Bioinformatic Approach of B and T Cell Epitopes of PLD and CP40 Proteins of Corynebacterium pseudotuberculosis ovis Mexican Isolate 2J-L towards a Peptide-Based Vaccine
Abstract
:1. Introduction
2. Results
2.1. B lymphocyte Epitope Predictions for PLD 2J-L and CP40 2J-L Proteins
2.2. T Lymphocyte Epitope Predictions for PLD 2J-L and CP40 2J-L Proteins
3. Discussion
3.1. B Lymphocyte Epitopes Prediction for PLD 2J-L and CP40 2J-L
3.2. T Lymphocyte Epitope Prediction of PLD 2J-L and CP40 2J-L
4. Materials and Methods
4.1. PLD 2J-L and CP40 2J-L Proteins Sequences and Structure
4.2. Linear and Conformational B Cell Epitope Prediction Tools
4.3. T Cell Epitope Prediction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dalsass, M.; Brozzi, A.; Medini, D.; Rappuoli, R. Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 2019, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Christensen, H.; Hickman, M.; Edmunds, W.J.; Trotter, C.L. Introducing vaccination against serogroup B meningococcal disease: An economic and mathematical modelling study of potential impact. Vaccine 2013, 31, 2638–2646. [Google Scholar] [CrossRef] [PubMed]
- De Groot, A.S.; Moise, L.; Terry, F.; Gutierrez, A.H.; Hindocha, P.; Richard, G.; Hoft, D.F.; Ross, T.M.; Noe, A.R.; Takahashi, Y.; et al. Better Epitope Discovery, Precision Immune Engineering, and Accelerated Vaccine Design Using Immunoinformatics Tools. Front. Immunol. 2020, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Trincado, J.L.; Gomez-Perosanz, M.; Reche, P.A. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J. Immunol. Res. 2017, 2017, 2680160. [Google Scholar] [CrossRef] [PubMed]
- Fleri, W.; Paul, S.; Dhanda, S.K.; Mahajan, S.; Xu, X.; Peters, B.; Sette, A. The Immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design. Front. Immunol. 2017, 8, 278. [Google Scholar] [CrossRef]
- Potocnakova, L.; Bhide, M.; Pulzova, L.B. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J. Immunol. Res. 2016, 2016, 6760830. [Google Scholar] [CrossRef]
- Hodgson, A.L.M.; Bird, P.; Nisbett, I.T. Cloning, nucleotide sequence, and expression in Escherichia coli of the phospholipase D gene from Corynebacterium pseudotuberculosis. J. Bacteriol. 1990, 172, 1256–1261. [Google Scholar] [CrossRef]
- Shadnezhad, A.; Naegeli, A.; Collin, M. CP40 from Corynebacterium pseudotuberculosis is a endo B-N- acetylglucosaminidase. BMC Microbiol. 2016, 63, 206–211. [Google Scholar] [CrossRef]
- Paule, B.J.A.; Azevedo, V.; Regis, L.F.; Carminati, R.; Bahia, R. Experimental Corynebacterium pseudotuberculosis primary infection in goats: Kinetics of IgG and interferon-γ production, IgG avidity and antigen recognition by Western blotting. Vet. Immunol. Immunopathol. 2003, 96, 129–139. [Google Scholar] [CrossRef]
- Seyffert, N.; Silva, R.F.; Jardin, J.; Silva, W.M.; Castro, T.L.; Tartaglia, N.R. Serological proteome analysis of Corynebacterium pseudotuberculosis isolated from different hosts reveals novel candidates for prophylactics to control caseous lymphadenitis. Vet. Microbiol. 2014, 174, 255–260. [Google Scholar] [CrossRef]
- Windsor, P.A.; Bush, R.D. Caseous lymphadenitis: Present and near forgotten from persistent vaccination? Small Rumin. Res. 2016, 142, 6–10. [Google Scholar] [CrossRef]
- Bastos, B.L.; Dias, P.R.W.; Dorella, F.A.; Ribeiro, D.; Seyffert, N. Corynebacterium pseudotuberculosis: Immunological responses in animal models and zoonotic potential. J. Clin. Cell Immunol. 2012, S4, 1–15. [Google Scholar] [CrossRef]
- de Pinho, R.B.; de Oliveira Silva, M.T.; Bezerra, F.S.B.; Borsuk, S. Vaccines for caseous lymphadenitis: Up-to-date and forward-looking strategies. Appl. Microbiol. Biotechnol. 2021, 105, 2287–2296. [Google Scholar] [CrossRef] [PubMed]
