2023, 97, 1723–1738. [Google Scholar] [CrossRef] [PubMed]
- López-García, J.; Lehocký, M.; Humpolíček, P.; Sáha, P. HaCaT Keratinocytes Response on Antimicrobial Atelocollagen Substrates: Extent of Cytotoxicity, Cell Viability and Proliferation. J. Funct. Biomater. 2014, 5, 43–57. [Google Scholar] [CrossRef]
- Benfeito, S.; Fernandes, C.; Chavarria, D.; Barreiro, S.; Cagide, F.; Sequeira, L.; Teixeira, J.; Silva, R.; Remião, F.; Oliveira, P.J.; et al. Modulating Cytotoxicity with Lego-like Chemistry: Upgrading Mitochondriotropic Antioxidants with Prototypical Cationic Carrier Bricks. J. Med. Chem. 2023, 66, 1835–1851. [Google Scholar] [CrossRef]
- Costa, F.M.S.; Saraiva, M.L.M.F.S.; Passos, M.L.C. Ionic Liquids and Organic Salts with Antimicrobial Activity as a Strategy against Resistant Microorganisms. J. Mol. Liq. 2022, 368, 120750. [Google Scholar] [CrossRef]
- Oliveira, C.; Cagide, F.; Teixeira, J.; Amorim, R.; Sequeira, L.; Mesiti, F.; Silva, T.; Garrido, J.; Remião, F.; Vilar, S.; et al. Hydroxybenzoic Acid Derivatives as Dual-Target Ligands: Mitochondriotropic Antioxidants and Cholinesterase Inhibitors. Front. Chem. 2018, 6, 126. [Google Scholar] [CrossRef]
- Meng, X.; Devemy, J.; Verney, V.; Gautier, A.; Husson, P.; Andanson, J.-M. Improving Cellulose Dissolution in Ionic Liquids by Tuning the Size of the Ions: Impact of the Length of the Alkyl Chains in Tetraalkylammonium Carboxylate. ChemSusChem 2017, 10, 1749–1760. [Google Scholar] [CrossRef]
- Oliveira, I.M.; Borges, A.; Borges, F.; Simões, M. Repurposing Ibuprofen to Control Staphylococcus aureus Biofilms. Eur. J. Med. Chem. 2019, 166, 197–205. [Google Scholar] [CrossRef]
- EN-1276; Chemical Disinfectants and Antiseptics—Quantitative Suspension Test for the Evaluation of Bactericidal Activity of Chemical Disinfectants and Antiseptics Used in Food, Industrial, Domestic and Institutional Areas—Test Method and Requirements (Phase 2, Step 1). CEN (Comité Européen de Normalisation, European Committee for Standardization): Brussels, Belgium, 2019. Available online: https://standards.iteh.ai/catalog/standards/cen/5b01722b-fe29-4d96-8608-7e5c9da8a80a/en-1276-2019 (accessed on 4 January 2023).
