IPSC-Derived Astrocytes Contribute to In Vitro Modeling of Parkinson’s Disease Caused by the GBA1 N370S Mutation
Abstract
:1. Introduction
2. Results
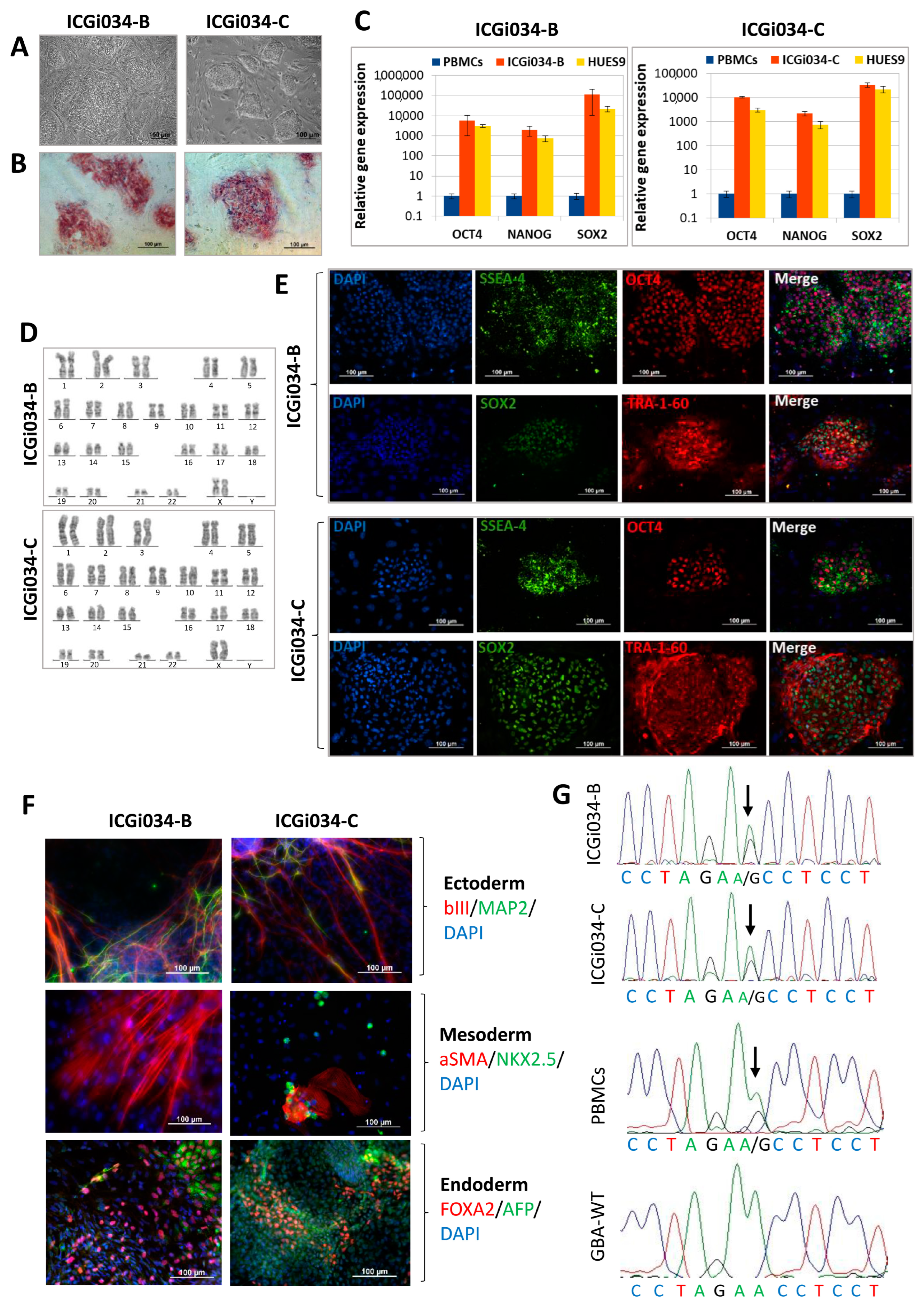
2.1. ICGi034-B and ICGi034-C Cell Lines Have a Typical Morphology for Induced Pluripotent Stem Cells (iPSCs) and Are AP-Positive
2.2. Lines ICGi034-B and ICGi034-C Demonstrate Expression of Pluripotency Markers
2.3. Lines Are Capable of Differentiating into Derivatives of Three Germ Layers
2.4. Lines ICGi034-B and ICGi034-C Have a Normal Karyotype and Are Genetically Identical to the Parental Peripheral Blood Mononuclear Cells (PBMCs)
2.5. ICGi034-B and ICGi034-C Lines Have Successfully Passed Additional Quality Control Tests
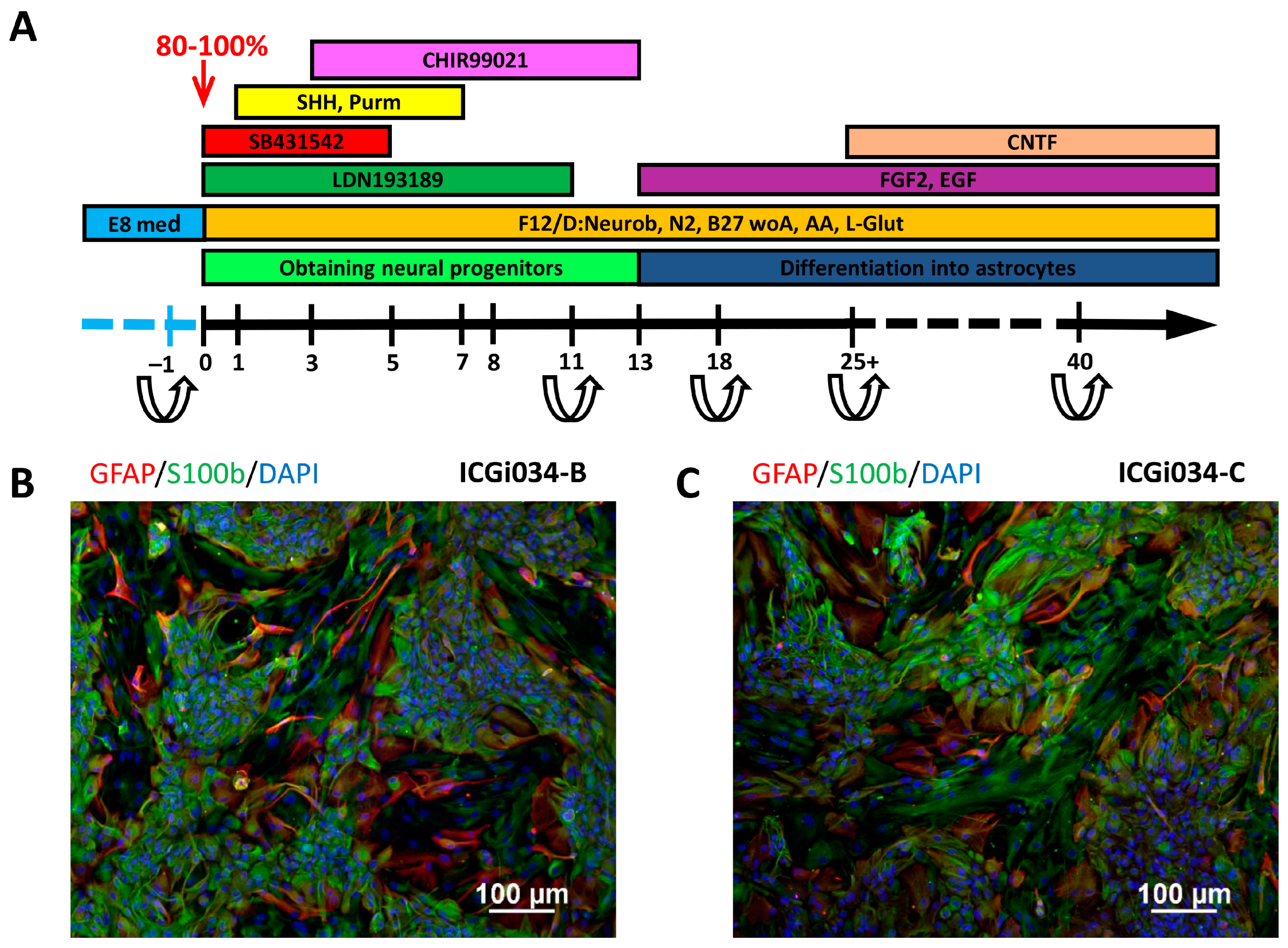
2.6. IPSCs ICGi034-B and ICGi034-C Successfully Differentiate into Astrocytes
3. Discussion
4. Materials and Methods
4.1. Obtaining and Culturing iPSCs
4.2. DNA Isolation
4.3. Detection of Mycoplasma and EBNA
4.4. Quantitative PCR
4.5. Sanger Sequencing
4.6. Karyotypic Analysis
4.7. Spontaneous Differentiation In Vitro
4.8. Immunofluorescence Analysis
4.9. Differentiation of iPSCs into Astrocytes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef] [PubMed]
- Riboldi, G.M.; Di Fonzo, A.B. GBA, Gaucher Disease, and Parkinson’s Disease: From Genetic to Clinic to New Therapeutic Approaches. Cells 2019, 8, 364. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, I.; Blumenreich, S.; Futerman, A.H. GBA mutations, glucosylceramide and Parkinson’s disease. Curr. Opin. Neurobiol. 2022, 72, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Krainc, D. Mechanisms of Glucocerebrosidase Dysfunction in Parkinson’s Disease. J. Mol. Biol. 2023, 435, 168023. [Google Scholar] [CrossRef] [PubMed]
- Senkevich, K.A.; Kopytova, A.E.; Usenko, T.S.; Emelyanov, A.K.; Pchelina, S.N. Parkinson’s Disease Associated with GBA Gene Mutations: Molecular Aspects and Potential Treatment Approaches. Acta Nat. 2021, 13, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Sidransky, E.; Lopez, G. The link between the GBA gene and parkinsonism. Lancet Neurol. 2012, 11, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Booth, H.D.E.; Hirst, W.D.; Wade-Martins, R. The role of astrocyte dysfunction in Parkinson’s disease pathogenesis. Trends Neurosci. 2017, 40, 358–370. [Google Scholar] [CrossRef]
