RanBP2/Nup358 Mediates Sumoylation of STAT1 and Antagonizes Interferon-α-Mediated Antiviral Innate Immunity
Abstract
:1. Introduction
2. Results
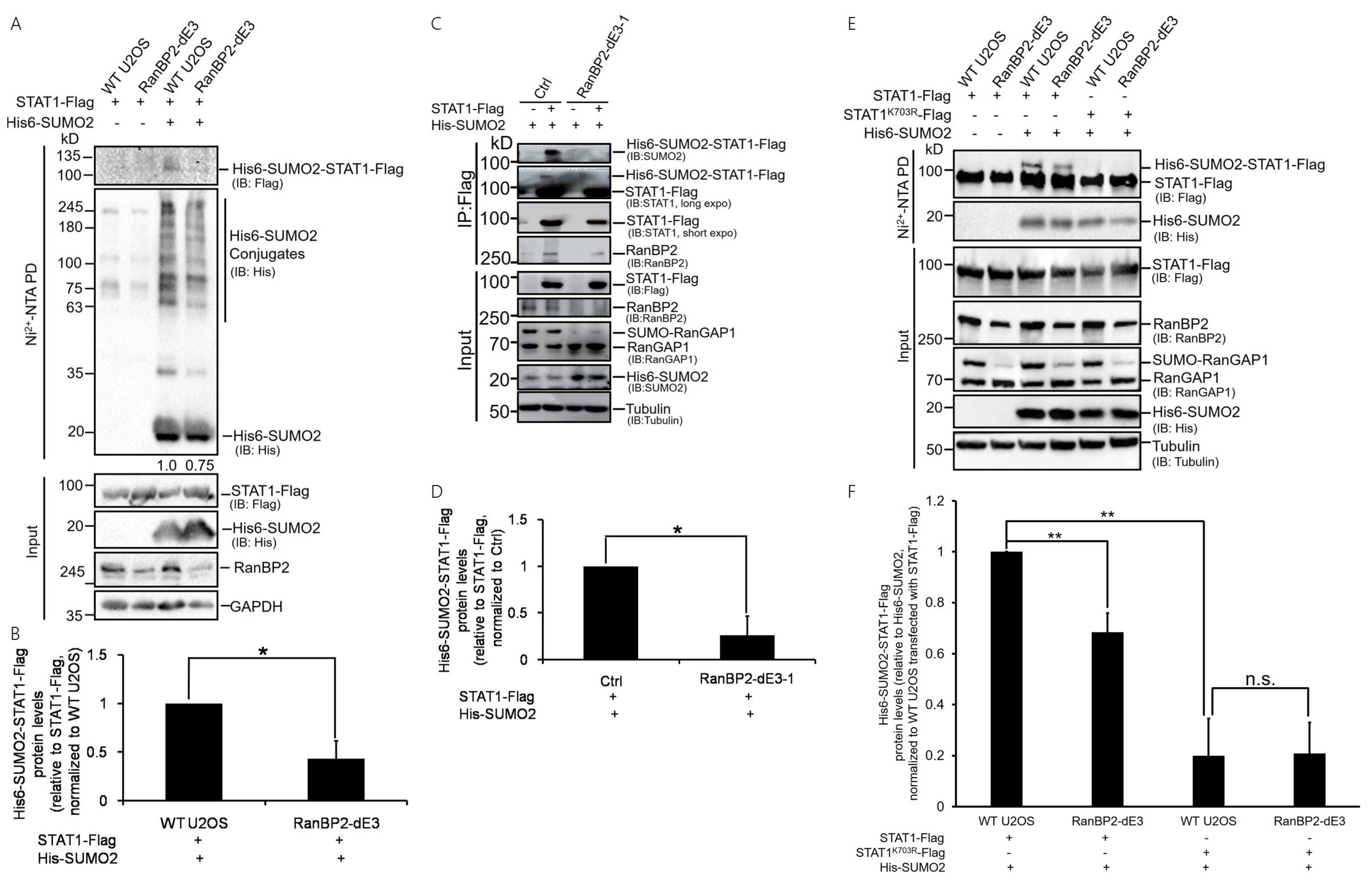
2.1. STAT1 Is a Sumoylation Substrate of RanBP2
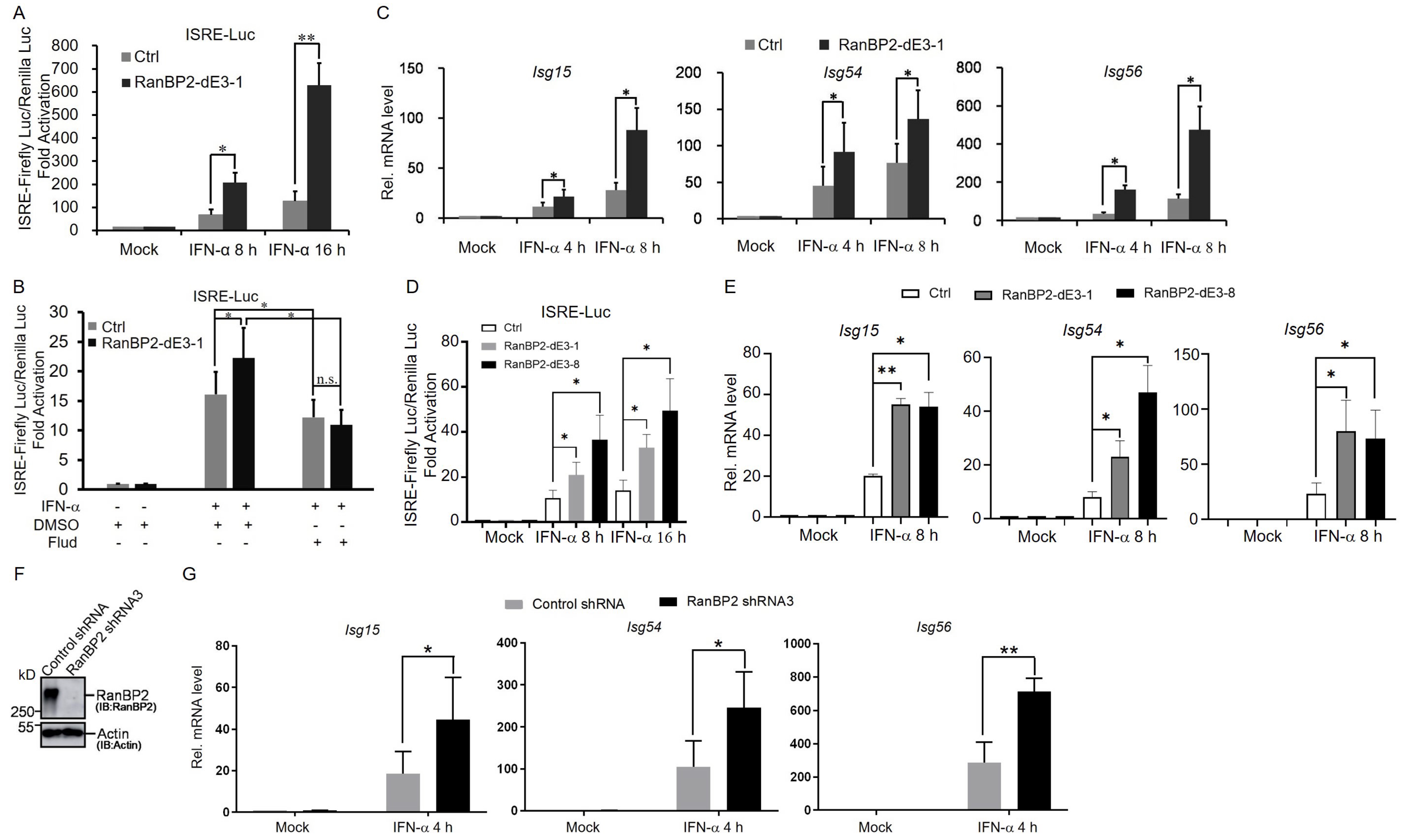
2.2. RanBP2 Inhibits the Interferon-α-Mediated Signaling Pathway
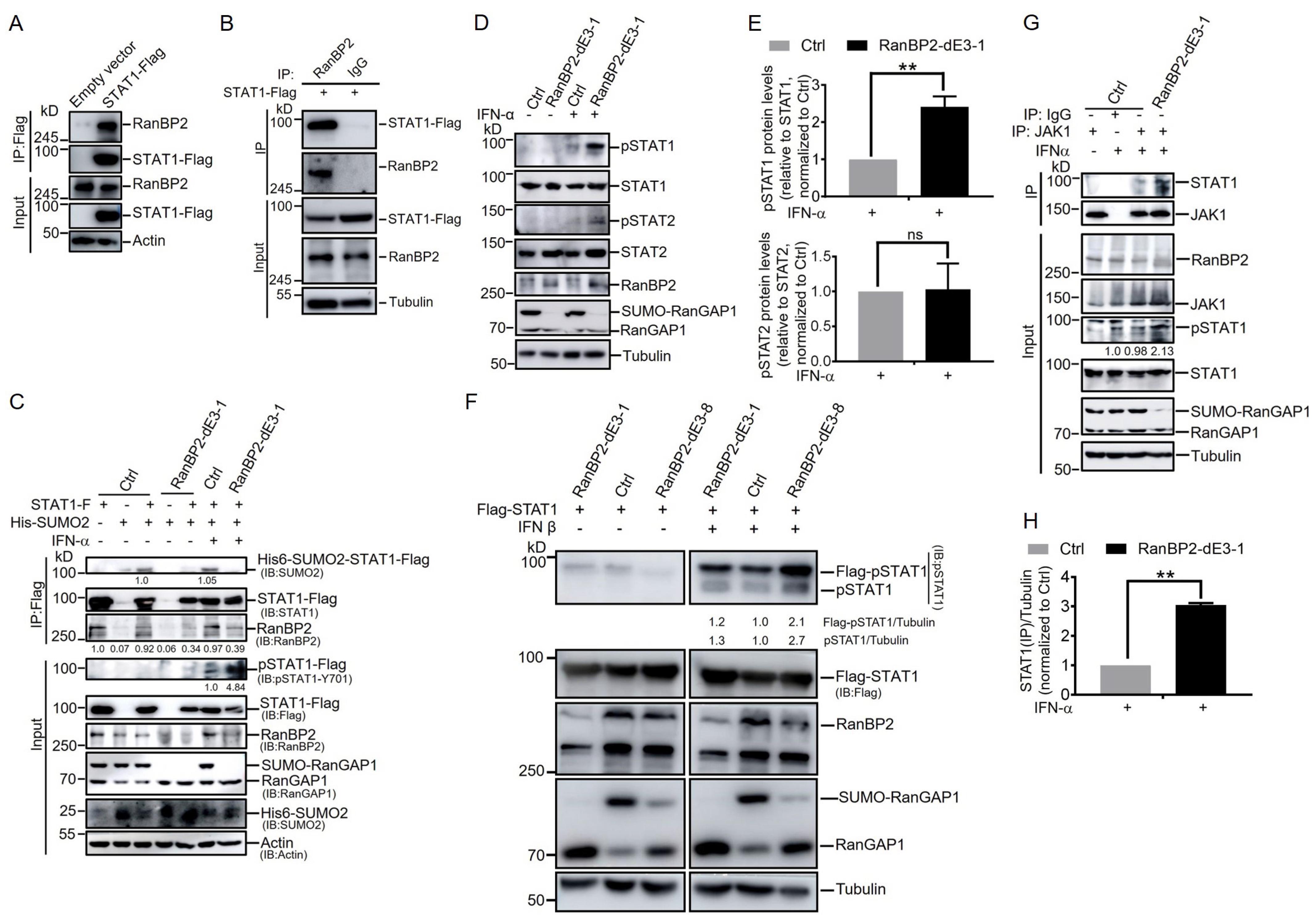
2.3. RanBP2 Inhibits STAT1 Phosphorylation after IFN-α Stimulation by Impairing the Interaction between STAT1 and JAK1
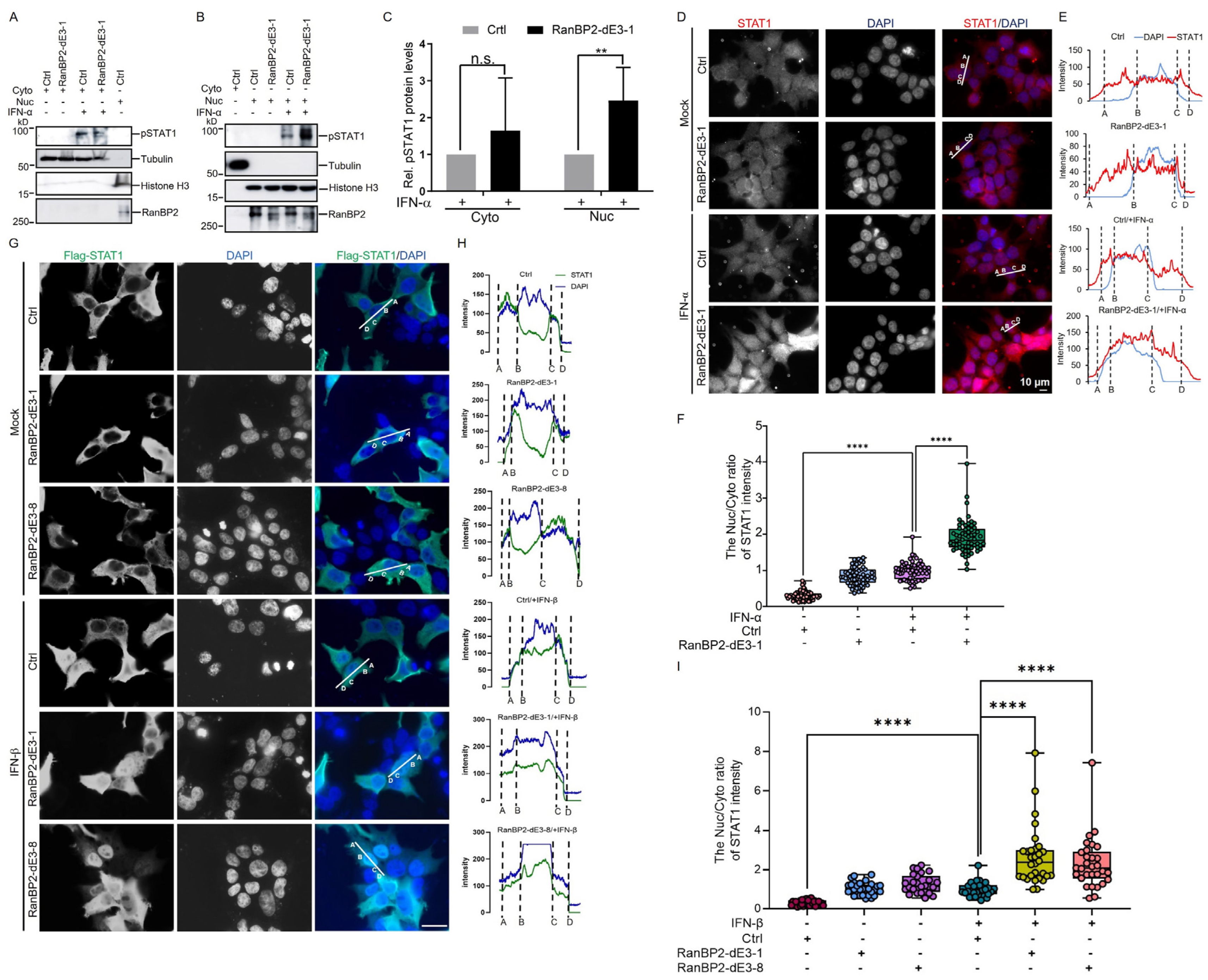
2.4. RanBP2 Inhibits the Nuclear Translocation of STAT1 upon IFN-α Stimulation
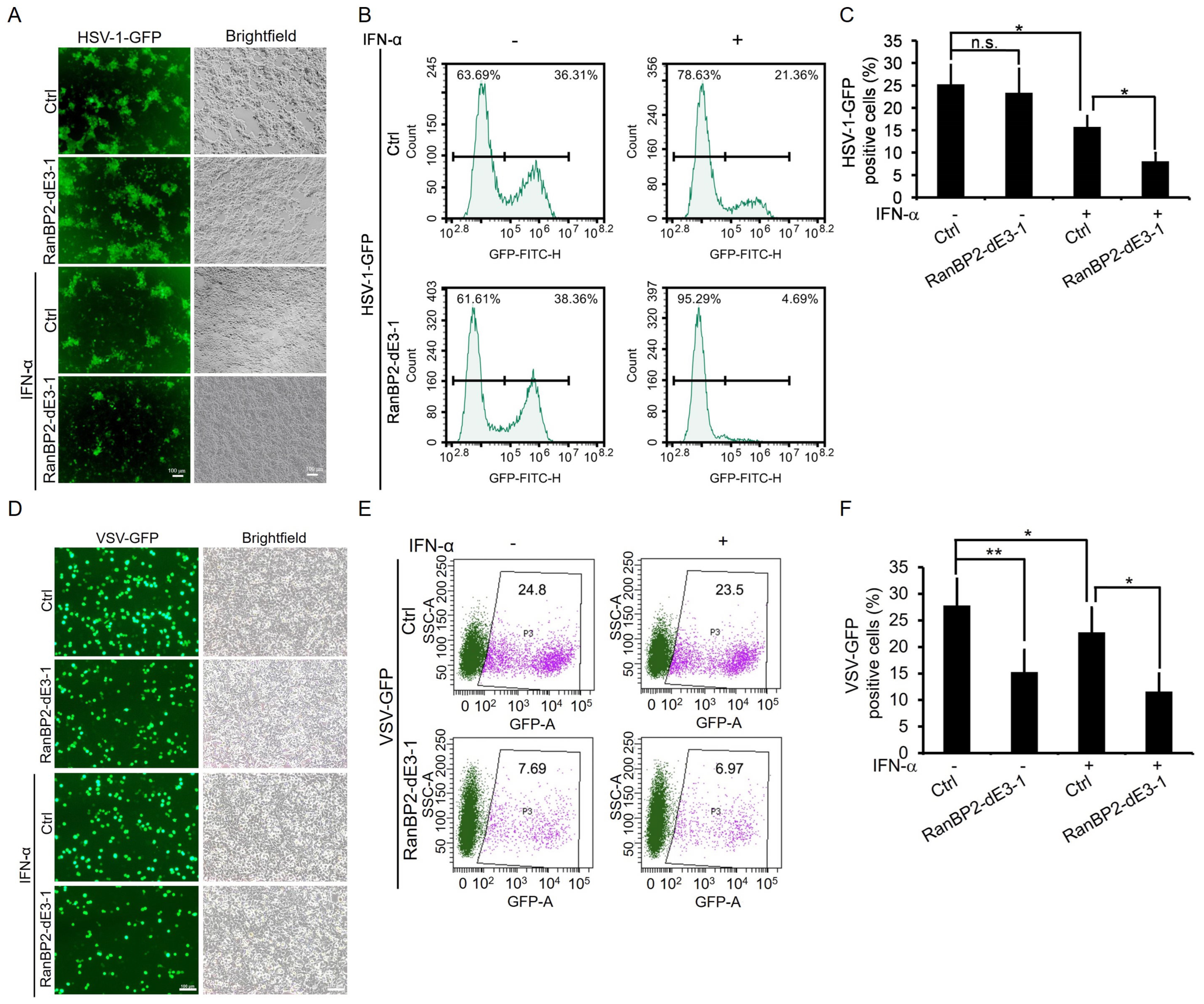
2.5. RanBP2 Promotes Viral Infection by Antagonizing Interferon-α-Mediated Antiviral Innate Immunity
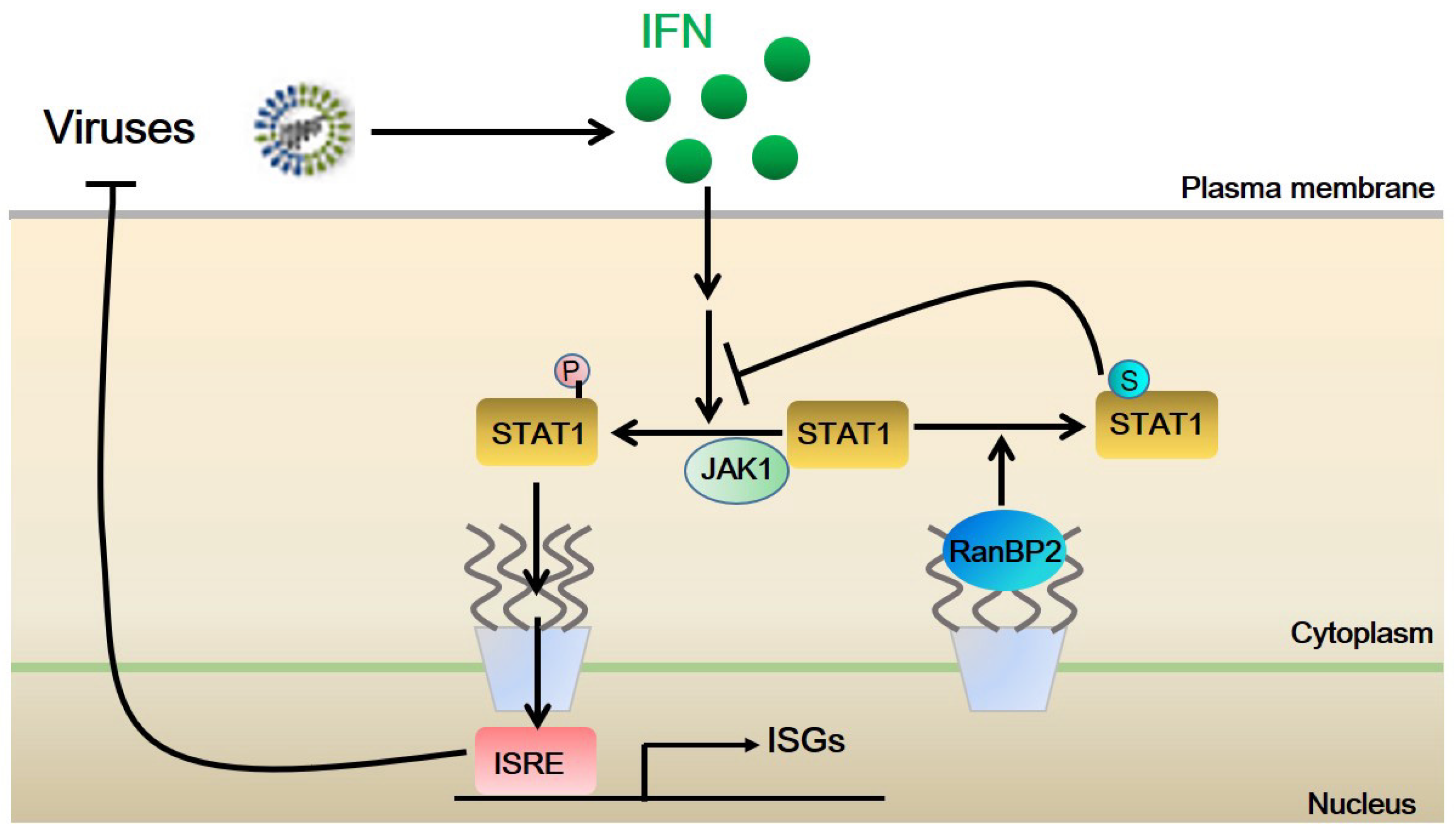
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Cell Transfection and Reagents
4.2. Plasmid Constructs
4.3. CRISPR-Cas9-Mediated Genome Editing
4.4. In Vivo Sumoylation Assay
4.5. Immunoblotting and Immunoprecipitation
4.6. Dual Luciferase Reporter Assay
4.7. RNA Isolation and Quantitative Real-Time PCR
4.8. Cell Fractionation Assays
4.9. Immunofluorescence Microscopy
4.10. Virus Infection and Quantification
4.11. Flow Cytometry
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yokoyama, N.; Hayashi, N.; Seki, T.; Panté, N.; Ohba, T.; Nishii, K.; Kuma, K.; Hayashida, T.; Miyata, T.; Aebi, U.; et al. A Giant Nucleopore Protein That Binds Ran/TC4. Nature 1995, 376, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Matunis, M.J.; Kraemer, D.; Blobel, G.; Coutavas, E. Nup358, a Cytoplasmically Exposed Nucleoporin with Peptide Repeats, Ran-GTP Binding Sites, Zinc Fingers, a Cyclophilin A Homologous Domain, and a Leucine-Rich Region. J. Biol. Chem. 1995, 270, 14209–14213. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Pickersgill, H.S.; Cordes, V.C.; Goldberg, M.W.; Allen, T.D.; Mattaj, I.W.; Fornerod, M. The Cytoplasmic Filaments of the Nuclear Pore Complex Are Dispensable for Selective Nuclear Protein Import. J. Cell Biol. 2002, 158, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, M.R.; Gaikwad, S.; Khuperkar, D.; Ashok, M.; Helen, M.; Yadav, S.K.; Singh, A.; Magre, I.; Deshmukh, P.; Dhanvijay, S.; et al. Nup358 Binds to AGO Proteins through Its SUMO-Interacting Motifs and Promotes the Association of Target mRNA with miRISC. EMBO Rep. 2017, 18, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Mavlyutov, T.A.; Cai, Y.; Ferreira, P.A. Identification of RanBP2- and Kinesin-Mediated Transport Pathways with Restricted Neuronal and Subcellular Localization. Traffic Cph. Den. 2002, 3, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Hampoelz, B.; Schwarz, A.; Ronchi, P.; Bragulat-Teixidor, H.; Tischer, C.; Gaspar, I.; Ephrussi, A.; Schwab, Y.; Beck, M. Nuclear Pores Assemble from Nucleoporin Condensates During Oogenesis. Cell 2019, 179, 671–686.e17. [Google Scholar] [CrossRef]