- Grobe, K.J.; Zahller, J.; Stewart, P.S. Role of Dose Concentration in Biocide Efficacy against Pseudomonas Aeruginosa Biofilms. J. Ind. Microbiol. Biotechnol. 2002, 29, 10–15. [Google Scholar] [CrossRef]
- Benfeito, S.; Oliveira, C.; Fernandes, C.; Cagide, F.; Teixeira, J.; Amorim, R.; Garrido, J.; Martins, C.; Sarmento, B.; Silva, R.; et al. Fine-Tuning the Neuroprotective and Blood-Brain Barrier Permeability Profile of Multi-Target Agents Designed to Prevent Progressive Mitochondrial Dysfunction. Eur. J. Med. Chem. 2019, 167, 525–545. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.; Martins, C.; Fonseca, A.; Nunes, R.; Matos, M.J.; Silva, R.; Garrido, J.; Sarmento, B.; Remião, F.; Otero-Espinar, F.J.; et al. PEGylated PLGA Nanoparticles as a Smart Carrier to Increase the Cellular Uptake of a Coumarin-Based Monoamine Oxidase B Inhibitor. ACS Appl. Mater. Interfaces 2018, 10, 39557–39569. [Google Scholar] [CrossRef] [PubMed]

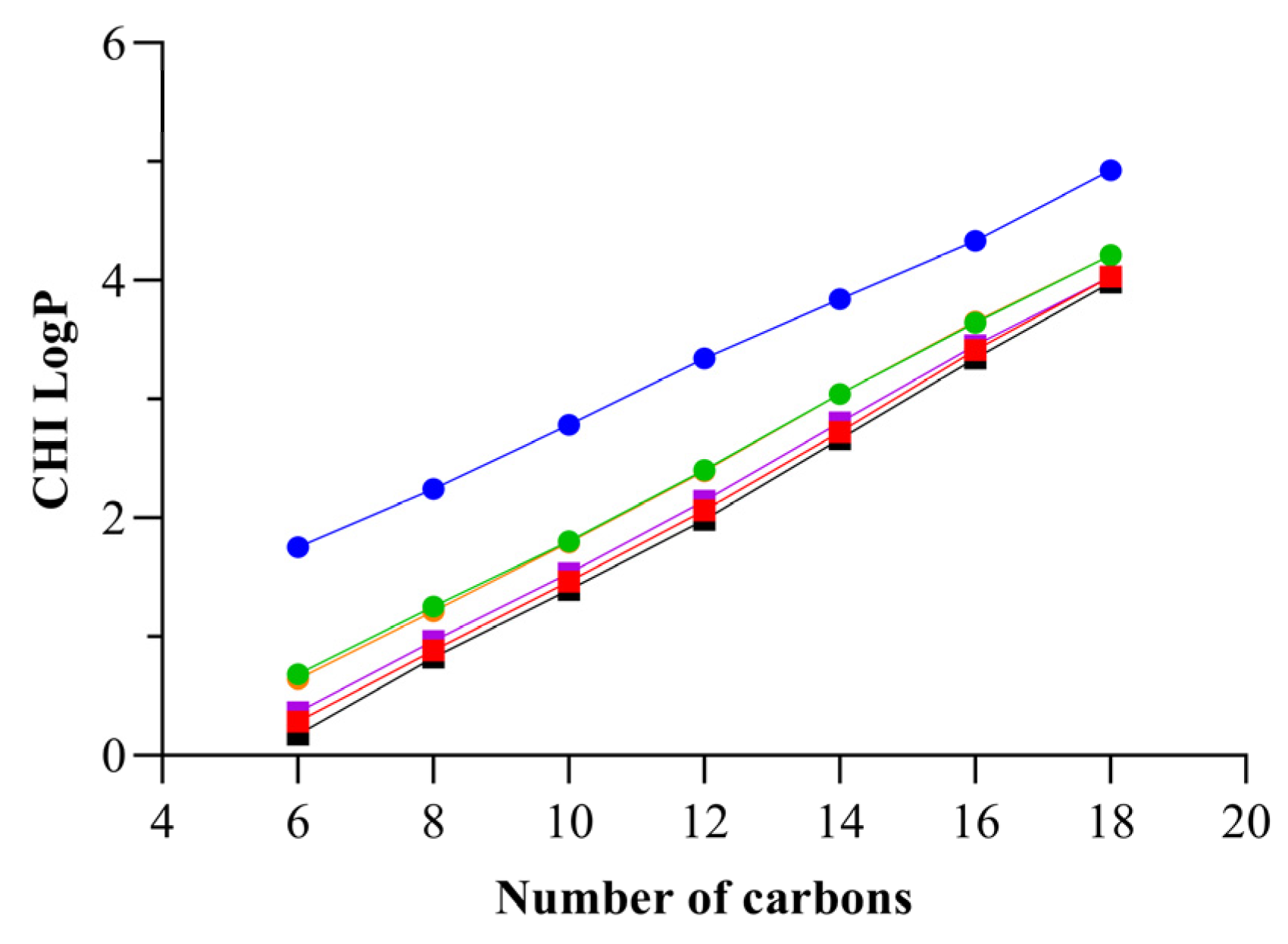
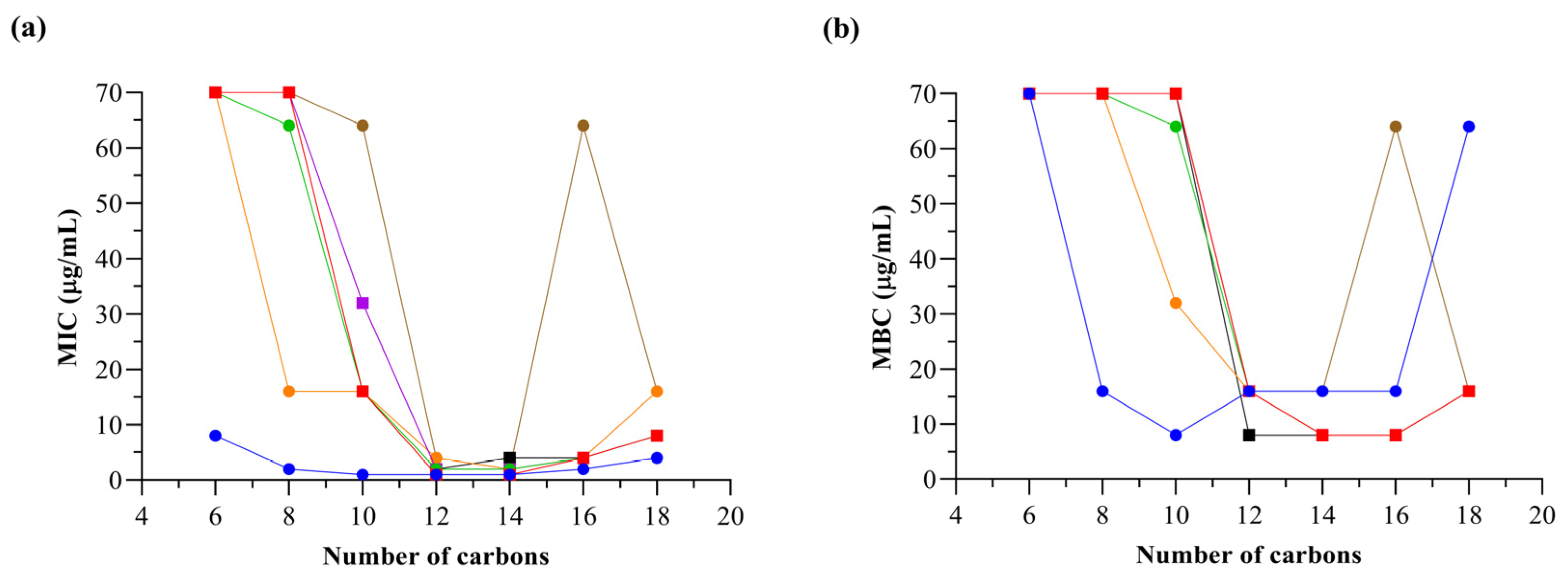

| Compound | tR/min | CHI 1 | CHI LogP 2 |
|---|---|---|---|
| 1a | 6.599 | 60.58 | 1.747 |
| 1b | 7.096 | 71.06 | 2.240 |
| 1c | 7.642 | 82.58 | 2.781 |
| 1d | 8.207 | 94.50 | 3.341 |
| 1e | 8.706 | 105.0 | 3.836 |
| 1f | 9.202 | 115.5 | 4.328 |
| 1g | 9.806 | 128.2 | 4.926 |
| 2a | 5.118 | 29.35 | 0.2794 |
| 2b | 5.727 | 42.19 | 0.8830 |
| 2c | 6.311 | 54.51 | 1.462 |
| 2d | 6.915 | 67.25 | 2.061 |
| 2e | 7.584 | 81.36 | 2.724 |
| 2f | 8.274 | 95.91 | 3.408 |
| 2g | 8.906 | 109.2 | 4.034 |
| 3a | 5.519 | 37.81 | 0.6769 |