- Zeng, Z.; Roussakis, A.A.; Lao-Kaim, N.P.; Piccini, P. Astrocytes in Parkinson’s disease: From preclinical assays to in vivo imaging and therapeutic probes. Neurobiol. Aging. 2020, 95, 264–270. [Google Scholar] [CrossRef]
- Patani, R.; Hardingham, G.E.; Liddelow, S.A. Functional roles of reactive astrocytes in neuroinflammation and neurodegeneration. Nat. Rev. Neurol. 2023, 19, 395–409. [Google Scholar] [CrossRef]
- Koprich, J.B.; Reske-Nielsen, C.; Mithal, P.; Isacson, O. Neuroinflammation mediated by IL-1β increases susceptibility of dopamine neurons to degeneration in an animal model of Parkinson’s disease. J. Neuroinflamm. 2008, 5, 8. [Google Scholar] [CrossRef]
- Grigor’eva, E.V.; Kopytova, A.E.; Yarkova, E.S.; Pavlova, S.V.; Sorogina, D.A.; Malakhova, A.A.; Malankhanova, T.B.; Baydakova, G.V.; Zakharova, E.Y.; Medvedev, S.P.; et al. Biochemical Characteristics of iPSC-Derived Dopaminergic Neurons from N370S GBA Variant Carriers with and without Parkinson’s Disease. Int. J. Mol. Sci. 2023, 24, 4437. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Yamakawa, T.; Matsumura, Y.; Sato, Y.; Amano, N.; Watanabe, A.; Goshima, N.; Yamanaka, S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 2013, 31, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Grigor’eva, E.V.; Drozdova, E.S.; Sorogina, D.A.; Malakhova, A.A.; Pavlova, S.V.; Vyatkin, Y.V.; Khabarova, E.A.; Rzaev, J.A.; Medvedev, S.P.; Zakian, S.M. Generation of induced pluripotent stem cell line, ICGi034-A, by reprogramming peripheral blood mononuclear cells from a patient with Parkinson’s disease associated with GBA mutation. Stem Cell Res. 2022, 59, 102651. [Google Scholar] [CrossRef] [PubMed]