- Marmor-Kollet, H.; Siany, A.; Kedersha, N.; Knafo, N.; Rivkin, N.; Danino, Y.M.; Moens, T.G.; Olender, T.; Sheban, D.; Cohen, N.; et al. Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. Mol. Cell 2020, 80, 876–891.e6. [Google Scholar] [CrossRef]
- Delphin, C.; Guan, T.; Melchior, F.; Gerace, L. RanGTP Targets P97 to RanBP2, a Filamentous Protein Localized at the Cytoplasmic Periphery of the Nuclear Pore Complex. Mol. Biol. Cell 1997, 8, 2379–2390. [Google Scholar] [CrossRef]
- Bley, C.J.; Nie, S.; Mobbs, G.W.; Petrovic, S.; Gres, A.T.; Liu, X.; Mukherjee, S.; Harvey, S.; Huber, F.M.; Lin, D.H.; et al. Architecture of the Cytoplasmic Face of the Nuclear Pore. Science 2022, 376, eabm9129. [Google Scholar] [CrossRef]
- Ben-Efraim, I.; Gerace, L. Gradient of Increasing Affinity of Importin Beta for Nucleoporins along the Pathway of Nuclear Import. J. Cell Biol. 2001, 152, 411–417. [Google Scholar] [CrossRef]
- Forler, D.; Rabut, G.; Ciccarelli, F.D.; Herold, A.; Köcher, T.; Niggeweg, R.; Bork, P.; Ellenberg, J.; Izaurralde, E. RanBP2/Nup358 Provides a Major Binding Site for NXF1-P15 Dimers at the Nuclear Pore Complex and Functions in Nuclear mRNA Export. Mol. Cell. Biol. 2004, 24, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Hutten, S.; Flotho, A.; Melchior, F.; Kehlenbach, R.H. The Nup358-RanGAP Complex Is Required for Efficient Importin Alpha/Beta-Dependent Nuclear Import. Mol. Biol. Cell 2008, 19, 2300–2310. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, N.; Sakamoto, C.; Hagiwara, M.; Agredano-Moreno, L.T.; Jiménez-García, L.F.; Nakao, M. The Distribution of Phosphorylated SR Proteins and Alternative Splicing Are Regulated by RANBP2. Mol. Biol. Cell 2012, 23, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Asally, M.; Yasuda, Y.; Oka, M.; Otsuka, S.; Yoshimura, S.H.; Takeyasu, K.; Yoneda, Y. Nup358, a Nucleoporin, Functions as a Key Determinant of the Nuclear Pore Complex Structure Remodeling during Skeletal Myogenesis. FEBS J. 2011, 278, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Um, J.W.; Min, D.S.; Rhim, H.; Kim, J.; Paik, S.R.; Chung, K.C. Parkin Ubiquitinates and Promotes the Degradation of RanBP2. J. Biol. Chem. 2006, 281, 3595–3603. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.A.; Nakayama, T.A.; Travis, G.H. Interconversion of Red Opsin Isoforms by the Cyclophilin-Related Chaperone Protein Ran-Binding Protein 2. Proc. Natl. Acad. Sci. USA 1997, 94, 1556–1561. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.A.; Nakayama, T.A.; Pak, W.L.; Travis, G.H. Cyclophilin-Related Protein RanBP2 Acts as Chaperone for Red/Green Opsin. Nature 1996, 383, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Aslanukov, A.; Bhowmick, R.; Guruju, M.; Oswald, J.; Raz, D.; Bush, R.A.; Sieving, P.A.; Lu, X.; Bock, C.B.; Ferreira, P.A. RanBP2 Modulates Cox11 and Hexokinase I Activities and Haploinsufficiency of RanBP2 Causes Deficits in Glucose Metabolism. PLoS Genet. 2006, 2, e177. [Google Scholar] [CrossRef]
- Grünwald, D.; Singer, R.H. In Vivo Imaging of Labelled Endogenous β-Actin mRNA during Nucleocytoplasmic Transport. Nature 2010, 467, 604–607. [Google Scholar] [CrossRef]
- Mahadevan, K.; Zhang, H.; Akef, A.; Cui, X.A.; Gueroussov, S.; Cenik, C.; Roth, F.P.; Palazzo, A.F. RanBP2/Nup358 Potentiates the Translation of a Subset of mRNAs Encoding Secretory Proteins. PLoS Biol. 2013, 11, e1001545. [Google Scholar] [CrossRef]
- Deshmukh, P.; Markande, S.; Fandade, V.; Ramtirtha, Y.; Madhusudhan, M.S.; Joseph, J. The miRISC Component AGO2 Has Multiple Binding Sites for Nup358 SUMO-Interacting Motif. Biochem. Biophys. Res. Commun. 2021, 556, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Wang, Y.E.; Truong, M.; Mahadevan, K.; Wu, J.J.; Zhang, H.; Li, J.; Smith, H.W.; Smibert, C.A.; Palazzo, A.F. RanBP2/Nup358 Enhances miRNA Activity by Sumoylating Argonautes. PLoS Genet. 2021, 17, e1009378. [Google Scholar] [CrossRef] [PubMed]
- Dawlaty, M.M.; Malureanu, L.; Jeganathan, K.B.; Kao, E.; Sustmann, C.; Tahk, S.; Shuai, K.; Grosschedl, R.; van Deursen, J.M. Resolution of Sister Centromeres Requires RanBP2-Mediated SUMOylation of Topoisomerase IIalpha. Cell 2008, 133, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Singh, B.B.; Aslanukov, A.; Zhao, H.; Ferreira, P.A. The Docking of Kinesins, KIF5B and KIF5C, to Ran-Binding Protein 2 (RanBP2) Is Mediated via a Novel RanBP2 Domain. J. Biol. Chem. 2001, 276, 41594–41602. [Google Scholar] [CrossRef]
- Hu, D.J.-K.; Baffet, A.D.; Nayak, T.; Akhmanova, A.; Doye, V.; Vallee, R.B. Dynein Recruitment to Nuclear Pores Activates Apical Nuclear Migration and Mitotic Entry in Brain Progenitor Cells. Cell 2013, 154, 1300–1313. [Google Scholar] [CrossRef]
- Joseph, J.; Liu, S.-T.; Jablonski, S.A.; Yen, T.J.; Dasso, M. The RanGAP1-RanBP2 Complex Is Essential for Microtubule-Kinetochore Interactions in Vivo. Curr. Biol. 2004, 14, 611–617. [Google Scholar] [CrossRef]
- Mahajan, R.; Delphin, C.; Guan, T.; Gerace, L.; Melchior, F. A Small Ubiquitin-Related Polypeptide Involved in Targeting RanGAP1 to Nuclear Pore Complex Protein RanBP2. Cell 1997, 88, 97–107. [Google Scholar] [CrossRef]
- Matunis, M.J.; Coutavas, E.; Blobel, G. A Novel Ubiquitin-like Modification Modulates the Partitioning of the Ran-GTPase-Activating Protein RanGAP1 between the Cytosol and the Nuclear Pore Complex. J. Cell Biol. 1996, 135, 1457–1470. [Google Scholar] [CrossRef]