- Dorella, F.A.; Pacheco, L.G.C.; Seyffert, N.; Portela, R.W.; Meyer, R.; Miyoshi, A.; Azevedo, V. Antigens of Corynebacterium pseudotuberculosis and prospects for vaccine development. Exp. Rev. Vaccines 2009, 8, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.; Jackson, H.J.; Wilson, M.J.; Eggleton, D.G.; Meeusen, E.N.T.; Brandon, M.R. Identification of a novel antigen from Corynebacterium pseudotuberculosis that protects sheep against caseous lymphadenitis. Infect. Immunol. 1994, 62, 2562–2567. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.; Wilson, M.J.; Brandon, M.R. Molecular and biochemical characterization of a protective 40-kilodalton antigen from Corynebacterium pseudotuberculosis. Infect. Immunol. 1995, 63, 206–211. [Google Scholar]
- Droppa-Almeida, D.; Vivas, W.L.; Silva, K.K.; Rezende, A.F.; Simionatto, S. Recombinant CP40 from Corynebacterium pseudotuberculosis confers protection in mice after challenge with a virulent strain. Vaccine 2016, 34, 1091–1096. [Google Scholar] [CrossRef]
- Silva, M.T.O.; Bezerra, F.S.B.; de Pinho, R.B.; Begnini, K.R.; Seixas, F.K.; Collares, T. Association of Corynebacterium pseudotuberculosis recombinant proteins rCP09720 or rCP01850 with rPLD as immunogens in caseous lymphadenitis immunoprophylaxis. Vaccine 2018, 36, 74–83. [Google Scholar] [CrossRef]
- Leal, K.S.; Silva, T.O.; Silva, A.F.R.; Brilhante, F.S.B.; Begnini, K.; Seixas, F.; Collares, T. Recombinant M. bovis BCG expressing the PLD protein promotes survival in mice challenged with a C. pseudotuberculosis virulent strain. Vaccine 2018, 36, 3578–3583. [Google Scholar]
- Silva, J.W.; Droppa-Almeida, D.; Borsuk, S.; Azevedo, V.; Portela, R.W. Corynebacterium pseudotuberculosis cp09 mutant and cp40 recombinant protein partially protect mice against caseous lymphadenitis. BMC Vet. Res. 2014, 10, 965. [Google Scholar] [CrossRef]
- Parise, D.; Parise, M.; Viana, M.V.C.; Muñoz, B.A.V.; Cortés-Pérez, Y.A.; Azevedo, V. First genome sequencing and comparative analyses of Corynebacterium pseudotuberculosis strains from Mexico. Stand. Genom. Sci. 2018, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Varela, G.J.A.; Montes de Oca, J.R.; Acosta, J.D.; Hernández, F.L.; Morales, E.V.; Monroy, S.G.H. First report of isolation and molecular characterization of the pathogenic Corynebacterium pseudotuberculosis from of sheep and goats in Mexico. Microb. Pathog. 2018, 117, 304–309. [Google Scholar]
- Rodríguez Domínguez, M.C.; Montes de Oca Jiménez, R.; Barbabosa-Pliego, A.; Díaz-Aparicio, E.; Varela-Guerrero, J.A.; Tenorio-Borroto, E. Isolation, cloning and phylogenetic analysis of pld and cp40, virulence factors of a mexican isolate of Corynebacterium pseudotuberculosis ovis. Trop. Subtrop. Agroecosyst. 2022, 25, 1–12. [Google Scholar] [CrossRef]
- Yao, B.; Zheng, D.; Liang, S.; Zhang, C. Conformational B-Cell Epitope Prediction on Antigen Protein Structures: A Review of current algorithms and comparison with common binding site prediction methods. PLoS ONE 2013, 8, e62249. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, A.L.M.; Krywult, J.; Corner, L.A.; Rothel, J.S.; Radford, A.J. Rational attenuation of Corynebacterium pseudotuberculosis: Potential cheesy gland vaccine and live delivery vehicle. Infect. Immunol. 1992, 60, 2900–2905. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, A.L.M.; Tachedjian, M.; Corner, L.A.; Radford, A.J. Protection of sheep against caseous lymphadenitis by use of a single oral dose of live recombinant Corynebacterium pseudotuberculosis. Infect. Immunol. 1994, 62, 5275–5280. [Google Scholar] [CrossRef]
- Hodgson, A.L.; Moore, R.J.; Rothel, L.; Krywult, J.; Radford, A.J.; Lund, K. Foreign gene expression in Corynebacterium pseudotuberculosis: Development of a live vaccine vector. Vaccine 1999, 18, 487–497. [Google Scholar]
- Barral, T.D.; Kalil, M.A.; Mariutti, R.B.; Arni, R.K.; Gismene, C.; Sousa, F.S.; Collares, T.; Seixas, F.K.; Borsuk, S.; Estrela-Lima, A.; et al. Immunoprophylactic properties of the Corynebacterium pseudotuberculosis-derived MBP:PLD:CP40 fusion protein. Appl. Microbiol. Biotechnol. 2022, 106, 8035–8051. [Google Scholar] [CrossRef]