- Cowan, C.A.; Klimanskaya, I.; McMahon, J.; Atienza, J.; Witmyer, J.; Zucker, J.P.; Wang, S.; Morton, C.C.; McMahon, A.P.; Powers, D.; et al. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004, 350, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- De Rus Jacquet, A. Preparation and Co-Culture of iPSC-Derived Dopaminergic Neurons and Astrocytes. Curr. Protoc. Cell Biol. 2019, 85, e98. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, E.; Garrido, A.; Scholz, S.W.; Poewe, W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol. 2021, 20, 385–397. [Google Scholar] [CrossRef]
- Wang, M.; Ling, K.H.; Tan, J.J.; Lu, C.B. Development and Differentiation of Midbrain Dopaminergic Neuron: From Bench to Bedside. Cells 2020, 9, 1489. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Okun, M.S. Diagnosis and Treatment of Parkinson Disease: A Review. JAMA 2020, 323, 548–560. [Google Scholar] [CrossRef]
- Kopytova, A.E.; Rychkov, G.N.; Cheblokov, A.A.; Grigor’eva, E.V.; Nikolaev, M.A.; Yarkova, E.S.; Sorogina, D.A.; Ibatullin, F.M.; Baydakova, G.V.; Izyumchenko, A.D.; et al. Potential Binding Sites of Pharmacological Chaperone NCGC00241607 on Mutant β-Glucocerebrosidase and Its Efficacy on Patient-Derived Cell Cultures in Gaucher and Parkinson’s Disease. Int. J. Mol. Sci. 2023, 24, 9105. [Google Scholar] [CrossRef]
- Herculano-Houzel, S. The human brain in numbers: A linearly scaled-up primate brain. Front. Hum. Neurosci. 2009, 3, 31. [Google Scholar] [CrossRef]
- Sofroniew, M.V.; Vinters, H.V. Astrocytes: Biology and pathology. Acta Neuropathol. 2010, 119, 7–35. [Google Scholar] [CrossRef] [PubMed]
- Labrador-Garrido, A.; Zhong, S.; Hughes, L.; Keshiya, S.; Kim, W.S.; Halliday, G.M.; Dzamko, N. Live cell in situ lysosomal GCase activity correlates to alpha-synuclein levels in human differentiated neurons with LRRK2 and GBA1 mutations. Front. Cell Neurosci. 2023, 17, 1229213. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Sun, Y.; Dettmer, U. Astrocytes in Parkinson’s Disease: From Role to Possible Intervention. Cells 2023, 12, 2336. [Google Scholar] [CrossRef] [PubMed]