- Matunis, M.J.; Wu, J.; Blobel, G. SUMO-1 Modification and Its Role in Targeting the Ran GTPase-Activating Protein, RanGAP1, to the Nuclear Pore Complex. J. Cell Biol. 1998, 140, 499–509. [Google Scholar] [CrossRef]
- Saitoh, H.; Pu, R.; Cavenagh, M.; Dasso, M. RanBP2 Associates with Ubc9p and a Modified Form of RanGAP1. Proc. Natl. Acad. Sci. USA 1997, 94, 3736–3741. [Google Scholar] [CrossRef]
- Swaminathan, S.; Kiendl, F.; Körner, R.; Lupetti, R.; Hengst, L.; Melchior, F. RanGAP1*SUMO1 Is Phosphorylated at the Onset of Mitosis and Remains Associated with RanBP2 upon NPC Disassembly. J. Cell Biol. 2004, 164, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, H.; Sparrow, D.B.; Shiomi, T.; Pu, R.T.; Nishimoto, T.; Mohun, T.J.; Dasso, M. Ubc9p and the Conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 1998, 8, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Reverter, D.; Lima, C.D. Insights into E3 Ligase Activity Revealed by a SUMO-RanGAP1-Ubc9-Nup358 Complex. Nature 2005, 435, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Sakin, V.; Richter, S.M.; Hsiao, H.-H.; Urlaub, H.; Melchior, F. Sumoylation of the GTPase Ran by the RanBP2 SUMO E3 Ligase Complex. J. Biol. Chem. 2015, 290, 23589–23602. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.R.; Haindl, M.; Nigg, E.A.; Muller, S. RanBP2 and SENP3 Function in a Mitotic SUMO2/3 Conjugation-Deconjugation Cycle on Borealin. Mol. Biol. Cell 2009, 20, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, Y.E.; Palazzo, A.F.; Shen, Q. Roles of Nucleoporin RanBP2/Nup358 in Acute Necrotizing Encephalopathy Type 1 (ANE1) and Viral Infection. Int. J. Mol. Sci. 2022, 23, 3548. [Google Scholar] [CrossRef]
- Neilson, D.E.; Adams, M.D.; Orr, C.M.D.; Schelling, D.K.; Eiben, R.M.; Kerr, D.S.; Anderson, J.; Bassuk, A.G.; Bye, A.M.; Childs, A.-M.; et al. Infection-Triggered Familial or Recurrent Cases of Acute Necrotizing Encephalopathy Caused by Mutations in a Component of the Nuclear Pore, RANBP2. Am. J. Hum. Genet. 2009, 84, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Sell, K.; Storch, K.; Hahn, G.; Lee-Kirsch, M.A.; Ramantani, G.; Jackson, S.; Neilson, D.; von der Hagen, M.; Hehr, U.; Smitka, M. Variable Clinical Course in Acute Necrotizing Encephalopathy and Identification of a Novel RANBP2 Mutation. Brain Dev. 2016, 38, 777–780. [Google Scholar] [CrossRef]
- Iyer, G.; Utage, P.; Bailur, S.; Utage, A.; Srirambhatla, A.; Hasan, Q. Familial Acute Necrotizing Encephalopathy: Evidence From Next Generation Sequencing of Digenic Inheritance. J. Child Neurol. 2020, 35, 393–397. [Google Scholar] [CrossRef]