| 3b | 6.098 | 50.02 | 1.251 |
| 3c | 6.648 | 61.62 | 1.796 |
| 3d | 7.258 | 74.48 | 2.401 |
| 3e | 7.898 | 87.98 | 3.035 |
| 3f | 8.508 | 100.8 | 3.640 |
| 3g | 9.087 | 113.1 | 4.214 |
| 4a | 5.200 | 31.08 | 0.3607 |
| 4b | 5.808 | 43.90 | 0.9633 |
| 4c | 6.381 | 55.99 | 1.531 |
| 4d | 6.996 | 68.96 | 2.141 |
| 4e | 7.657 | 82.90 | 2.796 |
| 4f | 8.315 | 96.77 | 3.448 |
| 4g | 8.902 | 109.2 | 4.030 |
| 5a | 5.480 | 36.98 | 0.6382 |
| 5b | 6.061 | 49.24 | 1.214 |
| 5c | 6.640 | 61.45 | 1.788 |
| 5d | 7.245 | 74.21 | 2.388 |
| 5e | 7.899 | 88.00 | 3.036 |
| 5f | 8.519 | 101.1 | 3.651 |
| 5g | 9.081 | 112.9 | 4.208 |
| 6a | 5.010 | 27.07 | 0.1723 |
| 6b | 5.660 | 40.78 | 0.8166 |
| 6c | 6.243 | 53.07 | 1.395 |
| 6d | 6.836 | 65.58 | 1.982 |
| 6e | 7.515 | 79.90 | 2.655 |
| 6f | 8.206 | 94.47 | 3.340 |
| 6g | 8.853 | 108.1 | 3.982 |
| Compound | CECT 976 | |
|---|---|---|
| MIC 1 | MBC 2 | |
| 1a | 8 | >64 |
| 1b | 2 | 16 |
| 1c | 1 | 8 |
| 1d | 1 | 16 |
| 1e | 1 | 16 |
| 1f | 2 | 16 |
| 1g | 4 | 64 |
| 2a | >64 | >64 |
| 2b | >64 | >64 |
| 2c | 16 | >64 |
| 2d | 1 | 16 |
| 2e | 1 | 8 |
| 2f | 4 | 8 |
| 2g | 8 | 16 |
| 3a | >64 | >64 |
| 3b | 64 | >64 |
| 3c | 16 | 64 |
| 3d | 2 | 16 |
| 3e | 2 | 8 |
| 3f | 4 | 8 |
| 3g | 8 | 16 |
| 4a | >64 | >64 |
| 4b | >64 | >64 |
| 4c | 32 | >64 |
| 4d | 2 | 16 |
| 4e | 2 | 8 |
| 4f | 4 | 8 |
| 4g | 8 | 16 |
| 5a | >64 | >64 |
| 5b | 16 | >64 |
| 5c | 16 | 32 |
| 5d | 4 | 16 |
| 5e | 2 | 8 |
| 5f | 4 | 8 |
| 5g | 16 | 16 |
| 6a | >64 | >64 |
| 6b | >64 | >64 |
| 6c | 32 | >64 |
| 6d | 2 | 8 |
