GLIS1, Correlated with Immune Infiltrates, Is a Potential Prognostic Biomarker in Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. Identification of tDEGs, tDEMs and tDEM-TGs in Prostate Cancer
2.2. GO and Pathway Enrichment Analysis
2.3. Identification of Key Genes and miRNAs
2.4. Diagnostic and Prognostic Values of GLIS1 in Pan-Cancer
2.5. GLIS1 Was Associated with the Prognosis of Prostate Cancer Patients
2.6. Functional Enrichment and Pathway Analysis of High and Low-GLIS1 Expression Samples
2.7. The Correlation between GLIS1 Expression and Immune Infiltration Levels
2.8. The Association between GLIS1, Chemokines and Chemokine Receptors
3. Discussion
4. Materials and Methods
4.1. GEO Data Extraction and DEGs/DEMs/tDEGs/tDEMs Analysis
4.2. Enrichment Analysis and Key Genes Acquisition
4.3. Expression and Prognosis of GLIS1 in Pan-Cancer
4.4. Analysis of DEGs between the High and Low GLIS1 Expression Groups in PRAD Patients
4.5. Gene Set Enrichment Analysis (GSEA)
4.6. Immune Infiltration in Tumor Tissues
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- De Nunzio, C.; Lombardo, R. Best of 2022 in prostate cancer and prostatic diseases. Prostate Cancer Prostatic. Dis. 2023, 26, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Taitt, H.E. Global Trends and Prostate Cancer: A Review of Incidence, Detection, and Mortality as Influenced by Race, Ethnicity, and Geographic Location. Am. J. Men’s Health 2018, 12, 1807–1823. [Google Scholar] [CrossRef] [PubMed]
- Pishgar, F.; Ebrahimi, H.; Saeedi Moghaddam, S.; Fitzmaurice, C.; Amini, E. Global, Regional and National Burden of Prostate Cancer, 1990 to 2015: Results from the Global Burden of Disease Study 2015. J. Urol. 2018, 199, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate cancer. Lancet 2021, 398, 1075–1090. [Google Scholar] [CrossRef]
- Attard, G.; Parker, C.; Eeles, R.A.; Schroder, F.; Tomlins, S.A.; Tannock, I.; Drake, C.G.; de Bono, J.S. Prostate cancer. Lancet 2016, 387, 70–82. [Google Scholar] [CrossRef]
- Schepisi, G.; Cursano, M.C.; Casadei, C.; Menna, C.; Altavilla, A.; Lolli, C.; Cerchione, C.; Paganelli, G.; Santini, D.; Tonini, G.; et al. CAR-T cell therapy: A potential new strategy against prostate cancer. J. Immunother. Cancer 2019, 7, 258. [Google Scholar] [CrossRef]
- Riva, A.; Chokshi, S. Immune checkpoint receptors: Homeostatic regulators of immunity. Hepatol. Int. 2018, 12, 223–236. [Google Scholar] [CrossRef]
- Isaacsson Velho, P.; Antonarakis, E.S. PD-1/PD-L1 pathway inhibitors in advanced prostate cancer. Expert. Rev. Clin. Pharmacol. 2018, 11, 475–486. [Google Scholar] [CrossRef]
