Technology Invention and Mechanism Analysis of Rapid Rooting of Taxus × media Rehder Branches Induced by Agrobacterium rhizogenes
Abstract
:1. Introduction
2. Results
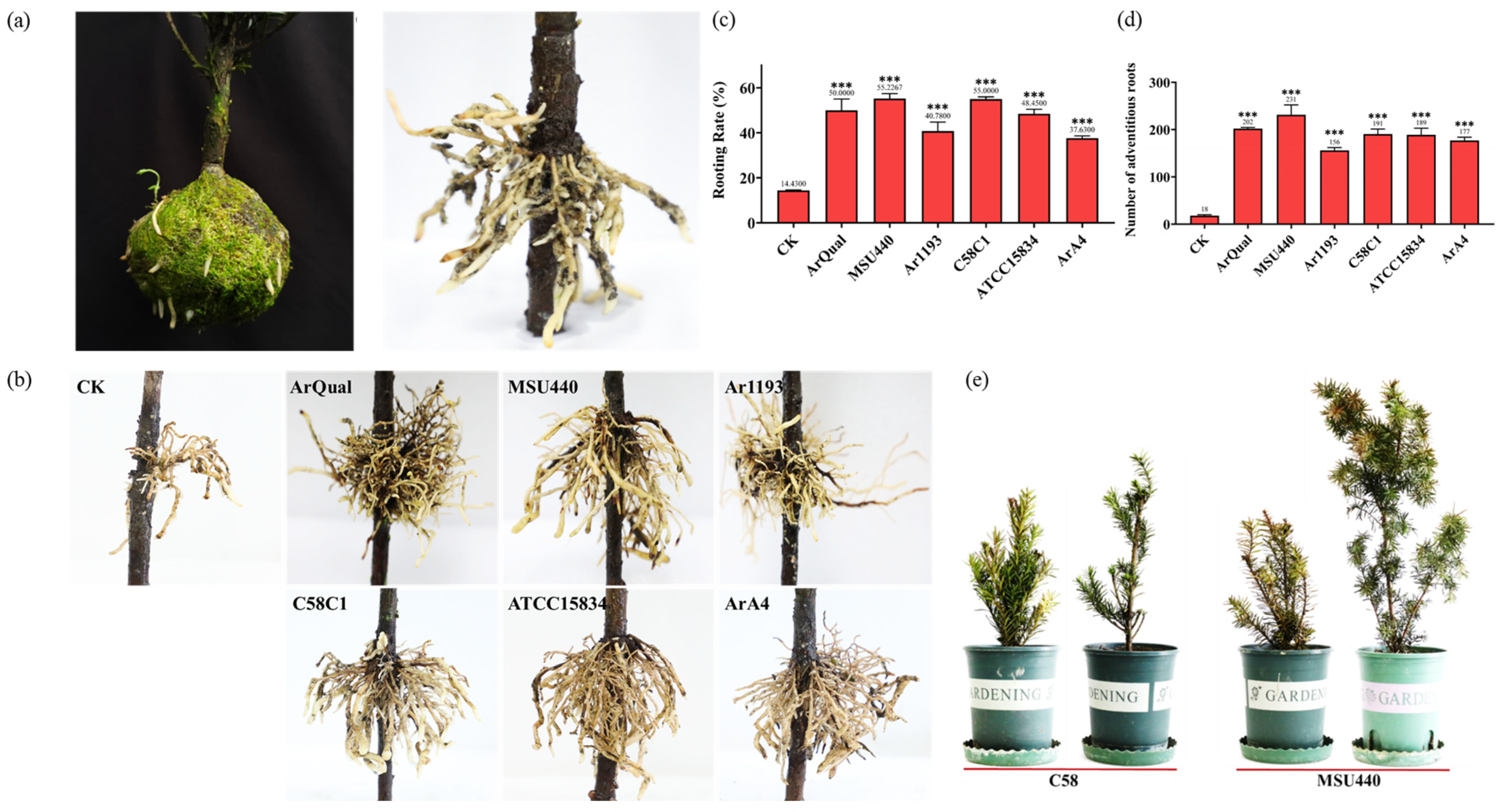
2.1. High-Efficiency Induction of T. media Branch Rooting Using A. rhizogenes
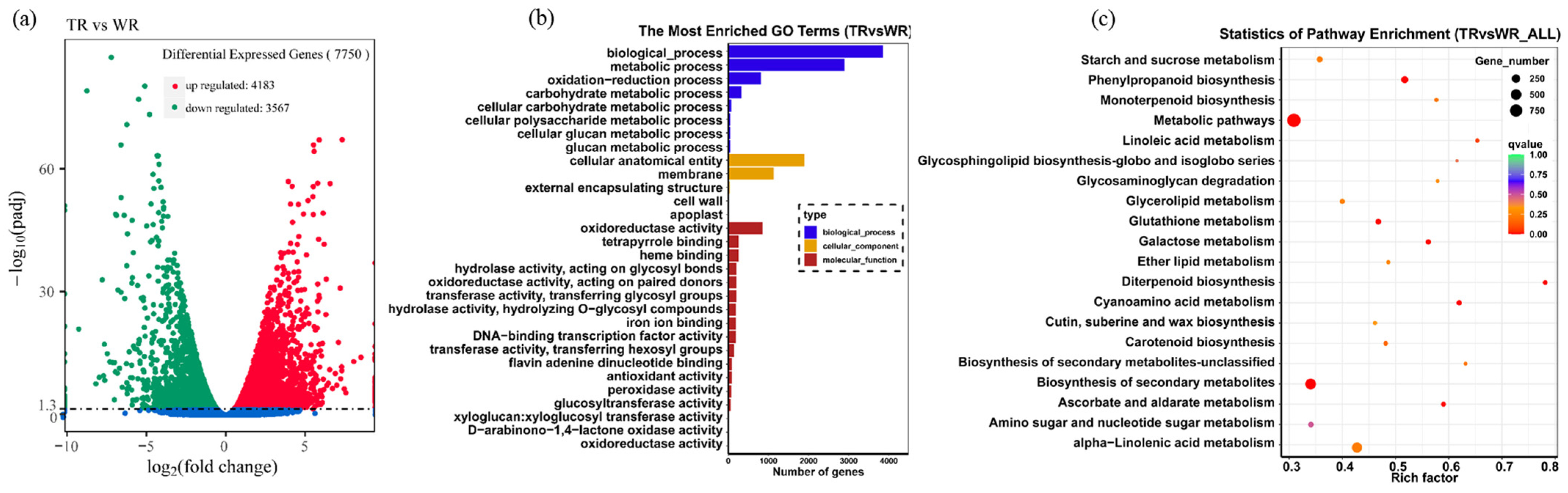
2.2. Analysis of the Gene Expression Profile in TR
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis of DEGs
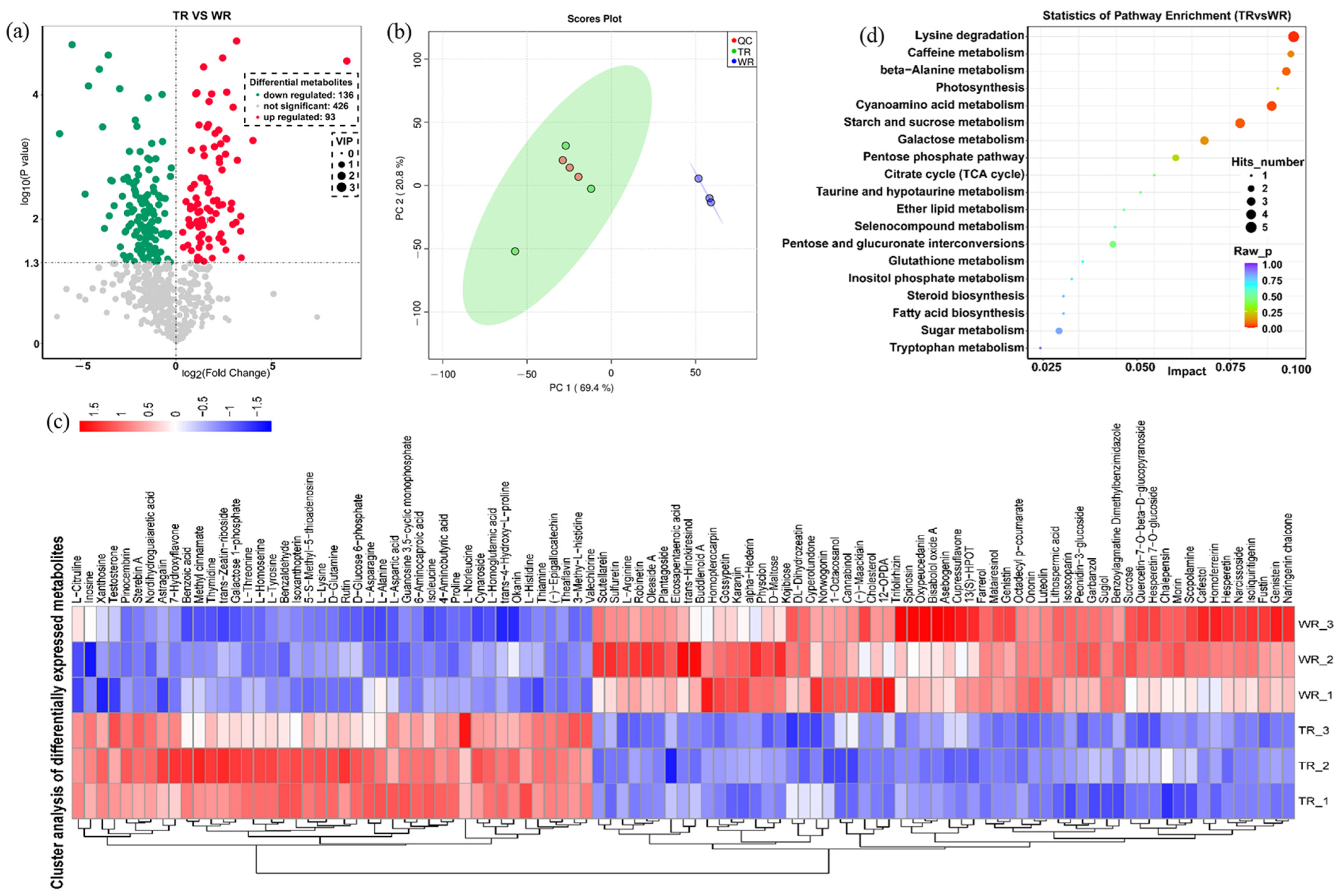
2.4. Analysis of Differentially Expressed Metabolites in TR
2.5. Combined Analysis of Transcriptome and Metabolome (DEGs/DEMs) in TR vs. WR
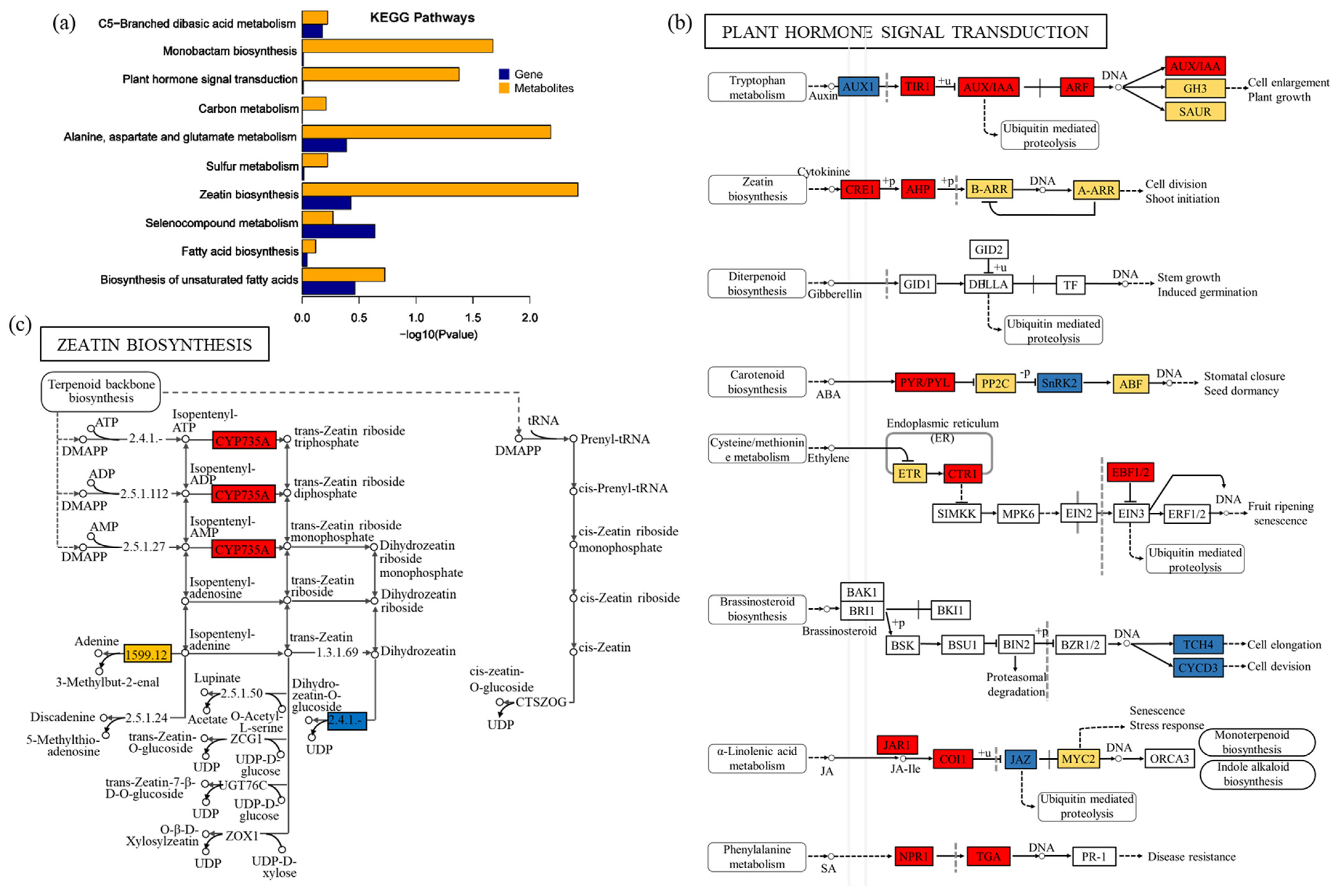
2.6. DEGs/DEMs Were Involved in Plant Hormone Signal Transduction
2.7. DEGs/DEMs Were Involved in Amino Acid Metabolism
2.8. DEGs/DEMs Were Involved in Zeatin Biosynthesis
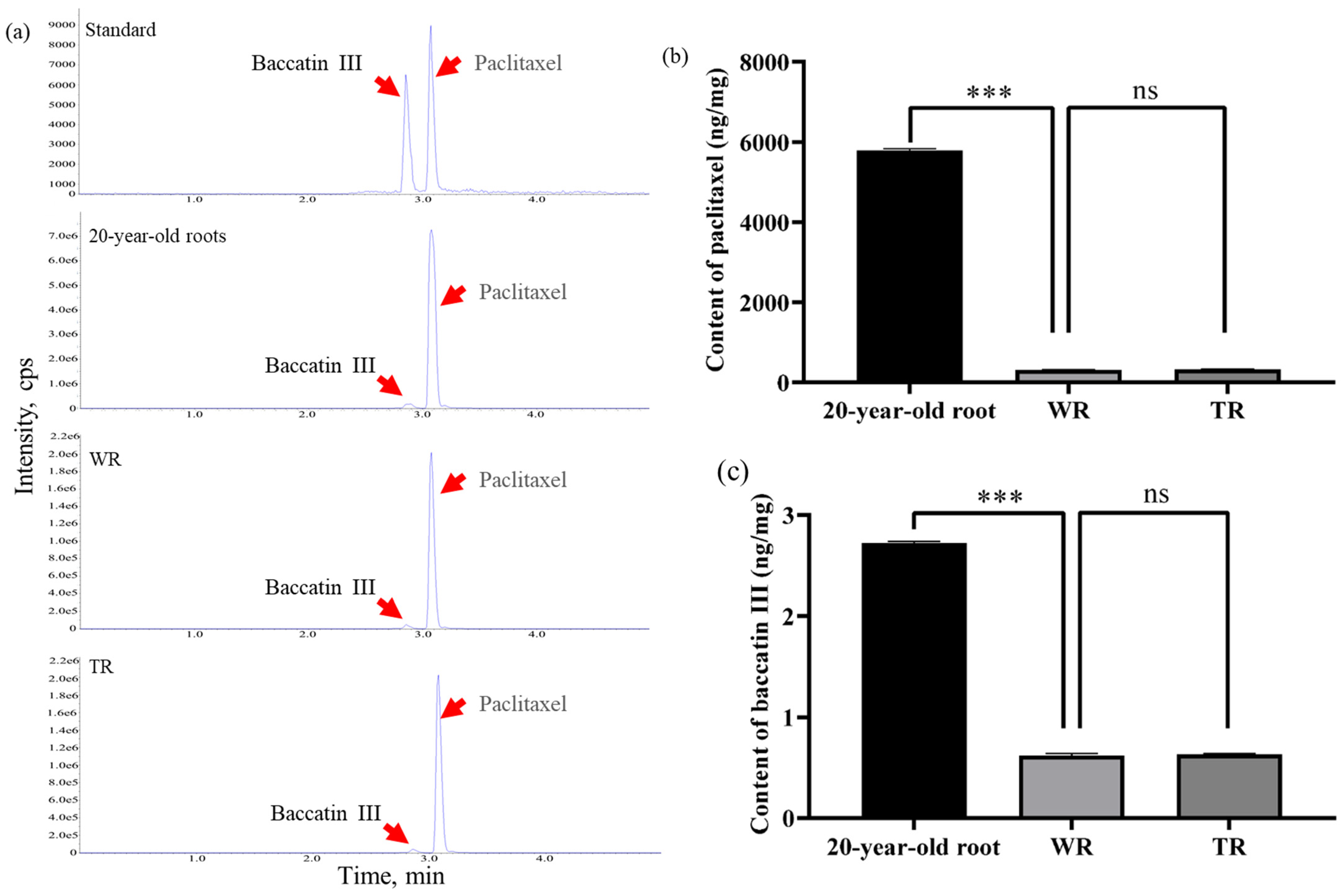
2.9. The Paclitaxel and Baccatin III Were Synthesized in TR and WR
3. Discussion
4. Materials and Methods
4.1. Plant and Substrate Materials
4.2. A. rhizogenes Strains