- de Pinho, R.B.; de Oliveira Silva, M.T.; Brenner, G.; Dié Alves, M.S.; Azevedo, V.; Dias Portela, R.; Borsuk, S. A novel approach for an immunogen against Corynebacterium pseudotuberculosis infection: An Escherichia coli bacterin expressing phospholipase D. Microb. Pathog. 2021, 151, 104746. [Google Scholar] [CrossRef]
- Droppa-Almeida, D.; Franceschi, E.; Padilha, F.F. Immune-informatic analysis and design of peptide vaccine from multi-epitopes against Corynebacterium pseudotuberculosis. Bioinform. Biol. Insights 2018, 12, 1177932218755337. [Google Scholar] [CrossRef]
- Kang, J.Y.; Nan, X.; Jin, M.S. Recognition of lipopeptide patterns by toll-like receptor 2-toll-like receptor 6 heterodimer. Immunity 2009, 31, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Droppa-Almeida, D.; da Silva, G.A.; Gaspar, L.M.D.A.C.; Pereyra, B.B.S.; Nascimento, R.J.M.; Borsuk, S.; Franceschi, E.; Padilha, F.F. Peptide vaccines designed with the aid of immunoinformatic against Caseous Lymphadenitis promotes humoral and cellular response induction in mice. PLoS ONE 2021, 16, e0256864. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, J.; Bui, H.H.; Li, W.; Fusseder, N.; Bourne, P.E. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Mukonyora, M. A Review of Important Discontinuous B-Cell Epitope Prediction Tools. J. Clin. Cell Immunol. 2015, 6, 358. [Google Scholar] [CrossRef]
- Stefanska, I.; Gierynska, M.; Rzewuska, M.; Binek, M. Survival of Corynebacterium pseudotuberculosis within macrophages and induction of phagocytes death. Pol. J. Vet. Sci. 2010, 13, 143–149. [Google Scholar] [PubMed]
- Martin, M.D.; Badovinac, V.P. Defining Memory CD8 T Cell. Front. Immunol. 2018, 9, 2692. [Google Scholar] [CrossRef] [PubMed]
- Neefjes, J.; Jongsma, M.L.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef]
- Rock, K.L.; Reits, E.; Neefjes, J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 2016, 37, 724–737. [Google Scholar] [CrossRef]
- Aflalo, A.; Boyle, L.H. Polymorphisms in MHC class I molecules influence their interactions with components of the antigen processing and presentation pathway. Int. J. Immunogenet. 2021, 48, 317–325. [Google Scholar] [CrossRef]
- Radwan, J.; Babik, W.; Kaufman, J.; Lenz, T.L.; Winternitz, J. Advances in the evolutionary understanding of MHC Polymorphism. Trends Genet. 2020, 36, 298–311. [Google Scholar] [CrossRef]
- Peters, B.; Nielsen, M.; Sette, A. T Cell Epitope Predictions. Annu. Rev. Immunol. 2020, 38, 123–145. [Google Scholar] [CrossRef] [PubMed]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Dukkipati, V.S.R.; Blair, H.T.; Garrick, D.J.; Murray, A. Ovar-Mhc—Ovine major histocompatibility complex: Role in genetic resistance to diseases. N. Z. Vet. J. 2006, 54, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, N.S.; Morgan, E.F.; Wetherall, J.D.; Stear, M.J.; Groth, D.M. A comprehensive mapping of the structure and gene organisation in the sheep MHC class I region. BMC Genom. 2015, 16, 810. [Google Scholar] [CrossRef] [PubMed]
- Rehermann, B.; Nascimbeni, M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 2005, 5, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Sercan, O.; Stoycheva, D.; Hämmerling, G.J.; Arnold, B.; Schüler, T. IFN-gamma receptor signaling regulates memory CD8+ T cell differentiation. J. Immunol. 2010, 184, 2855–2862. [Google Scholar] [CrossRef] [PubMed]
- Cottalorda, A.; Mercier, B.C.; Mbitikon-Kobo, F.M. TLR2 engagement on memory CD8 (+) T cells improves their cytokine-mediated proliferation and IFN-gamma secretion in the absence of Ag. Eur. J. Immunol. 2009, 39, 2673–2681. [Google Scholar] [CrossRef]
- Mercier, B.C.; Cottalorda, A.; Coupet, C.A.; Marvel, J.; Bonnefoy-Bérard, N. TLR2 engagement on CD8 T cells enables generation of functional memory cells in response to a suboptimal TCR signal. J. Immunol. 2009, 182, 1860–1867. [Google Scholar] [CrossRef]