- Kwon, E.D.; Drake, C.G.; Scher, H.I.; Fizazi, K.; Bossi, A.; van den Eertwegh, A.J.; Krainer, M.; Houede, N.; Santos, R.; Mahammedi, H.; et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): A multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014, 15, 700–712. [Google Scholar] [CrossRef]
- Beer, T.M.; Kwon, E.D.; Drake, C.G.; Fizazi, K.; Logothetis, C.; Gravis, G.; Ganju, V.; Polikoff, J.; Saad, F.; Humanski, P.; et al. Randomized, Double-Blind, Phase III Trial of Ipilimumab versus Placebo in Asymptomatic or Minimally Symptomatic Patients With Metastatic Chemotherapy-Naive Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2017, 35, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Ward, J.F.; Pettaway, C.A.; Shi, L.Z.; Subudhi, S.K.; Vence, L.M.; Zhao, H.; Chen, J.; Chen, H.; Efstathiou, E.; et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat. Med. 2017, 23, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Taverna, G.; Giusti, G.; Seveso, M.; Hurle, R.; Colombo, P.; Stifter, S.; Grizzi, F. Mast cells as a potential prognostic marker in prostate cancer. Dis. Markers 2013, 35, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Helfand, B.T.; Carneiro, B.A.; Qin, W.; Yang, X.J.; Lee, C.; Zhang, W.; Giles, F.J.; Cristofanilli, M.; Kuzel, T.M. Efficacy Against Human Prostate Cancer by Prostate-specific Membrane Antigen-specific, Transforming Growth Factor-beta Insensitive Genetically Targeted CD8(+) T-cells Derived from Patients with Metastatic Castrate-resistant Disease. Eur. Urol. 2018, 73, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Chen, H.; Luo, W.; Zhang, H.; Li, G.; Zeng, F.; Deng, F. The Landscape of Immune Cells Infiltrating in Prostate Cancer. Front. Oncol. 2020, 10, 517637. [Google Scholar] [CrossRef]
- Jetten, A.M. GLIS1-3 transcription factors: Critical roles in the regulation of multiple physiological processes and diseases. Cell Mol. Life Sci. 2018, 75, 3473–3494. [Google Scholar] [CrossRef]
- Scoville, D.W.; Kang, H.S.; Jetten, A.M. GLIS1-3: Emerging roles in reprogramming, stem and progenitor cell differentiation and maintenance. Stem Cell Investig. 2017, 4, 80. [Google Scholar] [CrossRef]
- Maekawa, M.; Yamaguchi, K.; Nakamura, T.; Shibukawa, R.; Kodanaka, I.; Ichisaka, T.; Kawamura, Y.; Mochizuki, H.; Goshima, N.; Yamanaka, S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 2011, 474, 225–229. [Google Scholar] [CrossRef]
- Maekawa, M.; Yamanaka, S. Glis1, a unique pro-reprogramming factor, may facilitate clinical applications of iPSC technology. Cell Cycle 2011, 10, 3613–3614. [Google Scholar] [CrossRef]