4.3. A. rhizogenes-Mediated Branch Rooting Transformation System
4.4. Sample Preparation for Multi-Omics Analysis
4.5. Metabolites Extraction
4.6. UHPLC-MS-MS Analysis
4.7. Metabolome Analysis
4.8. RNA-Seq
4.9. LC-MS-Targeted Quantitative Detection of Paclitaxel and Baccatin III
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, L.; Chen, L. Progress in research on paclitaxel and tumor immunotherapy. Cell. Mol. Biol. Lett. 2019, 24, 40. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Garg, M. A review of traditional use, phytoconstituents and biological activities of Himalayan yew, Taxus wallichiana. J. Integr. Med. 2015, 13, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-H.; Mao, J.-W.; Tan, X.-L. Research progress on the source, production, and anti-cancer mechanisms of paclitaxel. Chin. J. Nat. Med. 2020, 18, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Bahramnejad, B.; Naji, M.; Bose, R.; Jha, S. A critical review on use of Agrobacterium rhizogenes and their associated binary vectors for plant transformation. Biotechnol. Adv. 2019, 37, 107405. [Google Scholar] [CrossRef] [PubMed]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Less, H.; Angelovici, R.; Tzin, V.; Galili, G. Principal transcriptional regulation and genome-wide system interactions of the Asp-family and aromatic amino acid networks of amino acid metabolism in plants. Amino Acids. 2010, 39, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Frebort, I.; Kowalska, M.; Hluska, T.; Frebortova, J.; Galuszka, P. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Y.; Wang, P.; Wang, C.; Wang, J.; Wang, X.; Cheng, H. Highly efficient Agrobacterium rhizogenes-mediated hairy root transformation for gene editing analysis in cotton. Front. Plant Sci. 2022, 13, 1059404. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wang, D.; Fu, J.; Zhang, Z.; Qin, Y.; Hu, G.; Zhao, J. Agrobacterium rhizogenes-mediated hairy root transformation as an efficient system for gene function analysis in Litchi chinensis. Plant Methods 2021, 17, 103. [Google Scholar] [CrossRef] [PubMed]