- Matloubian, M.; Concepcion, R.J.; Ahmed, R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 1994, 68, 8056–8063. [Google Scholar] [CrossRef]
- Kelley, L.; Mezulis, S.; Yates, C. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Söding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 2007, 23, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Jefferys, B.R.; Kelley, L.A.; Sternberg, M.J. Protein folding requires crowd control in a simulated cell. J. Mol. Biol. 2010, 397, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R. Jmol—A paradigm shift in crystallographic visualization. J. Appl. Crystallogr. 2010, 43, 1250–1260. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic. Acids. Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Emini, E.A.; Hughes, J.V.; Perlow, D.S.; Boger, J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985, 55, 836–839. [Google Scholar] [CrossRef]
- Kolaskar, A.S.; Tongaonkar, P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef]
- Lundegaard, C.; Lamberth, K.; Harndahl, M.; Buus, S.; Lund, O.; Nielsen, M. NetMHC-3.0: Accurate web accessible predictions of Human, Mouse, and Monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008, 36, W509–W512. [Google Scholar] [CrossRef]
- Wang, P.; Sidney, J.; Dow, C.; Mothé, B.; Sette, A.; Peters, B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 2008, 4, e1000048. [Google Scholar] [CrossRef] [PubMed]
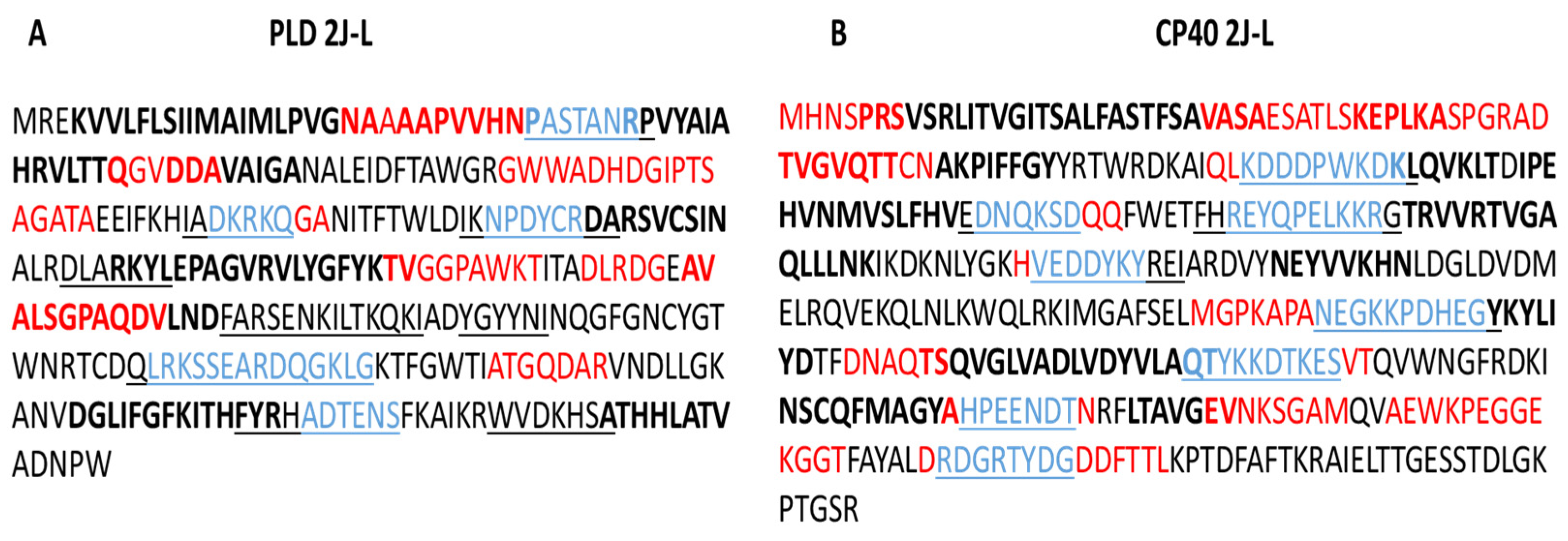

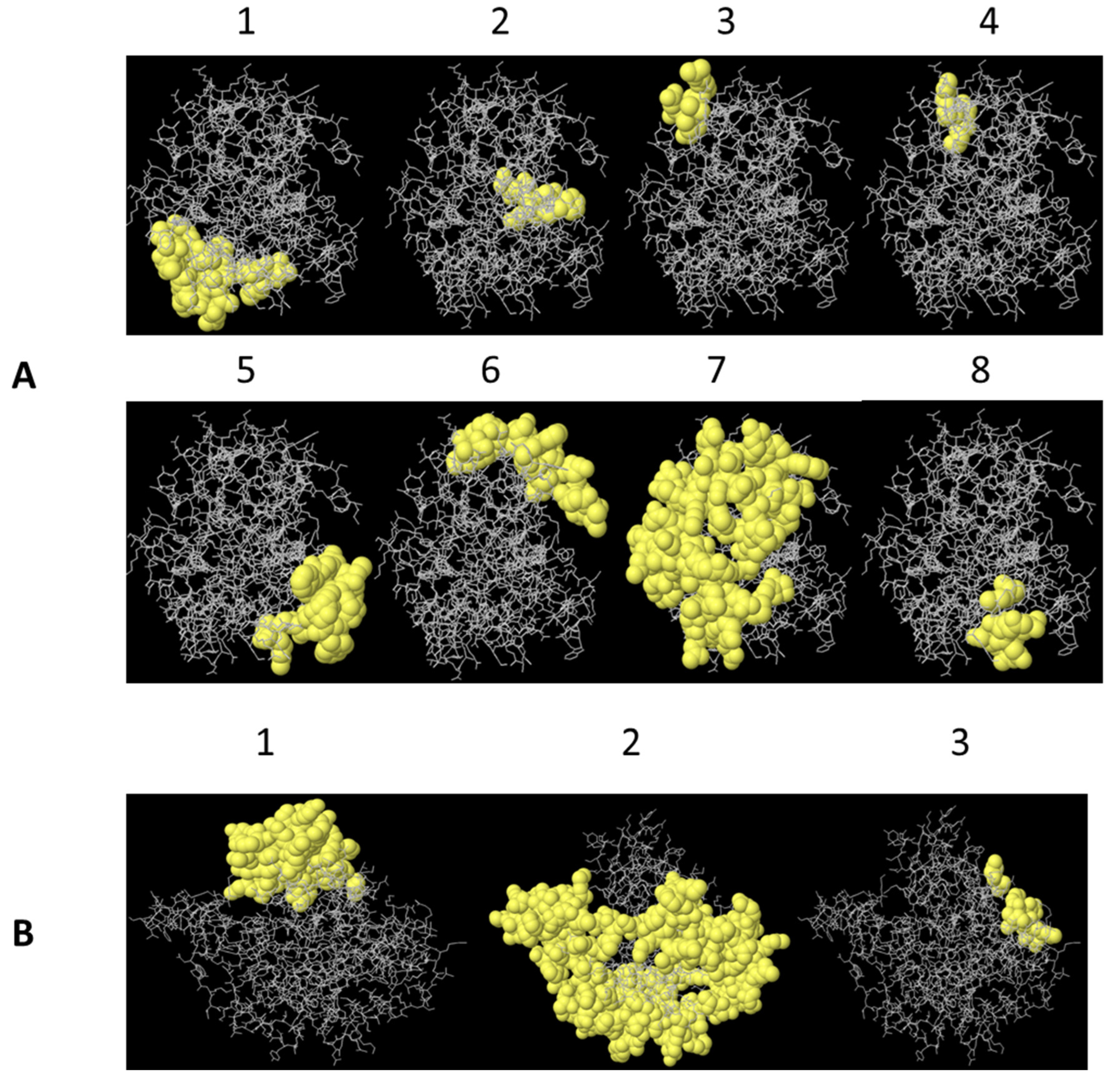
| Protein | No. | Epitopes | aa | Length | Score * |
|---|---|---|---|---|---|
| PLD 2J-L | 1 | AAPVVHNPASTAN | 24–36 | 13 | 0.511–0.639 |
| 2 | WWADHDGIPTSAGATAE | 74–90 | 17 | 0.510–0.656 | |
| 3 | DKRKQG | 98–103 | 6 | 0.519–0.538 | |
| 4 | DYCRDARSVC | 117–126 | 10 | 0.515–0.552 | |
| 5 | TVGGP | 153–157 | 5 | 0.509–0.527 | |
| 6 | QDVLNDFARSENKILTKQK | 179–197 | 19 | 0.502–0.582 | |
| 7 | YYNINQGFGNCYGTWNRTCDQLRKSSEARDQGKLG | 203–237 | 34 | 0.505–0.622 | |
| 8 | ATGQDAR | 245–251 | 7 | 0.501–0.549 | |
| 9 | THFYRHADTE | 271–280 | 10 | 0.507–0.591 | |
| 10 | DKHSATHHLATVA | 291–303 | 13 | 0.518–0.551 | |
| CP40 2J-L | 1 | PRSVS | 5–9 | 5 | 0.498–0.526 |
| 2 | AESATLSKEPLKASPGRADTVGVQTTCN | 31–58 | 28 | 0.508–0.670 | |
| 3 | RDKAIQLKDDDPWKDKLQVKLTD | 71–93 | 23 | 0.508–0.644 | |
| 4 | DNQKSDQQFWETFHRE | 108–123 | 16 | 0.532–0.594 | |
| 5 | LLLNKIKDKNLYGKHVEDDYK | 143–163 | 21 | 0.508–0.634 | |
| 6 | ELRQVEKQLNLKWQ | 189–202 | 14 | 0.501–0.644 | |
| 7 | LMGPKAPANEGKKPDHEG | 213–230 | 18 | 0.506–0.638 | |
| 8 | DNAQTSQ | 240–246 | 7 | 0.504–0.552 | |
| 9 | DTKESVTQVWNGFRDKINSCQF | 265–286 | 22 | 0.510–0.589 | |
| 10 | PEENDTNRFLTAVGEVNKSGAMQVAEWKPEGGE | 293–325 | 33 | 0.502–0.640 | |
| 11 | RTYDGDDFTTLKPTDFAF | 339–356 | 18 | 0.501–0.620 | |
| 12 | ELTTGESSTDLGKPT | 362–376 | 15 | 0.515–0.639 |
| Proteins | No. | aa Residues | Epitopes | Length | Score |
|---|---|---|---|---|---|
| PLD 2J-L | 1 | 9–33 | MREKVVLFLSIIMAIMLPVGNAAAAPVVHNPAS | 33 | 0.856 |
| 2 | 244–250 | IATGQDA | 7 | 0.721 | |
| 3 | 78–91 | HDGIPTSAGATAEE | 14 | 0.698 | |
| 4 | 211–226 | GNCYGTWNRTCDQLRK | 16 | 0.689 | |
| 5 | 275–285 | RHADTENSFKA | 11 | 0.684 | |
| 6 | 175–201 | SGPAQDVLNDFARSENKILTKQKIADY | 27 | 0.656 | |
| 7 | 132–142 | RDLARKYLEPA | 11 | 0.655 | |
| 8 | 116–125 | PDYCRDARSV | 10 | 0.642 | |
| 9 | 47–50 | LTTQ | 4 | 0.613 | |
| 10 | 69–76 | AWGRGWWA | 8 | 0.612 | |
| 11 | 160–168 | KTITADLRD | 9 | 0.583 | |
| 12 | 229–236 | EARDQGKL | 8 | 0.550 | |
| 13 | 289–292 | WVDK | 4 | 0.535 | |
| CP40 2J-L | 1 | 151–166 | KNLYGKHVEDDYKYRE | 16 | 0.807 |
| 2 | 220–229 | ANEGKKPDHE | 10 | 0.797 | |
| 3 | 9–46 | MHNSPRSVSRLITVGITSALFASTFSAVSASATLSKEPLKASPG | 46 | 0.749 | |
| 4 | 362–379 | ELTTGESSTDLGKPTGSR | 18 | 0.719 | |
| 5 | 337–355 | DGRTYDGDDFTTLKPTDFA | 19 | 0.707 | |
| 6 | 193–205 | VEKQLNLKWQLRK | 13 | 0.702 | |
| 7 | 294–315 | EENDTNRFLTAVGEVNKSGAMQ | 22 | 0.660 | |
| 8 | 262–281 | YKKDTKESVTQVWNGFRDKI | 20 | 0.638 | |