- Kim, M.J.; Jung, D.; Park, J.Y.; Lee, S.M.; An, H.J. GLIS1 in Cancer-Associated Fibroblasts Regulates the Migration and Invasion of Ovarian Cancer Cells. Int. J. Mol. Sci. 2022, 23, 2218. [Google Scholar] [CrossRef]
- Shimamoto, K.; Tanimoto, K.; Fukazawa, T.; Nakamura, H.; Kanai, A.; Bono, H.; Ono, H.; Eguchi, H.; Hirohashi, N. GLIS1, a novel hypoxia-inducible transcription factor, promotes breast cancer cell motility via activation of WNT5A. Carcinogenesis 2020, 41, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Vadnais, C.; Shooshtarizadeh, P.; Rajadurai, C.V.; Lesurf, R.; Hulea, L.; Davoudi, S.; Cadieux, C.; Hallett, M.; Park, M.; Nepveu, A. Autocrine Activation of the Wnt/beta-Catenin Pathway by CUX1 and GLIS1 in Breast Cancers. Biol. Open 2014, 3, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Rong, D.; Wang, Y.; Liu, L.; Cao, H.; Huang, T.; Liu, H.; Hao, X.; Sun, G.; Sun, G.; Zheng, Z.; et al. GLIS1 intervention enhances anti-PD1 therapy for hepatocellular carcinoma by targeting SGK1-STAT3-PD1 pathway. J. Immunother. Cancer 2023, 11, e005126. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.M.; Mohsen, M.T.; Malik, M.Z.; Bagabir, S.A.; Alkhanani, M.F.; Haque, S.; Serajuddin, M.; Bharadwaj, M. Identification of Potential Key Genes in Prostate Cancer with Gene Expression, Pivotal Pathways and Regulatory Networks Analysis Using Integrated Bioinformatics Methods. Genes 2022, 13, 655. [Google Scholar] [CrossRef]
- Fu, P.; Bu, C.; Cui, B.; Li, N.; Wu, J. Screening of differentially expressed genes and identification of AMACR as a prognostic marker in prostate cancer. Andrologia 2021, 53, e14067. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, M.; Chinambedu Dandapani, M.; SimonDuraiRaj; SandhyaSundaram; VenkatRamanan, S.; Ramachandran, I.; Venkatesan, V. Differentially expressed miR-20, miR-21, miR-100, miR-125a and miR-146a as a potential biomarker for prostate cancer. Mol. Biol. Rep. 2021, 48, 3349–3356. [Google Scholar] [CrossRef] [PubMed]
- Schimmelpfennig, C.; Rade, M.; Fussel, S.; Loffler, D.; Blumert, C.; Bertram, C.; Borkowetz, A.; Otto, D.J.; Puppel, S.H.; Honscheid, P.; et al. Characterization and evaluation of gene fusions as a measure of genetic instability and disease prognosis in prostate cancer. BMC Cancer 2023, 23, 575. [Google Scholar] [CrossRef] [PubMed]
- Meller, S.; Meyer, H.A.; Bethan, B.; Dietrich, D.; Maldonado, S.G.; Lein, M.; Montani, M.; Reszka, R.; Schatz, P.; Peter, E.; et al. Integration of tissue metabolomics, transcriptomics and immunohistochemistry reveals ERG- and gleason score-specific metabolomic alterations in prostate cancer. Oncotarget 2016, 7, 1421–1438. [Google Scholar] [CrossRef]