| 6e | 4 | 8 |
| 6f | 4 | 8 |
| 6g | 8 | 16 |
| 7a | >64 | >64 |
| 7b | >64 | >64 |
| 7c | 64 | >64 |
| 7d | 4 | 16 |
| 7e | 2 | 16 |
| 7f | 64 | 64 |
| 7g | 16 | 16 |
| [ERY] | 0.24 | ND |
| [TET] | 0.96 | ND |
| [CIP] | 1 | ND |
| [AMP] | 1.5 | ND |
| [OXA] | 0.48 | ND |
| Compound | XU212 | SA1199B | RN4220 | |||
|---|---|---|---|---|---|---|
| MIC 1 | MBC 2 | MIC 1 | MBC 2 | MIC 1 | MBC 2 | |
| 1e | 2 | 2 | 1 | 2 | 1 | 2 |
| 2e | 32 | 64 | 8 | 32 | 4 | 8 |
| 3e | 4 | 8 | 2 | 8 | 2 | 4 |
| 4e | 32 | 32 | 8 | 32 | 4 | 8 |
| 5e | 8 | 8 | 4 | 8 | 2 | 8 |
| 6e | 32 | 64 | 16 | 16 | 4 | 8 |
| 7e | 32 | 32 | 16 | 32 | 4 | 8 |
| [ERY] | ND | ND | ND | ND | 256 | ND |
| [TET] | 128 | ND | ND | ND | ND | ND |
| [CIP] | ND | ND | 128 | ND | ND | ND |
| Compound | Concentration (µg/mL) | |||
|---|---|---|---|---|
| 1 | 2 | 16 | 32 | |
| 1e | 53.86 ± 4.854% (****) | 72.84 ± 7.906% (****) | 12.29 ± 3.775% (****) | 13.78 ± 2.666% (****) |
| 2e | 90.88 ± 10.08% (**) | 86.71 ± 4.103% (****) | 12.85 ± 2.005% (****) | 13.64 ± 2.776% (****) |
| 3e | ND | 79.86 ± 9.893% (****) | 12.68 ± 2.385% (****) | 12.77 ± 1.368% (****) |
| 4e | ND | 92.62 ± 4.355% (*) | 14.45 ± 2.437% (****) | 21.52 ± 19.06% (****) |
| 5e | ND | 88.20 ± 3.101% (****) | 12.06 ± 2.988% (****) | 14.38 ± 4.045% (****) |
| 6e | ND | 94.21 ± 10.35% (ns) | 10.46 ± 3.129% (****) | 12.93 ± 3.760% (****) |
| 7e | ND | 86.24 ± 1.502% (****) | 18.51 ± 2.592% (****) | 16.59 ± 4.378% (****) |
| [ERY] | ND | 108.4 ± 4.998% (ns) | 107.1 ± 7.642% (ns) | 110.2 ± 9.546% (ns) |