- Amani, S.; Mohebodini, M.; Khademvatan, S.; Jafari, M. Agrobacterium rhizogenes mediated transformation of Ficus carica L. for the efficient production of secondary metabolites. J. Sci. Food Agric. 2020, 100, 2185–2197. [Google Scholar] [CrossRef] [PubMed]
- Leppyanen, I.V.; Kirienko, A.N.; Dolgikh, E.A. Agrobacterium rhizogenes—Mediated transformation of Pisum sativum L. roots as a tool for studying the mycorrhizal and root nodule symbioses. PeerJ 2019, 7, e6552. [Google Scholar] [CrossRef] [PubMed]
- Ketchum, R.E.B.; Wherland, L.; Croteau, R.B. Stable transformation and long-term maintenance of transgenic Taxus cell suspension cultures. Plant Cell Rep. 2007, 26, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Sykłowska-Baranek, K.; Sygitowicz, G.; Maciejak-Jastrzębska, A.; Pietrosiuk, A.; Szakiel, A. Application of Priming Strategy for Enhanced Paclitaxel Biosynthesis in Taxus × Media Hairy Root Cultures. Cells 2022, 11, 2062. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, M.I.; Atanas I., P.; Thomas, B. Hairy root type plant in vitro systems as sources of bioactive substances. Appl. Microbiol. Biotechnol. 2007, 74, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Srivastava, A.K. Hairy Root Culture for Mass-Production of High-Value Secondary Metabolites. Crit. Rev. Biotechnol. 2007, 27, 29–43. [Google Scholar] [CrossRef] [PubMed]
- NY 1428-2007; Water-Soluble Fertilizers Containing Micronutrients. Ministry of Agriculture and Rural Affairs of the People’s Republic of China: Beijing, China, 2007.
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. TrAC Trends Anal. Chem. 2008, 27, 251–260. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Gou, J.; Liao, Q.; Li, Y.; Zhou, Q.; Bi, G.; Li, C.; Du, R.; Wang, X.; Sun, T.; et al. The Taxus genome provides insights into paclitaxel biosynthesis. Nat. Plants 2021, 7, 1026–1036. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Luo, X.; Su, H.; Guan, G.; Liu, S.; Ren, M. Technology Invention and Mechanism Analysis of Rapid Rooting of Taxus × media Rehder Branches Induced by Agrobacterium rhizogenes. Int. J. Mol. Sci. 2024, 25, 375. https://doi.org/10.3390/ijms25010375
Wang Y, Luo X, Su H, Guan G, Liu S, Ren M. Technology Invention and Mechanism Analysis of Rapid Rooting of Taxus × media Rehder Branches Induced by Agrobacterium rhizogenes. International Journal of Molecular Sciences. 2024; 25(1):375. https://doi.org/10.3390/ijms25010375
Chicago/Turabian StyleWang, Ying, Xiumei Luo, Haotian Su, Ge Guan, Shuang Liu, and Maozhi Ren. 2024. "Technology Invention and Mechanism Analysis of Rapid Rooting of Taxus × media Rehder Branches Induced by Agrobacterium rhizogenes" International Journal of Molecular Sciences 25, no. 1: 375. https://doi.org/10.3390/ijms25010375
APA StyleWang, Y., Luo, X., Su, H., Guan, G., Liu, S., & Ren, M. (2024). Technology Invention and Mechanism Analysis of Rapid Rooting of Taxus × media Rehder Branches Induced by Agrobacterium rhizogenes. International Journal of Molecular Sciences, 25(1), 375. https://doi.org/10.3390/ijms25010375