- Ding, Y.; Wu, H.; Warden, C.; Steele, L.; Liu, X.; Iterson, M.V.; Wu, X.; Nelson, R.; Liu, Z.; Yuan, Y.C.; et al. Gene Expression Differences in Prostate Cancers between Young and Old Men. PLoS Genet. 2016, 12, e1006477. [Google Scholar] [CrossRef]
- Lefort, K.; Ostano, P.; Mello-Grand, M.; Calpini, V.; Scatolini, M.; Farsetti, A.; Dotto, G.P.; Chiorino, G. Dual tumor suppressing and promoting function of Notch1 signaling in human prostate cancer. Oncotarget 2016, 7, 48011–48026. [Google Scholar] [CrossRef][Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Lei, Y.; Li, J.K.; Du, W.X.; Li, R.G.; Yang, J.; Li, J.; Li, F.; Tan, H.B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020, 470, 126–133. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Andre, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Blery, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e13. [Google Scholar] [CrossRef] [PubMed]
- Abel, A.M.; Yang, C.; Thakar, M.S.; Malarkannan, S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018, 9, 1869. [Google Scholar] [CrossRef] [PubMed]
- Morvan, M.G.; Lanier, L.L. NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer 2016, 16, 7–19. [Google Scholar] [CrossRef]
- Gardner, A.; Ruffell, B. Dendritic Cells and Cancer Immunity. Trends Immunol. 2016, 37, 855–865. [Google Scholar] [CrossRef]
- Liang, P.; Henning, S.M.; Guan, J.; Grogan, T.; Elashoff, D.; Cohen, P.; Aronson, W.J. Effect of dietary omega-3 fatty acids on castrate-resistant prostate cancer and tumor-associated macrophages. Prostate Cancer Prostatic Dis. 2020, 23, 127–135. [Google Scholar] [CrossRef]
- Keeley, T.; Costanzo-Garvey, D.L.; Cook, L.M. Unmasking the Many Faces of Tumor-Associated Neutrophils and Macrophages: Considerations for Targeting Innate Immune Cells in Cancer. Trends Cancer 2019, 5, 789–798. [Google Scholar] [CrossRef]
- Morikawa, K.; Zhang, J.; Nonaka, M.; Morikawa, S. Modulatory effect of macrolide antibiotics on the Th1- and Th2-type cytokine production. Int. J. Antimicrob. Agents 2002, 19, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Ramamoorthi, G.; Albert, G.; Gallen, C.; Beyer, A.; Snyder, C.; Koski, G.; Disis, M.L.; Czerniecki, B.J.; Kodumudi, K. Differentiation and Regulation of T(H) Cells: A Balancing Act for Cancer Immunotherapy. Front. Immunol. 2021, 12, 669474. [Google Scholar] [CrossRef] [PubMed]