| 9 | 48–56 | ADTVGVQTT | 9 | 0.613 | |
| 10 | 318–326 | EWKPEGGEK | 9 | 0.604 | |
| 11 | 72–89 | DKAIQLKDDDPWKDKLQV | 18 | 0.588 | |
| 12 | 145–149 | LNKIK | 5 | 0.544 | |
| 13 | 240–245 | DNAQTS | 6 | 0.531 |
| Protein | No. | Epitopes | Lengh | Score |
|---|---|---|---|---|
| PLD 2J-L | 1 | _:M1, _:K4, _:V5, _:V6, _:L7, _:F8, _:L9, _:S10, _:I11, _:I12, _:M13, _:A14, _:I15, _:M16, _:L17, _:P18, _:V19, _:G20, _:N21, _:A22, _:A23, _:A24, _:A25, _:P26, _:V27, _:V28, _:H29, _:N30, _:P31, _:A32, _:S33, _:V290, _:D291, _:K292 | 34 | 0.822 |
| 2 | _:H276, _:A277, _:D278, _:T279, _:E280, _:N281, _:S282, _:F283 | 8 | 0.796 | |
| 3 | _:A135, _:R136, _:L139, _:E140, _:P141, _:A142 | 6 | 0.726 | |
| 4 | _:K101, _:Q102, _:G103 | 3 | 0.686 | |
| 5 | _:F210, _:G211, _:N212, _:C213, _:Y214, _:G215, _:T216, _:W217, _:N218, _:R219, _:T243, _:I244, _:A245, _:T246, _:G247, _:Q248, _:D249, _:A250, _:D254, _:G257, _:K258 | 21 | 0.675 | |
| 6 | _:L47, _:T48, _:T49, _:Q50, _:W75, _:A76, _:D77, _:D79, _:G80, _:I81, _:P82, _:T83, _:S84, _:A85, _:G86, _:A87, _:T88, _:E90, _:E91, _:D133, _:K137, _:Y138 | 22 | 0.662 | |
| 7 | _:A69, _:W70, _:G71, _:R72, _:G73, _:W74, _:N115, _:P116, _:D117, _:Y118, _:R120, _:D121, _:A122, _:S124, _:V125, _:I128, _:N129, _:R132, _:K152, _:T153, _:V154, _:G155, _:G156, _:P157, _:K160, _:T161, _:I162, _:T163, _:A164, _:D165, _:L166, _:R167, _:D168, _:S175, _:G176, _:P177, _:A178, _:Q179, _:D180, _:L182, _:N183, _:D184, _:A186, _:R187, _:S188, _:E189, _:N190, _:K191, _:I192, _:L193, _:T194, _:K195, _:Q196, _:K197, _:I198, _:A199, _:D200, _:Y201, _:G202, _:Y203, _:E229, _:A230, _:D232, _:Q233, _:G234, _:K235, _:L236 | 67 | 0.609 | |
| 8 | _:Q208, _:T220, _:C221, _:D222, _:Q223, _:R225, _:A259 | 7 | 0.529 | |
| CP40 2J-L | 1 | _:L145, _:N146, _:K147, _:I148, _:K149, _:D150, _:K151, _:N152, _:L153, _:Y154, _:G155, _:K156, _:H157, _:V158, _:E159, _:D160, _:D161, _:Y162, _:K163, _:Y164, _:R165, _:E166, _:R169, _:V193, _:E194, _:K195, _:Q196, _:L197, _:N198, _:L199, _:K200, _:W201, _:Q202, _:R204, _:K205, _:S245 | 36 | 0.721 |
| 2 | _:M1, _:H2, _:N3, _:S4, _:P5, _:R6, _:S7, _:V8, _:S9, _:R10, _:L11, _:I12, _:T13, _:V14, _:G15, _:I16, _:T17, _:S18, _:A19, _:L20, _:F21, _:A22, _:S23, _:T24, _:F25, _:S26, _:A27, _:V28, _:A29, _:A31, _:E32, _:S33, _:A34, _:T35, _:L36, _:S37, _:K38, _:E39, _:P40, _:L41, _:K42, _:A43, _:S44, _:P45, _:G46, _:R47, _:A48, _:D49, _:T50, _:V51, _:G52, _:V53, _:Q54, _:T55, _:T56, _:C57, _:D72, _:K73, _:A74, _:I75, _:Q76, _:L77, _:K78, _:D79, _:D80, _:D81, _:P82, _:W83, _:K84, _:D85, _:K86, _:L87, _:Q88, _:V89, _:Q110, _:K111, _:D113, _:Q114, _:R122, _:A220, _:N221, _:E222, _:G223, _:K224, _:K225, _:P226, _:D227, _:H228, _:E229, _:F239, _:D240, _:N241, _:A242, _:Q243, _:K263, _:K264, _:D265, _:T266, _:K267, _:E268, _:S269, _:V270, _:T271, _:Q272, _:V273, _:N282, _:P293, _:E294, _:E295, _:N296, _:D297, _:T298, _:N299, _:R300, _:F301, _:L302, _:T303, _:A304, _:V305, _:G306, _:E307, _:V308, _:N309, _:K310, _:S311, _:G312, _:M314, _:Q315, _:E318, _:W319, _:K320, _:P321, _:E322, _:G323, _:G324, _:E325, _:D337, _:G338, _:R339, _:T340, _:Y341, _:D342, _:G343, _:D344, _:D345, _:F346, _:T347, _:T348, _:L349, _:K350, _:P351, _:T352, _:D353, _:F354, _:A355, _:R359, _:T365, _:G366, _:E367, _:S368, _:S369, _:T370, _:D371, _:L372, _:G373, _:K374, _:P375, _:T376, _:G377, _:S378, _:R379 | 171 | 0.683 | |
| 3 | _:G248, _:L249, _:N275, _:G276, _:R278, _:D279, _:K280, _:I281 | 8 | 0.634 |
| PLD 2J-L aa | BepiPred 2.0 | Emini Surface Accessibility Scale | Antigenicity Kolaskar—Tongaonkar Scale | ElliPro Lineal | ElliPro Conformational |
|---|---|---|---|---|---|
| 1–50 | 24 AAPVVHNPASTAN 36 | 31 PASTANRP 38 | 4 KVVLFLSIIMAIMLPVGNA 22 24AAPVVHNP 31 36NRPVYAIAHRVLTTQ 50 | 1 MREKVVLFLSIIMAIMLPVGNAAAAPVVHNPAS 33 47 LTTQ 50 | 1 MKVVLFLSIIMAIMPVGNA AAAPVVHNPASVDK 292 47 LTTQWADDGIPTSAGATEEDKY 138 |