- Halliday, G.M.; Stevens, C.H. Glia: Initiators and progressors of pathology in Parkinson’s disease. Mov. Disord. 2011, 26, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Suk, J.-E.; Patrick, C.; Bae, E.-J.; Cho, J.-H.; Rho, S.; Hwang, D.; Masliah, E.; Lee, S.-J. Direct Transfer of Alpha-Synuclein from Neuron to Astroglia Causes Inflammatory Responses in Synucleinopathies. J. Biol. Chem. 2010, 285, 9262–9272. [Google Scholar] [CrossRef] [PubMed]
- Bo, R.X.; Li, Y.Y.; Zhou, T.T.; Chen, N.H.; Yuan, Y.H. The neuroinflammatory role of glucocerebrosidase in Parkinson’s disease. Neuropharmacology 2022, 207, 108964. [Google Scholar] [CrossRef]
- Ustyantseva, E.I.; Medvedev, S.P.; Vetchinova, A.S.; Minina, J.M.; Illarioshkin, S.N.; Zakian, S.M. A Platform for Studying Neurodegeneration Mechanisms Using Genetically Encoded Biosensors. Biochemistry 2019, 84, 299–309. [Google Scholar] [CrossRef]
- Gerasimova, T.; Stepanenko, E.; Novosadova, L.; Arsenyeva, E.; Shimchenko, D.; Tarantul, V.; Grivennikov, I.; Nenasheva, V.; Novosadova, E. Glial Cultures Differentiated from iPSCs of Patients with PARK2-Associated Parkinson’s Disease Demonstrate a Pro-Inflammatory Shift and Reduced Response to TNFα Stimulation. Int. J. Mol. Sci. 2023, 24, 2000. [Google Scholar] [CrossRef]
- Booth, H.D.E.; Wessely, F.; Connor-Robson, N.; Rinaldi, F.; Vowles, J.; Browne, C.; Evetts, S.G.; Hu, M.T.; Cowley, S.A.; Webber, C.; et al. RNA sequencing reveals MMP2 and TGFB1 downregulation in LRRK2 G2019S Parkinson’s iPSC-derived astrocytes. Neurobiol. Dis. 2019, 129, 56–66. [Google Scholar] [CrossRef]
- Banerjee, R.; Raj, A.; Potdar, C.; Pal, K.P.; Yadav, R.; Kamble, N.; Holla, V.; Datta, I. Astrocytes Differentiated from LRRK2-I1371V Parkinson’s-Disease-Induced Pluripotent Stem Cells Exhibit Similar Yield but Cell-Intrinsic Dysfunction in Glutamate Uptake and Metabolism, ATP Generation, and Nrf2-Mediated Glutathione Machinery. Cells 2023, 12, 1592. [Google Scholar] [CrossRef] [PubMed]
- Oberheim, N.A.; Goldman, S.A.; Nedergaard, M. Heterogeneity of astrocytic form and function. Methods Mol. Biol. 2012, 814, 23–45. [Google Scholar] [CrossRef] [PubMed]