- De Monte, L.; Reni, M.; Tassi, E.; Clavenna, D.; Papa, I.; Recalde, H.; Braga, M.; Di Carlo, V.; Doglioni, C.; Protti, M.P. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med. 2011, 208, 469–478. [Google Scholar] [CrossRef]
- Ziegler, A.; Heidenreich, R.; Braumuller, H.; Wolburg, H.; Weidemann, S.; Mocikat, R.; Rocken, M. EpCAM, a human tumor-associated antigen promotes Th2 development and tumor immune evasion. Blood 2009, 113, 3494–3502. [Google Scholar] [CrossRef] [PubMed]
- Fong, A.M.; Robinson, L.A.; Steeber, D.A.; Tedder, T.F.; Yoshie, O.; Imai, T.; Patel, D.D. Fractalkine and CX3CR1 mediate a novel mechanism of leukocyte capture, firm adhesion, and activation under physiologic flow. J. Exp. Med. 1998, 188, 1413–1419. [Google Scholar] [CrossRef] [PubMed]
- Aldinucci, D.; Borghese, C.; Casagrande, N. The CCL5/CCR5 Axis in Cancer Progression. Cancers 2020, 12, 1765. [Google Scholar] [CrossRef] [PubMed]
- Sawant, K.V.; Poluri, K.M.; Dutta, A.K.; Sepuru, K.M.; Troshkina, A.; Garofalo, R.P.; Rajarathnam, K. Chemokine CXCL1 mediated neutrophil recruitment: Role of glycosaminoglycan interactions. Sci. Rep. 2016, 6, 33123. [Google Scholar] [CrossRef]
- Bernardini, G.; Sciume, G.; Bosisio, D.; Morrone, S.; Sozzani, S.; Santoni, A. CCL3 and CXCL12 regulate trafficking of mouse bone marrow NK cell subsets. Blood 2008, 111, 3626–3634. [Google Scholar] [CrossRef]
- Ziegler, E.; Oberbarnscheidt, M.; Bulfone-Paus, S.; Forster, R.; Kunzendorf, U.; Krautwald, S. CCR7 signaling inhibits T cell proliferation. J. Immunol. 2007, 179, 6485–6493. [Google Scholar] [CrossRef]
- Bottcher, J.P.; Bonavita, E.; Chakravarty, P.; Blees, H.; Cabeza-Cabrerizo, M.; Sammicheli, S.; Rogers, N.C.; Sahai, E.; Zelenay, S.; Reis e Sousa, C. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 2018, 172, 1022–1037.e14. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]

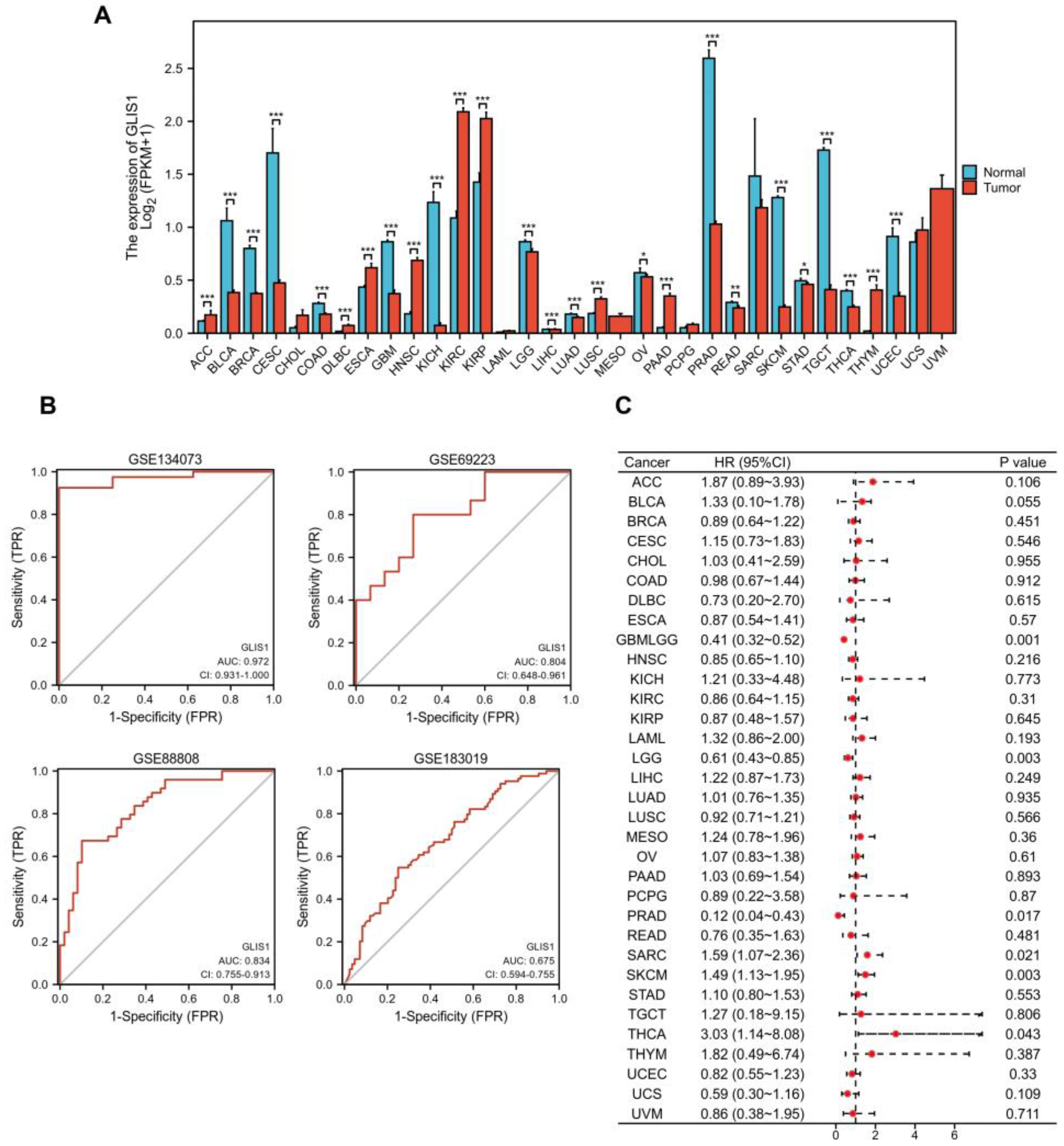
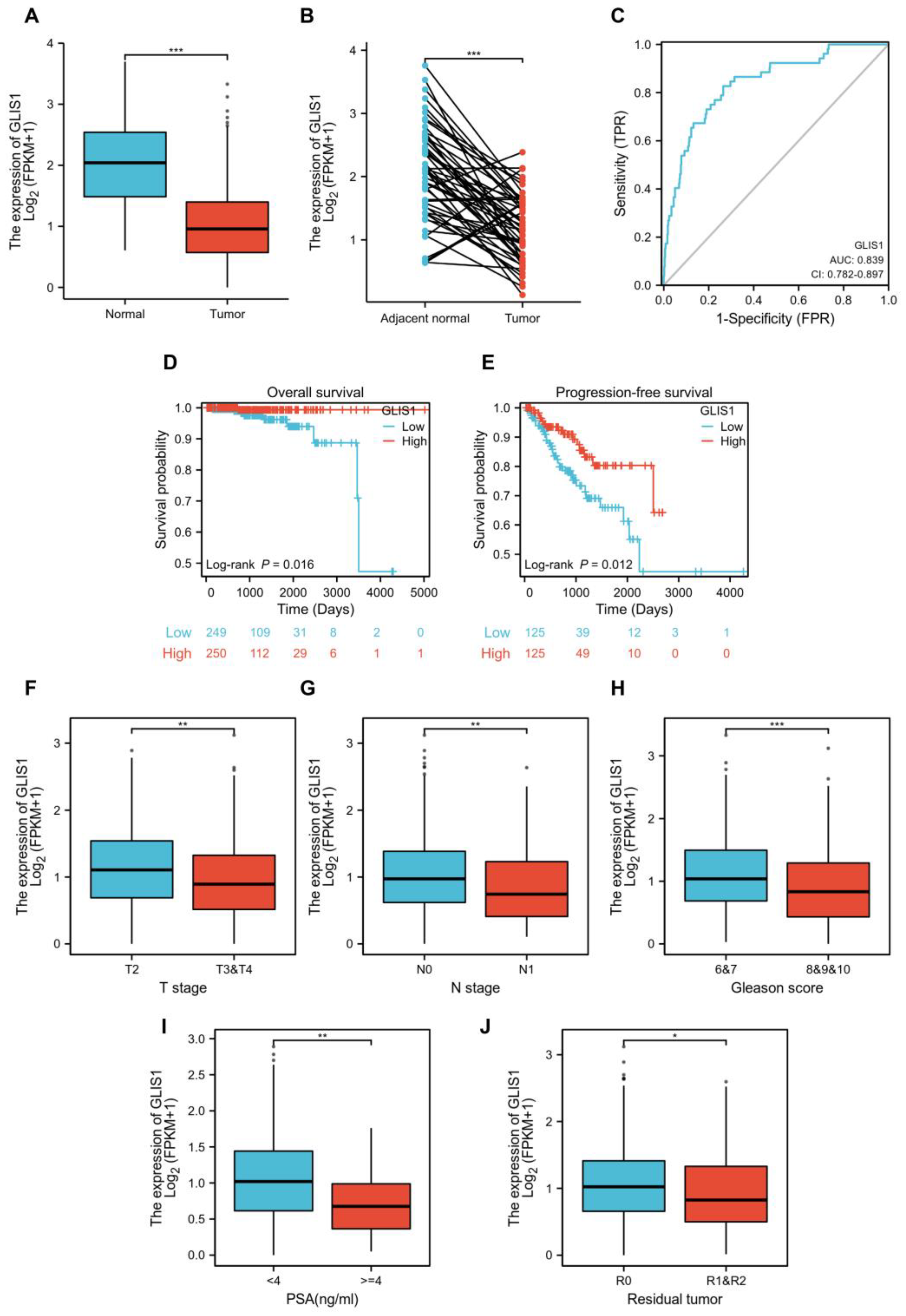


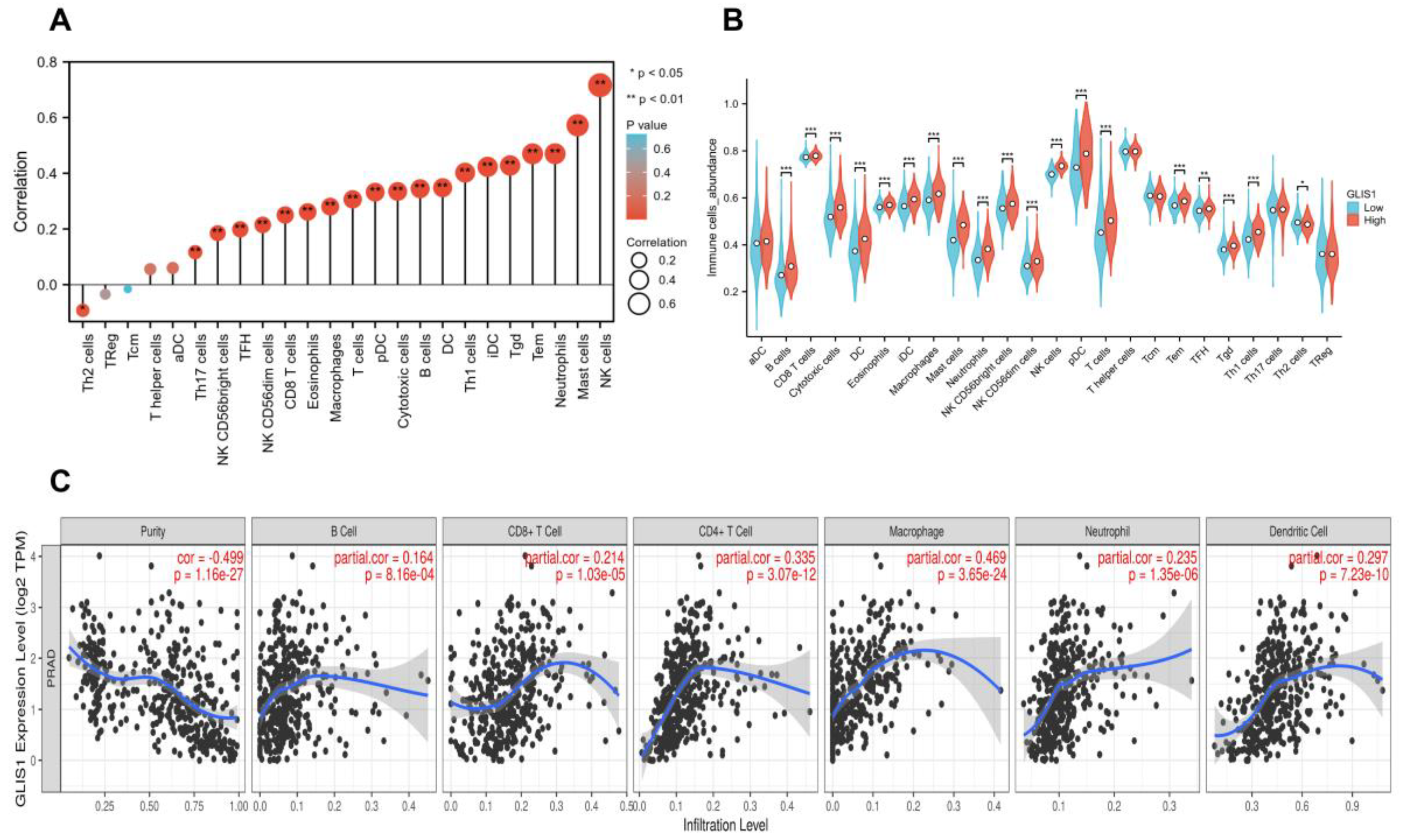
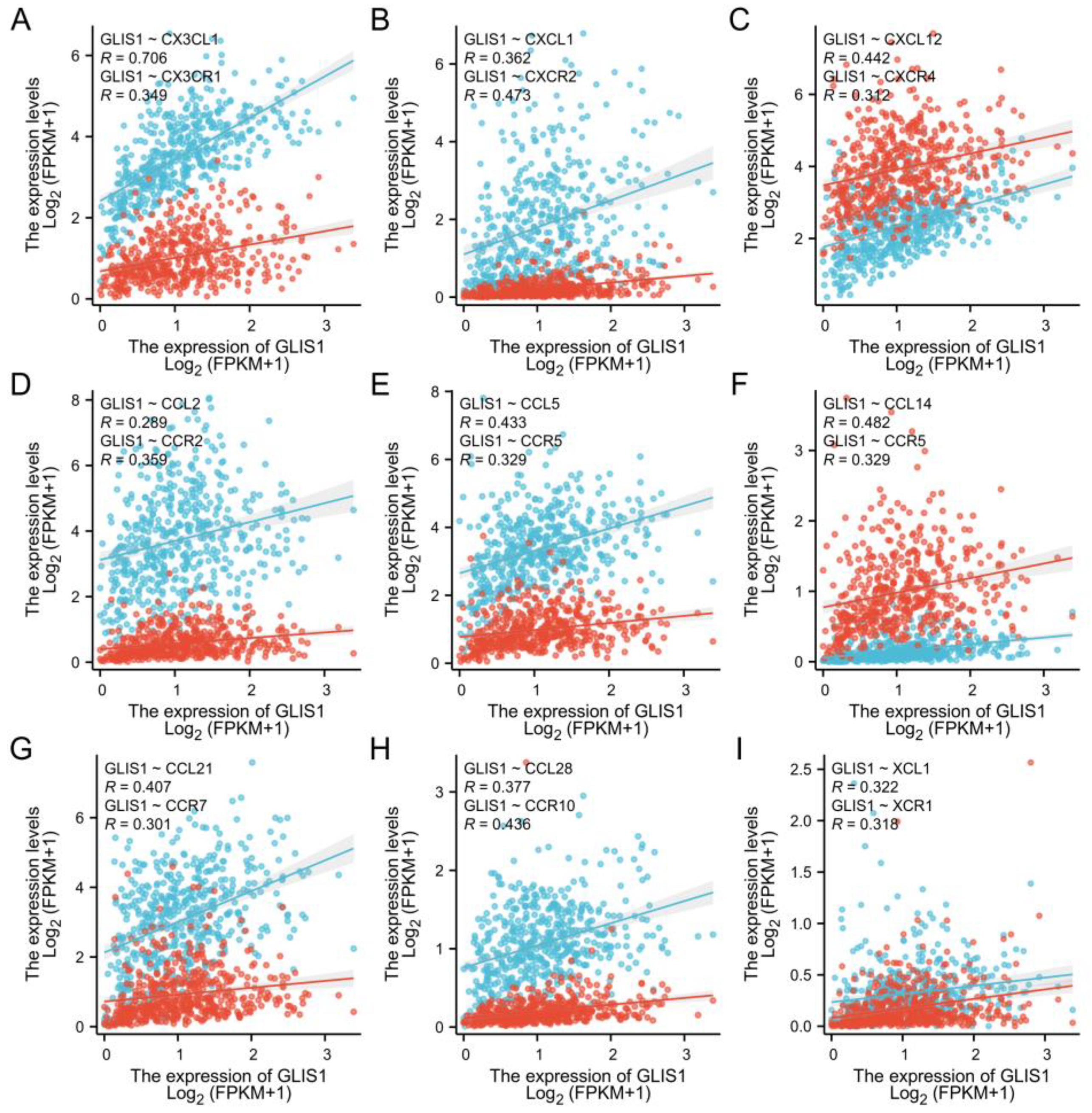
| Gene | Datasets | LogFC | p Value | adj. p Value | Expression |
|---|---|---|---|---|---|
| GLIS1 | GSE183019 | −0.63 | 0.000 | 0.012 | DOWN |
| GSE134073 | −2.09 | 0.000 | 0.000 | DOWN | |
| GSE69223 | −0.94 | 0.002 | 0.017 | DOWN | |
| GSE88808 | −0.77 | 0.000 | 0.000 | DOWN |
| Target Gene | miRNA | Datasets | LogFC | p Value | adj. p Value | Expression |
|---|---|---|---|---|---|---|
| GLIS1 | hsa-miR-663b | GSE60117 | 0.28 | 0.000 | 0.000 | UP |
| GSE89193 | 1.36 | 0.000 | 0.000 | UP | ||
| GLIS1 | hsa-miR-153 | GSE60117 | 0.35 | 0.003 | 0.015 | UP |