| 51–130 | 74 WWADHDGIPTSAG ATAE 90 98 DKRKQG 103 117 DYCRDARSVC 126 | 96 IADKRKQ 102 113 IKNPDYCRDA 122 | 53 DDAVAIGANALEID 66 116 PDYCRDARSVCSIN 129 | 69 AWGRGWWA 75 78 HDGIPTSAGATAEE 91 116 PDYCRDARSV 125 | 69 AWGRGWNPDYRDASVINRKTVGGPKTITADLRDSGPAQDLNDARSEN KILTKQKIADYGYEADQGKL 236 101 KQG 103 |
| 131–160 | 153 TVGGP 157 | 133 DLARKYL 139 | 131 LRDLARKYLEPAGVRVLYGFYKTVGGP 157 | 132 RDLARKYLEPA 142 | 135 ARLEPA 142 |
| 161–240 | 179 QDVLNDFARSENKILTKQK 197 203 YYNINQGFGNCYGTWNRTCDQLRKSSEARDQGKLG 237 | 185 FARSENKILTKQKI 198 223 QLRKSSEARDQGKLG 237 | 171 AVALSGPAQDVLND 184 198 IADYGYY 204 | 160 KTITADLRD 168 175 SGPAQDVLNDFARSENKILTKQKIADY 201 211 GNCYGTWNRTCDQLRK 226 229 EARDQGKL 236 | 208 QTCDQRA 259 210 FGNCYGTWNRTIATGQDADGK 258 |
| 240–260 | 245 ATGQDAR 251 | NP | 252 VNDLLGKA 259 | 244 IATGQDA 250 | * |
| 261–290 | 271 THFYRHADTE 280 | 273 YRHADTENS 282 | 261 VDGLIFGFKITHFYRH 276 | 275 RHADTENSFKA 285 | 276 HADTENSF 283 |
| 291–307 | 291 DKHSATHHLATVA 303 | 289 WVDKHSA 295 | 295 ATHHLATVA 303 | 289 WVDK 292 | * |
| CP40 2J-L aa | BepiPred 2.0 | Emini Surface Accessibility Scale | Antigenicity Kolaskar—Tongaonkar scale | ElliPro (Lineal) | ElliPro (Discontinuous) |
| 1–70 | 5 PRSVS 9 31 AESATLSKEPLKAS PGRADTVGVQTTCN 58 | NP | 5 PRSVSRLITVGITSALFASTFSAVASA 31 33 SATLSKEPLKAS 44 50 TVGVQTTCNAKPIFFGY 66 | 1 MHNSPRSVSRLITVGIT SALFASTFSAVASAESATLSKE PLKASPG 46 | 1MHNSPRSVSRLITVGITSALFASTFSAAAESATLSKEPLKA SPGRADTVGVQTTCDKAIQLKDDDPWKDKLQVQKDQRANEGKKPDHEFDNAQKKDTKESVTQVNPEENDTNRFLTA-VGEVNKSGMQEKPEGGE DGRTYD-GDDFTTLKPTDFART-GESSTDLGKPTGSR 379 |
| 71–170 | 71 RDKAIQLKDDDPWKDKLQVKLTD 93 108 DNQKSDQQFWETFHRE 123 143 LLLNKIKDKNLYGKHVEDDYK 163 | 78 KDDDPWKDKL 87 107 EDNQKSD 113 158 VEDDYKYREI 167 | 86 KLQVKLTDIPEHVNMVSLFHVE 107 133 TRVVRTVGAQLLLNK 147 | 72 DKAIQLKDDDPWKDKLQV 89 145 LNKIK 149 151 KNLYGKHVEDDYKYRE 166 | 145 LNKIKDKNLYGKHVEDDYKYRERVEKQLNLKWQRKS 245 |
| 171–260 | 189 ELRQVEKQLNLKWQ 202 213 LMGPKAPANEGKKPDHEG 230 240 DNAQTSQ 246 | 221 NEGKKP DHEGY 231 | 172 YNEYVVKHNL 181 189 ELRQVEKQLNLK 200 231 YKYLIYD 237 244 TSQVGLVADLVDYVLAQT 261 | 193 VEKQLNLKWQLRK 205 220 ANEGKKPDHE 229 240 DNAQTS 245 | * |
| 261–325 | 265 DTKESVTQVWNGFRDKINSCQF 286 293 PEENDTNRFLTAVGEVNKSGAMQVAEWKPEGGE 325 | 260 QTYKKDTKES 269 292 HPEENDT 298 | 281 INSCQFMAGYA 291 302 LTAVGEV 308 | 262 YKKDTKESVTQVWNGFRDKI 281 294 EENDTNRFLTAVGEVNKSGAMQ 315 318 EWKPEGGEK 326 | 248 GLNGRDKI 281 * |
| 326–379 | 339 RTYDGDDFTTLKPTDFAF 356 362 ELTTGESSTDLGKPT 376 | 336 RDGRTYDG 343 | 337 DGRTYDGDDFTTLKPTDFA 355 362 ELTTGESSTDLGKPTGSR 379 | * | |
| Protein PLD 2J-L | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No | Allele | Start | End | Epitope | Score | BepiPred 2.0 | Emini Surface Accessibility Scale | Antigenicity Kolaskar Tongaonkar Scale | Ellipro Lineal | Ellipro Conformational |
| 1 | H-2-Kb | 146 | 154 | VLYGFYKTV | 0.916 | 153′TV′154 | NP | 146′VLYGFYKTV′154 | NP | 152′KTV′154 |
| 2 | H-2-Kk | 279 | 286 | TENSFKAI | 0.87 | 279′TE′280 | 279′TEN′281 | NP | 279′TENSFKA′285 | 279′TENSF′283 |
| 3 | H-2-Kb | 104 | 111 | ANITFTWL | 0.822 | NP | NP | NP | NP | NP |
| 4 | H-2-Kk | 89 | 96 | AEEIFKHI | 0.791 | NP | 96′I | NP | 89′AEE′91 | 290′VDK′292 |
| 5 | H-2-Kd | 39 | 47 | VYAIAHRVL | 0.77 | NP | NP | 39′VYAIAHRVL′47 | 47′L | 47′L |
| 6 | H-2-Db | 56 | 65 | VAIGANALEI | 0.715 | NP | NP | 56′VAIGANALEI′65 | NP | NP |
| 7 | H-2-Kk | 139 | 147 | LEPAGVRVL | 0.715 | NP | NP | 139′LEPAGVRVL′147 | 139′LEPA′142 | 139′LEPA′142 |
| 8 | H-2-Kd | 202 | 210 | GYYNINQGF | 0.628 | 203′YYNINQGF′210 | 202′GYYNI′206 | 202′GYY′204 | NP | 202′GY203′208′Q 210′F |
| 9 | H-2-Kb | 4 | 11 | KVVLFLSI | 0.62 | NP | NP | 4′KVVLFLSI′11 | 4′KVVLFLSI′11 | 4′KVVLFLSI′11 |
| 10 | H-2-Kd | 137 | 146 | KYLEPAGVRV | 0.609 | NP | 137′KYL′139 | 137′KYLEPAGVRV′146 | 137′KYLEPA′142 | 137′KYLEPA′142 |
| 11 | H-2-Kb | 199 | 206 | ADYGYYNI | 0.553 | 203′YYNI′206 | 201′YGYYNI′206 | 199′ADYGYY′204 | 199′ADY′201 | 199′ADYGY′203 |