- Choppa, P.C.; Vojdani, A.; Tagle, C.; Andrin, R.; Magtoto, L. Multiplex PCR for the detection of Mycoplasma fermentans, M. hominis and M. penetrans in cell cultures and blood samples of patients with chronic fatigue syndrome. Mol. Cell Probes 1998, 12, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Grigor’eva, E.V.; Malankhanova, T.B.; Surumbayeva, A.; Pavlova, S.V.; Minina, J.M.; Kizilova, E.A.; Suldina, L.A.; Morozova, K.N.; Kiseleva, E.; Sorokoumov, E.D.; et al. Generation of GABAergic striatal neurons by a novel iPSC differentiation protocol enabling scalability and cryopreservation of progenitor cells. Cytotechnology 2020, 72, 649–663. [Google Scholar] [CrossRef]
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography Bright field | Normal | Figure 1A |
| Pluripotency status | Qualitative analysis: Alkaline phosphatase staining | Positive | Figure 1B |
| Qualitative analysis: Immunocytochemistry | Positive staining for pluripotency markers: OCT3/4, SOX2, TRA-1-60, SSEA-4 | Figure 1E | |
| Quantitative analysis: RT-qPCR | Expression of pluripotency markers: NANOG, OCT4, SOX2 | Figure 1C | |
| Genotype | Karyotype (G-banding) | 46,XX | Figure 1D |
| Mutation analysis | Sanger sequencing of DNA from patient’s PBMCs and iPSCs | Heterozygous p.N370S in GBA1 | Figure 1G |
| Differentiation potential | Embryoid body formation | Positive staining for germ layer markers: ɑSMA and NKX2.5 (mesoderm); MAP2 and TUBB3/TUJ1 (ectoderm); FOXA2/AFP (endoderm) | Figure 1F |
| Specific pathogen-free status | Mycoplasma | Negative | Supplementary Figure S1 |
| Antibodies Used for Immunocytochemistry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Rabbit IgG anti-OCT4 | 1:200 | Abcam, Cambridge, UK, Cat # ab18976, RRID:AB_444714 |
| Mouse IgG3 anti-SSEA4 | 1:200 | Abcam, Cambridge, UK, Cat # ab16287, RRID:AB_778073 | |
| Mouse IgM anti-TRA-1-60 | 1:200 | Abcam, Cambridge, UK, Cat # ab16288, RRID:AB_778563 | |
| Rabbit IgG anti-SOX2 | 1:500 | Cell Signaling, Danvers, MA, USA, Cat # 3579, RRID:AB_2195767 | |
| Differentiation Markers | Mouse IgG2a anti-αSMA | 1:100 | Dako, Glostrup, Denmark, Cat # M0851, RRID:AB_2223500 |
| Rabbit IgG anti-NKX2.5 (H-114) | 1:100 | Santa Cruz Biotechnology, Dallas, TX, USA, Cat # sc-14033, RRID:AB_650281 | |