- Liu, Y.; Trnka, M.J.; Guan, S.; Kwon, D.; Kim, D.-H.; Chen, J.-J.; Greer, P.A.; Burlingame, A.L.; Correia, M.A. A Novel Mechanism for NF-κB-Activation via IκB-Aggregation: Implications for Hepatic Mallory-Denk-Body Induced Inflammation. Mol. Cell. Proteom. MCP 2020, 19, 1968–1985. [Google Scholar] [CrossRef]
- Flotho, A.; Melchior, F. Sumoylation: A Regulatory Protein Modification in Health and Disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.S. Protein Modification by SUMO. Annu. Rev. Biochem. 2004, 73, 355–382. [Google Scholar] [CrossRef] [PubMed]
- Lowrey, A.J.; Cramblet, W.; Bentz, G.L. Viral Manipulation of the Cellular Sumoylation Machinery. Cell Commun. Signal. CCS 2017, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, X.; Zhang, L.; Zong, Z.; Wang, F.; Huang, J.; Zeng, L.; Zhang, C.; Yan, H.; Zhang, L.; et al. SUMOylation in Viral Replication and Antiviral Defense. Adv. Sci. 2022, 9, 2104126. [Google Scholar] [CrossRef]
- Imbert, F.; Langford, D. Viruses, SUMO, and Immunity: The Interplay between Viruses and the Host SUMOylation System. J. Neurovirol. 2021, 27, 531–541. [Google Scholar] [CrossRef]
- He, Y.; Yang, Z.; Zhao, C.; Xiao, Z.; Gong, Y.; Li, Y.-Y.; Chen, Y.; Du, Y.; Feng, D.; Altman, A.; et al. T-Cell Receptor (TCR) Signaling Promotes the Assembly of RanBP2/RanGAP1-SUMO1/Ubc9 Nuclear Pore Subcomplex via PKC-θ-Mediated Phosphorylation of RanGAP1. eLife 2021, 10, e67123. [Google Scholar] [CrossRef]
- Dutrieux, J.; Portilho, D.M.; Arhel, N.J.; Hazan, U.; Nisole, S. TRIM5α Is a SUMO Substrate. Retrovirology 2015, 12, 28. [Google Scholar] [CrossRef]
- Lukic, Z.; Goff, S.P.; Campbell, E.M.; Arriagada, G. Role of SUMO-1 and SUMO Interacting Motifs in Rhesus TRIM5α-Mediated Restriction. Retrovirology 2013, 10, 10. [Google Scholar] [CrossRef]
- Arriagada, G.; Muntean, L.N.; Goff, S.P. SUMO-Interacting Motifs of Human TRIM5α Are Important for Antiviral Activity. PLoS Pathog. 2011, 7, e1002019. [Google Scholar] [CrossRef]
- Maarifi, G.; Fernandez, J.; Portilho, D.M.; Boulay, A.; Dutrieux, J.; Oddos, S.; Butler-Browne, G.; Nisole, S.; Arhel, N.J. RanBP2 Regulates the Anti-Retroviral Activity of TRIM5α by SUMOylation at a Predicted Phosphorylated SUMOylation Motif. Commun. Biol. 2018, 1, 193. [Google Scholar] [CrossRef]
- Bowie, A.G.; Unterholzner, L. Viral Evasion and Subversion of Pattern-Recognition Receptor Signalling. Nat. Rev. Immunol. 2008, 8, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Stetson, D.B.; Medzhitov, R. Type I Interferons in Host Defense. Immunity 2006, 25, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Antiviral Actions of Interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Darnell, J.E. The JAK-STAT Pathway at Twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, J.; Liu, S.; Xia, M.; Zhang, X.; Han, D.; Jiang, Y.; Wang, C.; Cao, X. Methyltransferase SETD2-Mediated Methylation of STAT1 Is Critical for Interferon Antiviral Activity. Cell 2017, 170, 492–506.e14. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Feng, Q.; Jin, L.; Huang, F.; Miao, Y.; Liu, J.; Xu, Y.; Chen, X.; Zhang, H.; Guo, T.; et al. Regulation of the Linear Ubiquitination of STAT1 Controls Antiviral Interferon Signaling. Nat. Commun. 2020, 11, 1146. [Google Scholar] [CrossRef]
- Rogers, R.S.; Horvath, C.M.; Matunis, M.J. SUMO Modification of STAT1 and Its Role in PIAS-Mediated Inhibition of Gene Activation. J. Biol. Chem. 2003, 278, 30091–30097. [Google Scholar] [CrossRef]
- Ungureanu, D.; Vanhatupa, S.; Kotaja, N.; Yang, J.; Aittomäki, S.; Jänne, O.A.; Palvimo, J.J.; Silvennoinen, O. PIAS Proteins Promote SUMO-1 Conjugation to STAT1. Blood 2003, 102, 3311–3313. [Google Scholar] [CrossRef]
- Ungureanu, D.; Vanhatupa, S.; Grönholm, J.; Palvimo, J.J.; Silvennoinen, O. SUMO-1 Conjugation Selectively Modulates STAT1-Mediated Gene Responses. Blood 2005, 106, 224–226. [Google Scholar] [CrossRef]
- Droescher, M.; Begitt, A.; Marg, A.; Zacharias, M.; Vinkemeier, U. Cytokine-Induced Paracrystals Prolong the Activity of Signal Transducers and Activators of Transcription (STAT) and Provide a Model for the Regulation of Protein Solubility by Small Ubiquitin-like Modifier (SUMO). J. Biol. Chem. 2011, 286, 18731–18746. [Google Scholar] [CrossRef]
- Al Shehri, T.; Gilmour, K.; Gothe, F.; Loughlin, S.; Bibi, S.; Rowan, A.D.; Grainger, A.; Mohanadas, T.; Cant, A.J.; Slatter, M.A.; et al. Novel Gain-of-Function Mutation in Stat1 Sumoylation Site Leads to CMC/CID Phenotype Responsive to Ruxolitinib. J. Clin. Immunol. 2019, 39, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Begitt, A.; Droescher, M.; Knobeloch, K.-P.; Vinkemeier, U. SUMO Conjugation of STAT1 Protects Cells from Hyperresponsiveness to IFNγ. Blood 2011, 118, 1002–1007. [Google Scholar] [CrossRef]
- Song, L.; Bhattacharya, S.; Yunus, A.A.; Lima, C.D.; Schindler, C. Stat1 and SUMO Modification. Blood 2006, 108, 3237–3244. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Gao, X.; Jin, C.; Zhu, M.; Wang, X.; Shaw, A.; Wen, L.; Yao, X.; Xue, Y. Systematic Study of Protein Sumoylation: Development of a Site-Specific Predictor of SUMOsp 2.0. Proteomics 2009, 9, 3409–3412. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.A.; Mahajan, S.; Ritz, J. Fludarabine-Induced Immunosuppression Is Associated with Inhibition of STAT1 Signaling. Nat. Med. 1999, 5, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Zhang, Y.; Yeruva, L.; Marinov, A.; Prantner, D.; Wyrick, P.B.; Lupashin, V.; Nagarajan, U.M. The DNA Sensor, Cyclic GMP–AMP Synthase, Is Essential for Induction of IFN-β during Chlamydia Trachomatis Infection. J. Immunol. 2014, 193, 2394–2404. [Google Scholar] [CrossRef]