| 12 | H-2-Kk | 139 | 146 | LEPAGVRV | 0.545 | NP | 139′L | 139′LEPAGVRV′146 | 139′LEPA′142 | 139′LEPA′142 |
| 13 | H-2-Kk | 63 | 70 | LEIDFTAW | 0.526 | NP | NP | 63′LEID′70 | 69′AW′70 | 69′AW′70 |
| 14 | H-2-Kb | 56 | 63 | VAIGANAL | 0.524 | NP | NP | 56′VAIGANAL′63 | NP | NP |
| 15 | H-2-Kb | 266 | 273 | FGFKITHF | 0.522 | 271′THF′273 | 273′F | 266′FGFKIT -HF′273 | NP | NP |
| 16 | H-2-Qa1 | 138 | 146 | YLEPAGVRV | 0.516 | NP | 138′YL′139 | 138′YLEPAGVRV′146 | 138′YLEPA′142 | 139′LEPA′142 |
| Protein CP40 2J-L | ||||||||||
| 1 | H-2-Kk | 173 | 181 | NEYVVKHNL | 0.943 | NP | NP | 173′NEYVVKHNL′181 | NP | NP |
| 2 | H-2-Kk | 294 | 302 | EENDTNRFL | 0.927 | 194′EENDTNRFL′202 | 194′EENDT′298 | 302′L | 194′EENDTNRFL′202 | 194′EENDTNRFL′202 |
| 3 | H-2-Kk | 158 | 167 | VEDDYKYREI | 0.694 | 158′VEDDYK′163 | 158′VEDDYKYREI′167 | NP | 158′VEDDYKYRE′166 | 158′VEDDYKYRE′166 |
| 4 | H-2-Kb | 205 | 213 | KIMGAFSEL | 0.672 | NP | NP | NP | 205′K | NP |
| 5 | H-2-Qa1 | 137 | 145 | RTVGAQLLL | 0.664 | 143′LL′145 | NP | 137′RTVGAQLLL′145 | 145′L | 145′L |
| 6 | H-2-Kb | 206 | 213 | IMGAFSEL | 0.654 | 213′L | NP | NP | NP | NP |
| 7 | H-2-Kk | 228 | 235 | HEGYKYLI | 0.651 | 228′HEG′230 | 228′HEGY′231 | 231′YKYLI′235 | 228′HE′229 | 228′HE′229, |
| 8 | H-2-Qa1 | 333 | 341 | ALDRDGRTY | 0.645 | 339′RTY′341 | 336′RDGRTY′341 | NP | 337′DGRTY′341 | 339′R |
| 9 | H-2-Kk | 172 | 181 | YNEYVVKHNL | 0.61 | NP | NP | 172′YNEYVVKHNL′181 | NP | NP |
| 10 | H-2-Kd | 163 | 171 | KYREIARDV | 0.58 | 163′K | 163′KYREI′167 | NP | 163′KYRE′166 | 163′KYRE′166 169′R 171′V |
| 11 | H-2-Kk | 294 | 301 | EENDTNRF | 0.568 | 294′EENDTNRF′301 | 294′EENDT′298 | NP | 294′EENDTNRF′301 | 294′EENDTNRF′301 |
| 12 | H-2-Qa1 | 205 | 213 | KIMGAFSEL | 0.539 | 213′L | NP | NP | 205′K | NP |
| 13 | H-2-Kb | 339 | 349 | RTYDGDDFTTL | 0.53 | 339′RTYDGDDFTTL′349 | 339′MRTYDG’ 343 | NP | 339′RTYDGDDFTTL′349 | 339′RTYDGDDFTTL′349 |
| 14 | H-2-Kb | 135 | 143 | VVRTVGAQL | 0.52 | 143′L | NP | 135′VVRTVGAQL′143 | NP | NP |
| 15 | H-2-Kb | 17 | 25 | TSALFASTF | 0.502 | NP | NP | 17′TSALFASTF′25 | 17′TSALFASTF′25 | 17′TSALFASTF′25 |
| Protein PLD 2J-L | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No | Allele | Start | End | Epitope | Score | BepiPred 2.0 | Emini Surface Accessibility Scale | Antigenicity Kolaskar Tongaonkar Scale | Ellipro Lineal | Ellipro Conformational |
| 1 | H2-IAb | 24 | 38 | AAPVVHNPASTANRP | 0.6864 | 24′AAPVVHNPASTAN′36 | 31′PASTANRP′38 | 24′AAPVVHNP′31, 36′NRP′38 | 24′AAPVVHNPAS′33 | 24′AAPVVHNPAS′33 |
| 2 | H2-IAb | 25 | 39 | APVVHNPASTANRPV | 0.6886 | 25′APVVHNPASTAN′36 | 31′PASTANRP′38 | 36′NRPV′39 | 25′APVVHNPAS′33 | 25′APVVHNPAS′33 |
| 3 | H2-IAb | 20 | 34 | GNAAAAPVVHNPAST | 0.662 | 24′AAPVVHNPAST′34 | 31′PAST′34 | 24′AAPVVHNP′31 | 20′GNAAAAPVVHNPAS′33 | 20′GNAAAAPVVHNPAS′33 |
| 4 | H2-IAb | 271 | 285 | THFYRHADTENSFKA | 0.6544 | 271′THFYRHADTE′280 | 273′YRHADTENS′282 | 271′THFYRH′276 | 275′RHADTENSFKA′285 | 276′HADTENSF′283 |
| 5 | H2-IAb | 19 | 33 | VGNAAAAPVVHNPAS | 0.6458 | 24′AAPVVHNPAS′33 | 31′PAS′33 | 24′AAPVVHNP′31 | 19′VGNAAAAPVVHNPAS′33 | 19′VGNAAAAPVVHNPAS′33 |
| 6 | H2-IAb | 270 | 284 | ITHFYRHADTENSFK | 0.6179 | 271′THFYRHADTE′280 | 273′YRHADTENS′282 | 270′ITHFYRH′276 | 275′RHADTENSFK′284 | 276′HADTENSF′283 |
| 7 | H2-IAb | 18 | 32 | PVGNAAAAPVVHNPA | 0.546 | 24′AAPVVHNPA′32 | 31′PA′32 | 24′AAPVVHNP′31 | 20′PVGNAAAAPVVHNPA′32 | 18′PVGNAAAAPVVHNPA′32 |
| 8 | H2-IAb | 23 | 37 | AAAPVVHNPASTANR | 0.5471 | 24′AAPVVHNPASTAN′36 | 31′PASTANR′37 | 24′AAPVVHNP′31, 36′NR′37 | 23′AAAPVVHNPAS′33 | 23′AAAPVVHNPAS′33 |
| 9 | H2-IAd | 167 | 181 | RDGEAVALSGPAQDV | 0.6131 | 179’ QDV’ 181 | NP | 171 AVALSGPAQDV′181 | 167′RD′168, 175′SGPAQDV′181 | 167′RD′168, 175′SGPAQD′180 |
| 10 | H2-IAd | 166 | 180 | LRDGEAVALSGPAQD | 0.5894 | 179’ QD’ 180 | NP | 171 AVALSGPAQD′180 | 166′DLRD′168 | 166′LRD′168, 175′SGPAQD′180 |
| Protein CP40 2J-L | ||||||||||
| 1 | H2-IAb | 210 | 224 | FSELMGPKAPANEGK | 0.7198 | 213′LMGPKAPANEGK′224 | 221′NEGK′224 | NP | 220′ANEGK′224 | 220′ANEGK′224 |