| Mouse IgG2a anti-AFP | 1:250 | Sigma-Aldrich, Darmstadt, Germany, Cat # A8452, RRID:AB_258392 | |
| Mouse IgG2a anti-Tubulin β 3 (TUBB3)/Clone: TUJ1 | 1:1000 | BioLegend, San Diego, CA, USA, Cat # 801,201, RRID:AB_2313773 | |
| Chicken IgG anti MAP2 | 1:1000 | Abcam, Cambridge, UK, Cat # ab5392, RRID:AB_2138153 | |
| Mouse IgG1 anti-HNF3b (FOXA2) | 1:50 | Santa Cruz Biotechnology, Dallas, TX, USA, Cat # sc-374,376, RRID:AB_10989742 | |
| Mouse IgG1 anti-S100β | 1:400 | Sigma-Aldrich, Darmstadt, Germany, Cat # S2532, RRID:AB_477499 | |
| Rabbit IgG anti-GFAP | 1:200 | Dako, Glostrup, Denmark, Cat # Z0334 | |
| Secondary antibodies | Goat anti-Mouse IgG (H + L) Secondary Antibody, Alexa Fluor 488 | 1:400 | Thermo Fisher Scientific, Waltham, MA, USA, Cat # A11029, RRID:AB_2534088 |
| Goat anti-Mouse IgG (H + L) Secondary Antibody, Alexa Fluor 568 | 1:400 | Thermo Fisher Scientific, Waltham, MA, USA, Cat # A11031, RRID:AB_144696 | |
| Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | 1:400 | Thermo Fisher Scientific, Waltham, MA, USA, Cat # A11008, RRID:AB_143165 | |
| Goat anti-Rabbit IgG (H + L) Alexa Fluor 568 | 1:400 | Thermo Fisher Scientific, Waltham, MA, USA, Cat # A11011, RRID:AB_143157 | |
| Goat anti-Mouse IgG1 Alexa Fluor 568 | 1:400 | Thermo Fisher Scientific, Waltham, MA, USA, Cat # A21124, RRID:AB_2535766 | |
| Goat anti-Mouse IgG2a Alexa Fluor 488 | 1:400 | Thermo Fisher Scientific, Waltham, MA, USA, Cat # A21131, RRID:AB_2535771 | |
| Goat anti-Chicken IgY (H + L) Alexa Fluor 488 | 1:400 | Abcam, Cambridge, UK, Cat # ab150173, RRID:AB_2827653 | |
| Primers | |||
| Target | Size of band | Forward/Reverse primer (5′-3′) | |
| Episomal plasmid vector detection | EBNA-1 | 61 bp | TTCCACGAGGGTAGTGAACC/ TCGGGGGTGTTAGAGACAAC |
| Mycoplasma detection | 16S ribosomal RNA gene | 280 bp | GGGAGCAAACAGGATTAGATACCCT/ TGCACCATCTGTCACTCTGTTAACCTC |
| House-keeping gene (RT-qPCR) | beta-2-microglobulin | 280 bp | TAGCTGTGCTCGCGCTACT/ TCTCTGCTGGATGACGTGAG |
| Pluripotency marker (RT-qPCR) | NANOG | 116 bp | TTTGTGGGCCTGAAGAAAACT/ AGGGCTGTCCTGAATAAGCAG |