| GSE89193 | 2.22 | 0.000 | 0.000 | UP | ||
| GLIS1 | hsa-miR-483-5p | GSE60117 | −0.62 | -0.010 | 0.039 | DOWN |
| GSE89193 | −1.10 | 0.001 | 0.006 | DOWN |
| Clinicopathologic Variables | No. of Cases | GLIS1 Expression Level | χ | p | |
|---|---|---|---|---|---|
| Low | High | ||||
| All cases | 499 | 249(44.3%) | 250(55.7%) | ||
| Age | 0.019 | 0.889 | |||
| ≤60 | 224 (44.9%) | 111 (22.2%) | 113 (22.6%) | ||
| >60 | 275 (55.1%) | 138 (27.7%) | 137 (27.5%) | ||
| Clinical T stage | 0.623 | 0.430 | |||
| T1 + T2 | 351 (86.5%) | 165 (40.6%) | 186 (45.8%) | ||
| T3 + T4 | 55 (13.5%) | 29 (7.1%) | 26 (6.4%) | ||
| Pathologic T stage | 8.256 | 0.004 | |||
| T2 | 189 (38.4%) | 79 (16.1%) | 110 (22.4%) | ||
| T3 + T4 | 303 (61.6%) | 167 (33.9%) | 136 (27.6%) | ||
| Lymph nodes status | 6.477 | 0.011 | |||
| Negative | 347 (81.5%) | 169 (39.7%) | 178 (41.8%) | ||
| Positive | 79 (18.5%) | 51 (12.0%) | 28 (6.6%) | ||
| PSA (ng/mL) | 7.179 | 0.007 | |||
| <4 | 415 (93.9%) | 197 (44.6%) | 218 (49.3%) | ||
| ≥4 | 27 (6.1%) | 20 (4.5%) | 7 (1.6%) | ||
| Gleason score | 6.674 | 0.010 | |||
| 6–7 | 293 (58.7%) | 132 (26.5%) | 161 (32.3%) | ||
| 8–10 | 206 (41.3%) | 117 (23.4%) | 89 (17.8%) | ||
| Survival Status | 6.564 | 0.010 | |||
| Live | 489 (98.0%) | 240 (48.1%) | 249 (49.9%) | ||
| Dead | 10 (2.0%) | 9 (1.8%) | 1 (0.2%) | ||
| Characteristics | Total (N) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| Hazard Ratio (95% CI) | p Value | Hazard Ratio (95% CI) | p Value | ||
| Age | 499 | ||||
| ≤60 | 224 | Reference | |||
| >60 | 275 | 1.577 (0.440–5.648) | 0.484 | ||
| T stage | 492 | ||||
| T2 | 189 | Reference | |||
| T3 & T4 | 303 | 3.294 (0.612–17.727) | 0.165 | ||
| N stage | 426 | ||||
| N0 | 347 | Reference | |||
| N1 | 79 | 3.516 (0.778–15.896) | 0.102 | ||
| PSA (ng/mL) | 442 | ||||
| <4 | 415 | Reference | |||
| ≥4 | 27 | 10.479 (2.471–44.437) | 0.001 | 3.411 (0.735–15.826) | 0.117 |
| Gleason score | 499 | ||||
| 6 & 7 | 293 | Reference | |||
| 8 & 9 & 10 | 206 | 6.664 (1.373–32.340) | 0.019 | 5.372 (0.843–34.209) | 0.075 |
| Residual tumor | 468 | ||||
| R0 | 315 | Reference | |||
| R1 & R2 | 153 | 2.598 (0.696–9.694) | 0.155 | ||
| GLIS1 | 499 | ||||
| Low | 249 | Reference | |||
| High | 250 | 0.122 (0.015–0.965) | 0.046 | 0.157 (0.018–1.355) | 0.092 |
| Characteristics | Total (N) | Odds Ratio (OR) | p Value |
|---|---|---|---|
| Age (>60 vs. ≤60) | 499 | 1.007 (0.708–1.434) | 0.968 |
| T stage (T3 & T4 vs. T2) | 492 | 0.585 (0.404–0.843) | 0.004 |
| N stage (N1 vs. N0) | 426 | 0.521 (0.311–0.859) | 0.012 |