| 2 | H2-IAb | 209 | 223 | AFSELMGPKAPANEG | 0.6642 | 213′LMGPKAPANEG′223 | 221′NEG′223 | NP | 220′ANEG′223 | 220′ANEG′223 |
| 3 | H2-IAb | 208 | 222 | GAFSELMGPKAPANE | 0.5786 | 213′LMGPKAPANEG′222 | 221′NE′222 | NP | 220′ANE′222 | 220′ANE′222 |
| 4 | H2-IAb | 286 | 300 | FMAGYAHPEENDTNR | 0.5793 | 286′F, 293′PEENDTNR′300 | 292 HPEENDT 298 | 286′FMAGYA′291 | 294′EENDTNR′300 | 293′PEENDTNR′300 |
| 5 | H2-IAb | 211 | 225 | SELMGPKAPANEGKK | 0.5847 | 213′LMGPKAPANEGKK 225 | 221′NEGKK′225 | NP | 220′ANEGK′225 | 220′ANEGK′224 |
| 6 | H2-IAb | 283 | 297 | SCQFMAGYAHPEEND | 0.5853 | 283′SCQF′286, 293′PEEND′297 | 292′HPEEND’ 297 | 283′NSCQFMAGYA′291 | 294′EEND′297 | 293′PEEND′297 |
| 7 | H2-IAb | 287 | 301 | MAGYAHPEENDTNRF | 0.5647 | 293′PEENDTNRF′301 | 292 HPEENDT 298 | 287′FMAGYA′291 | 294′EENDTNRF′301 | 293′PEENDTNRF′301 |
| 8 | H2-IAd | 243 | 257 | QTSQVGLVADLVDYV | 0.6316 | 243′QTSQ′246 | NP | 244′TSQVGLVADLVDY′256 | 243′QTS′245 | 243′Q, 245′S, 248′GL′249 |
| 9 | H2-IAd | 242 | 256 | AQTSQVGLVADLVDY | 0.6365 | 243′AQTSQ′246 | NP | 244′TSQVGLVADLVDYV′258 | 243′QTS′245 | 242′AQ′243, 245′S, 248′GL′249 |
| 10 | H2-IAb | 285 | 299 | QFMAGYAHPEENDTN | 0.5379 | 285′QF′286, 293′PEENDTN′325 | 292′HPEENDT′298 | 285′QFMAGYA′291 | 294′EENDTN′299 | 293′PEENDTN′299 |
| 11 | H2-IAd | 265 | 279 | DTKESVTQVWNGFRD | 0.612 | 265′DTKESVTQVWNGFRD′279 | 265′DTKES′269 | NP | 265′DTKESVTQVWNGFRD′279 | 265′DTKESVTQV′273 |
| 12 | H2-IAb | 285 | 299 | QFMAGYAHPEENDTN | 0.5379 | 285′QF′286, 293′PEENDTN′299 | 292′HPEENDT′298 | 285′QFMAGYA′291 | 294′EENDTN′299 | 293′PEENDTN′299 |
| 13 | H2-IAd | 265 | 279 | DTKESVTQVWNGFRD | 0.612 | 265′DTKESVTQVWNGFRD′279 | 265′DTKES′269 | NP | 265′DTKESVTQVWNGFRD′279 | 265′DTKESVTQV′273 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Domínguez, M.C.; Montes-de-Oca-Jiménez, R.; Vázquez-Chagoyán, J.C.; Rivadeneira-Barreiro, P.E.; Zambrano-Rodríguez, P.C.; Ruiz-Riva-Palacio, M.E.; Gutiérrez-Castillo, A.d.C.; de-Castro-Soares, S.; Vieyra-Reyes, P.; Arteaga-Troncoso, G. Bioinformatic Approach of B and T Cell Epitopes of PLD and CP40 Proteins of Corynebacterium pseudotuberculosis ovis Mexican Isolate 2J-L towards a Peptide-Based Vaccine. Int. J. Mol. Sci. 2024, 25, 270. https://doi.org/10.3390/ijms25010270
Rodríguez-Domínguez MC, Montes-de-Oca-Jiménez R, Vázquez-Chagoyán JC, Rivadeneira-Barreiro PE, Zambrano-Rodríguez PC, Ruiz-Riva-Palacio ME, Gutiérrez-Castillo AdC, de-Castro-Soares S, Vieyra-Reyes P, Arteaga-Troncoso G. Bioinformatic Approach of B and T Cell Epitopes of PLD and CP40 Proteins of Corynebacterium pseudotuberculosis ovis Mexican Isolate 2J-L towards a Peptide-Based Vaccine. International Journal of Molecular Sciences. 2024; 25(1):270. https://doi.org/10.3390/ijms25010270
Chicago/Turabian StyleRodríguez-Domínguez, Maria Carla, Roberto Montes-de-Oca-Jiménez, Juan Carlos Vázquez-Chagoyán, Pilar Eliana Rivadeneira-Barreiro, Pablo Cleomenes Zambrano-Rodríguez, Martha Elba Ruiz-Riva-Palacio, Adriana del Carmen Gutiérrez-Castillo, Siomar de-Castro-Soares, Patricia Vieyra-Reyes, and Gabriel Arteaga-Troncoso. 2024. "Bioinformatic Approach of B and T Cell Epitopes of PLD and CP40 Proteins of Corynebacterium pseudotuberculosis ovis Mexican Isolate 2J-L towards a Peptide-Based Vaccine" International Journal of Molecular Sciences 25, no. 1: 270. https://doi.org/10.3390/ijms25010270
APA StyleRodríguez-Domínguez, M. C., Montes-de-Oca-Jiménez, R., Vázquez-Chagoyán, J. C., Rivadeneira-Barreiro, P. E., Zambrano-Rodríguez, P. C., Ruiz-Riva-Palacio, M. E., Gutiérrez-Castillo, A. d. C., de-Castro-Soares, S., Vieyra-Reyes, P., & Arteaga-Troncoso, G. (2024). Bioinformatic Approach of B and T Cell Epitopes of PLD and CP40 Proteins of Corynebacterium pseudotuberculosis ovis Mexican Isolate 2J-L towards a Peptide-Based Vaccine. International Journal of Molecular Sciences, 25(1), 270. https://doi.org/10.3390/ijms25010270