| OCT4 | 94 bp | CTTCTGCTTCAGGAGCTTGG/ GAAGGAGAAGCTGGAGCAAA | |
| SOX2 | 100 bp | GCTTAGCCTCGTCGATGAAC/ AACCCCAAGATGCACAACTC | |
| Targeted mutation analysis | GBA1 | 600 bp | CTGTTGCTACCTAGTCACTTCC/ CCCTATCTTCCCTTTCCTTCAC |
| Substance | Company | Concentration | Days |
|---|---|---|---|
| LDN193189 | Sigma-Aldrich, Darmstadt, Germany | 100 nM | 0–11 |
| SB431542 | Abcam, Cambridge, UK | 10 µM | 0–5 |
| Purmorphamine | Tocris, Ellisville, MO, USA | 2 µM | 1–7 |
| SHH C25II | PeproTech, Cranbury, NJ, USA | 100 ng/mL | 1–7 |
| CHIR99021 | Sigma-Aldrich, Darmstadt, Germany | 3 µM | 3–13 |
| bFGF | SCI Store, Moscow, Russia | 20 ng/mL | 13–… |
| EGF | PeproTech, Cranbury, NJ, USA | 20 ng/mL | 13–… |
| CNTF | PeproTech, Cranbury, NJ, USA | 10 ng/mL | 25–… |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yarkova, E.S.; Grigor’eva, E.V.; Medvedev, S.P.; Pavlova, S.V.; Zakian, S.M.; Malakhova, A.A. IPSC-Derived Astrocytes Contribute to In Vitro Modeling of Parkinson’s Disease Caused by the GBA1 N370S Mutation. Int. J. Mol. Sci. 2024, 25, 327. https://doi.org/10.3390/ijms25010327
Yarkova ES, Grigor’eva EV, Medvedev SP, Pavlova SV, Zakian SM, Malakhova AA. IPSC-Derived Astrocytes Contribute to In Vitro Modeling of Parkinson’s Disease Caused by the GBA1 N370S Mutation. International Journal of Molecular Sciences. 2024; 25(1):327. https://doi.org/10.3390/ijms25010327
Chicago/Turabian StyleYarkova, Elena S., Elena V. Grigor’eva, Sergey P. Medvedev, Sophia V. Pavlova, Suren M. Zakian, and Anastasia A. Malakhova. 2024. "IPSC-Derived Astrocytes Contribute to In Vitro Modeling of Parkinson’s Disease Caused by the GBA1 N370S Mutation" International Journal of Molecular Sciences 25, no. 1: 327. https://doi.org/10.3390/ijms25010327
APA StyleYarkova, E. S., Grigor’eva, E. V., Medvedev, S. P., Pavlova, S. V., Zakian, S. M., & Malakhova, A. A. (2024). IPSC-Derived Astrocytes Contribute to In Vitro Modeling of Parkinson’s Disease Caused by the GBA1 N370S Mutation. International Journal of Molecular Sciences, 25(1), 327. https://doi.org/10.3390/ijms25010327