| [TET] | ND | 109.0 ± 5.251% (ns) | 107.5 ± 9.694% (ns) | 106.8 ± 6.699% (ns) |
| [CIP] | ND | 106.7 ± 11.16% (ns) | 96.82 ± 5.237% (ns) | 92.35 ± 6.807% (ns) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunes, B.; Cagide, F.; Fernandes, C.; Borges, A.; Borges, F.; Simões, M. Efficacy of Novel Quaternary Ammonium and Phosphonium Salts Differing in Cation Type and Alkyl Chain Length against Antibiotic-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2024, 25, 504. https://doi.org/10.3390/ijms25010504
Nunes B, Cagide F, Fernandes C, Borges A, Borges F, Simões M. Efficacy of Novel Quaternary Ammonium and Phosphonium Salts Differing in Cation Type and Alkyl Chain Length against Antibiotic-Resistant Staphylococcus aureus. International Journal of Molecular Sciences. 2024; 25(1):504. https://doi.org/10.3390/ijms25010504
Chicago/Turabian StyleNunes, Bárbara, Fernando Cagide, Carlos Fernandes, Anabela Borges, Fernanda Borges, and Manuel Simões. 2024. "Efficacy of Novel Quaternary Ammonium and Phosphonium Salts Differing in Cation Type and Alkyl Chain Length against Antibiotic-Resistant Staphylococcus aureus" International Journal of Molecular Sciences 25, no. 1: 504. https://doi.org/10.3390/ijms25010504
APA StyleNunes, B., Cagide, F., Fernandes, C., Borges, A., Borges, F., & Simões, M. (2024). Efficacy of Novel Quaternary Ammonium and Phosphonium Salts Differing in Cation Type and Alkyl Chain Length against Antibiotic-Resistant Staphylococcus aureus. International Journal of Molecular Sciences, 25(1), 504. https://doi.org/10.3390/ijms25010504