- Desmyter, J.; Melnick, J.L.; Rawls, W.E. Defectiveness of Interferon Production and of Rubella Virus Interference in a Line of African Green Monkey Kidney Cells (Vero). J. Virol. 1968, 2, 955–961. [Google Scholar] [CrossRef]
- Osada, N.; Kohara, A.; Yamaji, T.; Hirayama, N.; Kasai, F.; Sekizuka, T.; Kuroda, M.; Hanada, K. The Genome Landscape of the African Green Monkey Kidney-Derived Vero Cell Line. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2014, 21, 673–683. [Google Scholar] [CrossRef]
- Tan, X.; Sun, L.; Chen, J.; Chen, Z.J. Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu. Rev. Microbiol. 2018, 72, 447–478. [Google Scholar] [CrossRef]
- Singh, B.B.; Patel, H.H.; Roepman, R.; Schick, D.; Ferreira, P.A. The Zinc Finger Cluster Domain of RanBP2 Is a Specific Docking Site for the Nuclear Export Factor, Exportin-1. J. Biol. Chem. 1999, 274, 37370–37378. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Wang, Y.E.; Palazzo, A.F. Crosstalk between Nucleocytoplasmic Trafficking and the Innate Immune Response to Viral Infection. J. Biol. Chem. 2021, 297, 100856. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Yoon, D.; Qiu, S.; Danziger, Z.; Grill, W.M.; Wetsel, W.C.; Ferreira, P.A. Loss of Ranbp2 in Motoneurons Causes Disruption of Nucleocytoplasmic and Chemokine Signaling, Proteostasis of hnRNPH3 and Mmp28, and Development of Amyotrophic Lateral Sclerosis-like Syndromes. Dis. Model. Mech. 2017, 10, 559–579. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Herrera, C.F.D.L.; Shire, K.; Siddiqi, U.Z.; Frappier, L. A Genome-Wide Screen of Epstein-Barr Virus Proteins That Modulate Host SUMOylation Identifies a SUMO E3 Ligase Conserved in Herpesviruses. PLoS Pathog. 2018, 14, e1007176. [Google Scholar] [CrossRef] [PubMed]
- Vertegaal, A.C.O.; Andersen, J.S.; Ogg, S.C.; Hay, R.T.; Mann, M.; Lamond, A.I. Distinct and Overlapping Sets of SUMO-1 and SUMO-2 Target Proteins Revealed by Quantitative Proteomics. Mol. Cell. Proteom. MCP 2006, 5, 2298–2310. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved Vectors and Genome-Wide Libraries for CRISPR Screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.-Y.; Siu, K.-L.; Lin, H.; Chan, C.-P.; Yeung, M.L.; Jin, D.-Y. SARS-CoV-2 NSP13 Helicase Suppresses Interferon Signaling by Perturbing JAK1 Phosphorylation of STAT1. Cell Biosci. 2022, 12, 36. [Google Scholar] [CrossRef]