| PSA (ng/mL) (≥4 vs. <4) | 442 | 0.316 (0.122–0.731) | 0.011 |
| Gleason score (8 & 9 & 10 vs. 6 & 7) | 499 | 0.645 (0.450–0.922) | 0.017 |
| Race (Black or African American & White vs. Asian) | 484 | 3.130 (0.921–14.237) | 0.090 |
| Residual tumor (R1 & R2 vs. R0) | 468 | 0.671 (0.454–0.990) | 0.045 |
| Gene Markers | PRAD | ||||
|---|---|---|---|---|---|
| None | Purity | ||||
| Correlation | p | Correlation | p | ||
| CD8+ T cell | CD8A | 0.423 | 4.54 × 10−23 | 0.238 | 9.38 × 10−7 |
| CD8B | 0.275 | 4.27 × 10−10 | 0.158 | 1.24 × 10−3 | |
| T cell (general) | CD3D | 0.329 | 4.71 × 10−14 | 0.115 | 1.91 × 10−2 |
| CD3E | 0.405 | 4.8 × 10−21 | 0.203 | 2.98 × 10−5 | |
| CD2 | 0.366 | 2.94 × 10−17 | 0.181 | 2.15 × 10−4 | |
| B cell | CD19 | 0.247 | 2.42 × 10−8 | 0.11 | 2.45 × 10−2 |
| CD79A | 0.221 | 6.67 × 10−7 | 0.077 | 1.18 × 10−1 | |
| Monocyte | CD86 | 0.284 | 1.00 × 10−10 | 0.131 | 7.67 × 10−3 |
| CD115 (CSF1R) | 0.481 | 3.08 × 10−30 | 0.344 | 5.46 × 10−13 | |
| TAM | CCL2 | 0.302 | 5.37 × 10−12 | 0.149 | 2.38 × 10−3 |
| CD68 | −0.311 | 7.92 × 10−11 | 0.08 | 1.04 × 10−1 | |
| IL10 | 0.29 | 4.42 × 10−11 | 0.188 | 1.17 × 10−4 | |
| M1 macrophage | INOS (NOS2) | 0.29 | 4.42 × 10−11 | 0.188 | 1.17 × 10−4 |
| IRF5 | 0.133 | 2.95 × 10−3 | 0.064 | 1.91 × 10−1 | |
| COX2 (PTGS2) | 0.353 | 4.81 × 10−16 | 0.254 | 1.46 × 10−7 | |
| M2 macrophage | CD163 | 0.185 | 3.15 × 10−5 | 0.061 | 2.15 × 10−1 |
| VSIG4 | 0.29 | 4.12 × 10−11 | 0.159 | 1.18 × 10−3 | |
| MS4A4A | 0.224 | 4.57 × 10−7 | 0.082 | 9.54 × 10−2 | |
| Neutrophils | CD66b (CEACAM8) | 0.038 | 3.99 × 10−1 | 0.007 | 8.79 × 10−1 |
| CD11B (ITGAM) | 0.387 | 3.08 × 10−19 | 0.241 | 6.59 × 10−7 | |
| CCR7 | 0.306 | 2.97 × 10−12 | 0.089 | 7.11 × 10−2 | |
| Natural killer cell | KIR2DL1 | 0.046 | 3.02 × 10−1 | −0.048 | 3.25 × 10−1 |
| KIR2DL3 | 0.118 | 8.44 × 10−3 | 0.115 | 1.89 × 10−2 | |
| KIR2DL4 | 0.116 | 9.66 × 10−3 | 0.007 | 8.82 × 10−1 | |
| KIR3DL1 | 0.101 | 2.44 × 10−2 | −0.006 | 8.96 × 10−1 | |
| KIR3DL2 | 0.094 | 3.54 × 10−2 | 0.055 | 2.65 × 10−1 | |
| KIR3DL3 | 0.004 | 9.35 × 10−1 | 0.063 | 1.99 × 10−1 | |
| KIR2DS4 | 0.148 | 9.07 × 10−4 | 0.092 | 5.98 × 10−2 | |
| Dendritic cell | HLA-DPB1 | 0.49 | 1.75 × 10−31 | 0.326 | 9.84 × 10−12 |
| HLA-DQB1 | 0.264 | 2.08 × 10−9 | 0.132 | 6.87 × 10−3 | |
| HLA-DRA | 0.351 | 7.66 × 10−16 | 0.161 | 1.00 × 10−3 | |
| HLA-DPA1 | 0.423 | 4.35 × 10−23 | 0.271 | 1.93 × 10−8 | |
| BDCA-1 (CD1C) | 0.442 | 3.4 × 10−25 | 0.268 | 2.70 × 10−8 | |
| BDCA-4 (NRP1) | −0.004 | 9.27 × 10−1 | −0.023 | 6.44 × 10−1 | |
| CD11c (ITGAX) | 0.244 | 3.67 × 10−8 | 0.087 | 7.61 × 10−2 | |
| Th1 | T-bet (TBX21) | 0.332 | 2.9 × 10−14 | 0.154 | 1.63 × 10−3 |
| STAT4 | 0.354 | 3.94 × 10−16 | 0.178 | 2.56 × 10−4 | |
| STAT1 | 0.087 | 5.35 × 10−2 | −0.01 | 8.43 × 10−1 | |
| IFN-γ (IFNG) | 0.091 | 4.19 × 10−2 | −0.002 | 9.75 × 10−1 | |
| TNF-α (TNF) | 0.214 | 1.52 × 10−6 | 0.096 | 4.92 × 10−2 | |
| Th2 | GATA3 | 0.643 | 1.62 × 10−59 | 0.545 | 1.33 × 10−33 |
| STAT6 | 0.437 | 1.13 × 10−24 | 0.356 | 7.61 × 10−14 | |
| STAT5A | 0.622 | 1.33 × 10−54 | 0.499 | 1.30 × 10−27 | |
| IL13 | 0.058 | 1.95 × 10−1 | 0.014 | 7.79 × 10−1 | |
| Tfh | BCL6 | 0.288 | 6.22 × 10−11 | 0.146 | 2.90 × 10−3 |
| IL21 | 0.053 | 2.39 × 10−1 | 0.008 | 8.73 × 10−1 | |
| Th17 | STAT3 | 0.255 | 7.41 × 10−9 | 0.148 | 2.53 × 10−3 |
| IL17A | 0.159 | 3.56 × 10−4 | 0.023 | 6.47 × 10−1 | |
| Treg | FOXP3 | 0.188 | 2.41 × 10−5 | 0.075 | 1.27 × 10−1 |
| CCR8 | 0.125 | 5.06 × 10−3 | 0.025 | 6.11 × 10−1 | |
| STAT5B | 0.5 | 6.45 × 10−33 | 0.432 | 2.33 × 10−20 | |
| TGF-β (TGFB1) | 0.532 | 1.08 × 10−37 | 0.412 | 1.94 × 10−18 | |
| T cell exhaustion | PD-1 (PDCD1) | 0.317 | 4.27 × 10−13 | 0.128 | 8.92 × 10−3 |
| CTLA4 | 0.146 | 1.08 × 10−3 | −0.025 | 6.17 × 10−1 | |
| TIM-3 (HAVCR2) | 0.275 | 4.25 × 10−10 | 0.122 | 1.30 × 10−2 | |
| GZMB | 0.222 | 5.57 × 10−7 | 0.023 | 6.37 × 10−1 | |
| LAG3 | 0.466 | 3.62 × 10−28 | 0.346 | 3.67 × 10−13 | |
| PDL1 (CD274) | 0.286 | 7.63 × 10−11 | 0.158 | 1.23× 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Q.; Xie, T.; Wang, Y.; Ho, V.W.-S.; Teoh, J.Y.-C.; Chiu, P.K.-F.; Ng, C.-F. GLIS1, Correlated with Immune Infiltrates, Is a Potential Prognostic Biomarker in Prostate Cancer. Int. J. Mol. Sci. 2024, 25, 489. https://doi.org/10.3390/ijms25010489
Peng Q, Xie T, Wang Y, Ho VW-S, Teoh JY-C, Chiu PK-F, Ng C-F. GLIS1, Correlated with Immune Infiltrates, Is a Potential Prognostic Biomarker in Prostate Cancer. International Journal of Molecular Sciences. 2024; 25(1):489. https://doi.org/10.3390/ijms25010489
Chicago/Turabian StylePeng, Qiang, Tingting Xie, Yuliang Wang, Vincy Wing-Sze Ho, Jeremy Yuen-Chun Teoh, Peter Ka-Fung Chiu, and Chi-Fai Ng. 2024. "GLIS1, Correlated with Immune Infiltrates, Is a Potential Prognostic Biomarker in Prostate Cancer" International Journal of Molecular Sciences 25, no. 1: 489. https://doi.org/10.3390/ijms25010489
APA StylePeng, Q., Xie, T., Wang, Y., Ho, V. W.-S., Teoh, J. Y.-C., Chiu, P. K.-F., & Ng, C.-F. (2024). GLIS1, Correlated with Immune Infiltrates, Is a Potential Prognostic Biomarker in Prostate Cancer. International Journal of Molecular Sciences, 25(1), 489. https://doi.org/10.3390/ijms25010489