- Wong, L.-Y.R.; Ye, Z.-W.; Lui, P.-Y.; Zheng, X.; Yuan, S.; Zhu, L.; Fung, S.-Y.; Yuen, K.-S.; Siu, K.-L.; Yeung, M.-L.; et al. Middle East Respiratory Syndrome Coronavirus ORF8b Accessory Protein Suppresses Type I IFN Expression by Impeding HSP70-Dependent Activation of IRF3 Kinase IKKε. J. Immunol. 2020, 205, 1564–1579. [Google Scholar] [CrossRef]
- Yang, H.; Xiong, F.; Song, Y.-G.; Jiang, H.-F.; Qin, H.-B.; Zhou, J.; Lu, S.; Grieco, S.F.; Xu, X.; Zeng, W.-B.; et al. HSV-1 H129-Derived Anterograde Neural Circuit Tracers: Improvements, Production, and Applications. Neurosci. Bull. 2021, 37, 701–719. [Google Scholar] [CrossRef]
- Zeng, W.-B.; Jiang, H.-F.; Gang, Y.-D.; Song, Y.-G.; Shen, Z.-Z.; Yang, H.; Dong, X.; Tian, Y.-L.; Ni, R.-J.; Liu, Y.; et al. Anterograde Monosynaptic Transneuronal Tracers Derived from Herpes Simplex Virus 1 Strain H129. Mol. Neurodegener. 2017, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, R.; Qiao, S.; Wang, X.; Zhang, W.; Ruan, W.; Dai, L.; Han, P.; Gao, G.F. Omicron SARS-CoV-2 Neutralization from Inactivated and ZF2001 Vaccines. N. Engl. J. Med. 2022, 387, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Pichler, A.; Gast, A.; Seeler, J.S.; Dejean, A.; Melchior, F. The Nucleoporin RanBP2 Has SUMO1 E3 Ligase Activity. Cell 2002, 108, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Gareau, J.R.; Reverter, D.; Lima, C.D. Determinants of Small Ubiquitin-like Modifier 1 (SUMO1) Protein Specificity, E3 Ligase, and SUMO-RanGAP1 Binding Activities of Nucleoporin RanBP2. J. Biol. Chem. 2012, 287, 4740–4751. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Su, L.; Jiang, J.; Wang, Y.E.; Ling, Y.; Qiu, Y.; Yu, H.; Huang, Y.; Wu, J.; Jiang, S.; et al. RanBP2/Nup358 Mediates Sumoylation of STAT1 and Antagonizes Interferon-α-Mediated Antiviral Innate Immunity. Int. J. Mol. Sci. 2024, 25, 299. https://doi.org/10.3390/ijms25010299
Li J, Su L, Jiang J, Wang YE, Ling Y, Qiu Y, Yu H, Huang Y, Wu J, Jiang S, et al. RanBP2/Nup358 Mediates Sumoylation of STAT1 and Antagonizes Interferon-α-Mediated Antiviral Innate Immunity. International Journal of Molecular Sciences. 2024; 25(1):299. https://doi.org/10.3390/ijms25010299
Chicago/Turabian StyleLi, Jiawei, Lili Su, Jing Jiang, Yifan E. Wang, Yingying Ling, Yi Qiu, Huahui Yu, Yucong Huang, Jiangmin Wu, Shan Jiang, and et al. 2024. "RanBP2/Nup358 Mediates Sumoylation of STAT1 and Antagonizes Interferon-α-Mediated Antiviral Innate Immunity" International Journal of Molecular Sciences 25, no. 1: 299. https://doi.org/10.3390/ijms25010299
APA StyleLi, J., Su, L., Jiang, J., Wang, Y. E., Ling, Y., Qiu, Y., Yu, H., Huang, Y., Wu, J., Jiang, S., Zhang, T., Palazzo, A. F., & Shen, Q. (2024). RanBP2/Nup358 Mediates Sumoylation of STAT1 and Antagonizes Interferon-α-Mediated Antiviral Innate Immunity. International Journal of Molecular Sciences, 25(1), 299. https://doi.org/10.3390/ijms25010299





