Effects of Adipose Tissue-Specific Knockout of Delta-like Non-Canonical Notch Ligand 1 on Lipid Metabolism in Mice
Abstract
:1. Introduction
2. Results
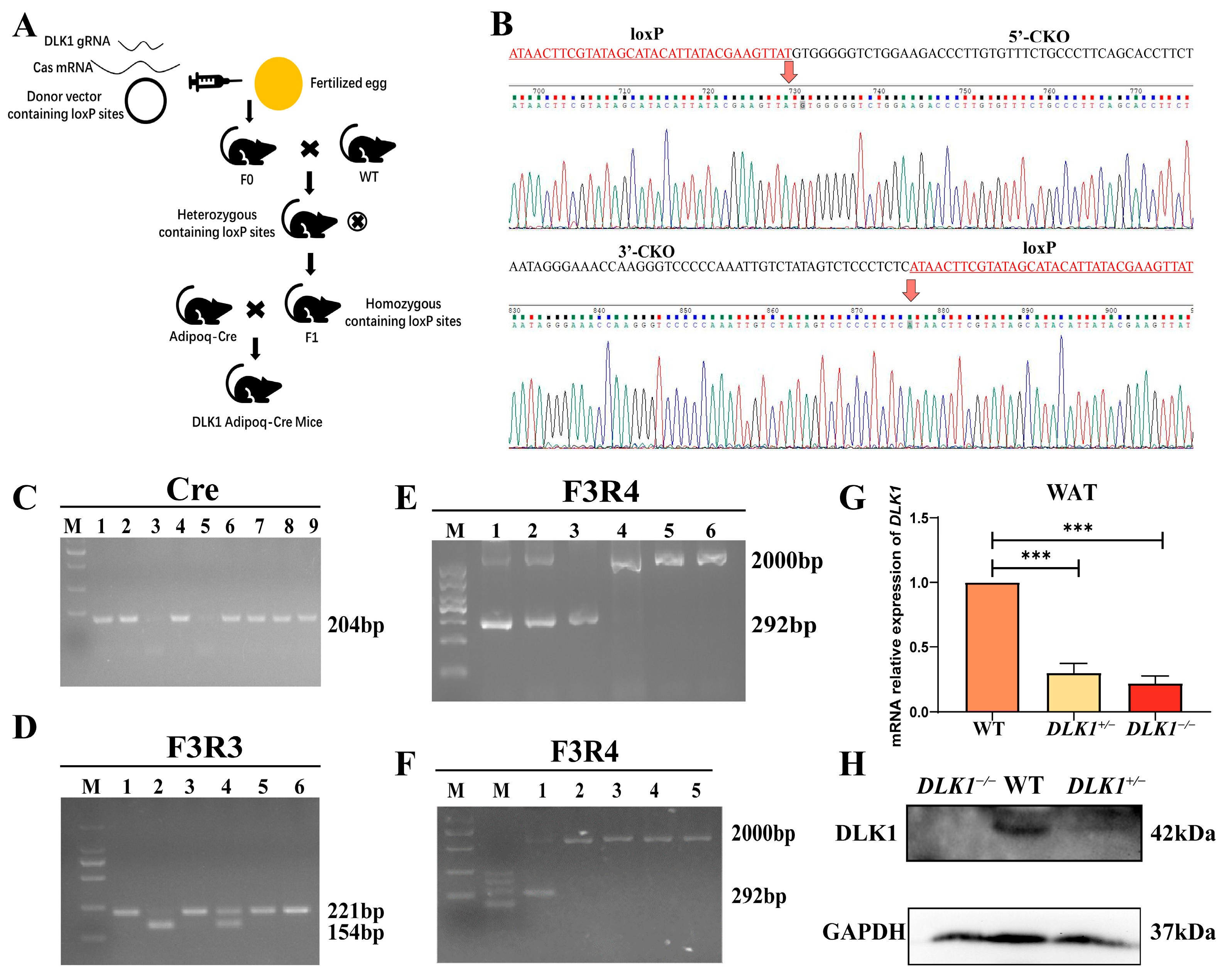
2.1. The Identification of Adipose Tissue-Specific Knockout of DLK1 Mice (DLK1−/− Mice)
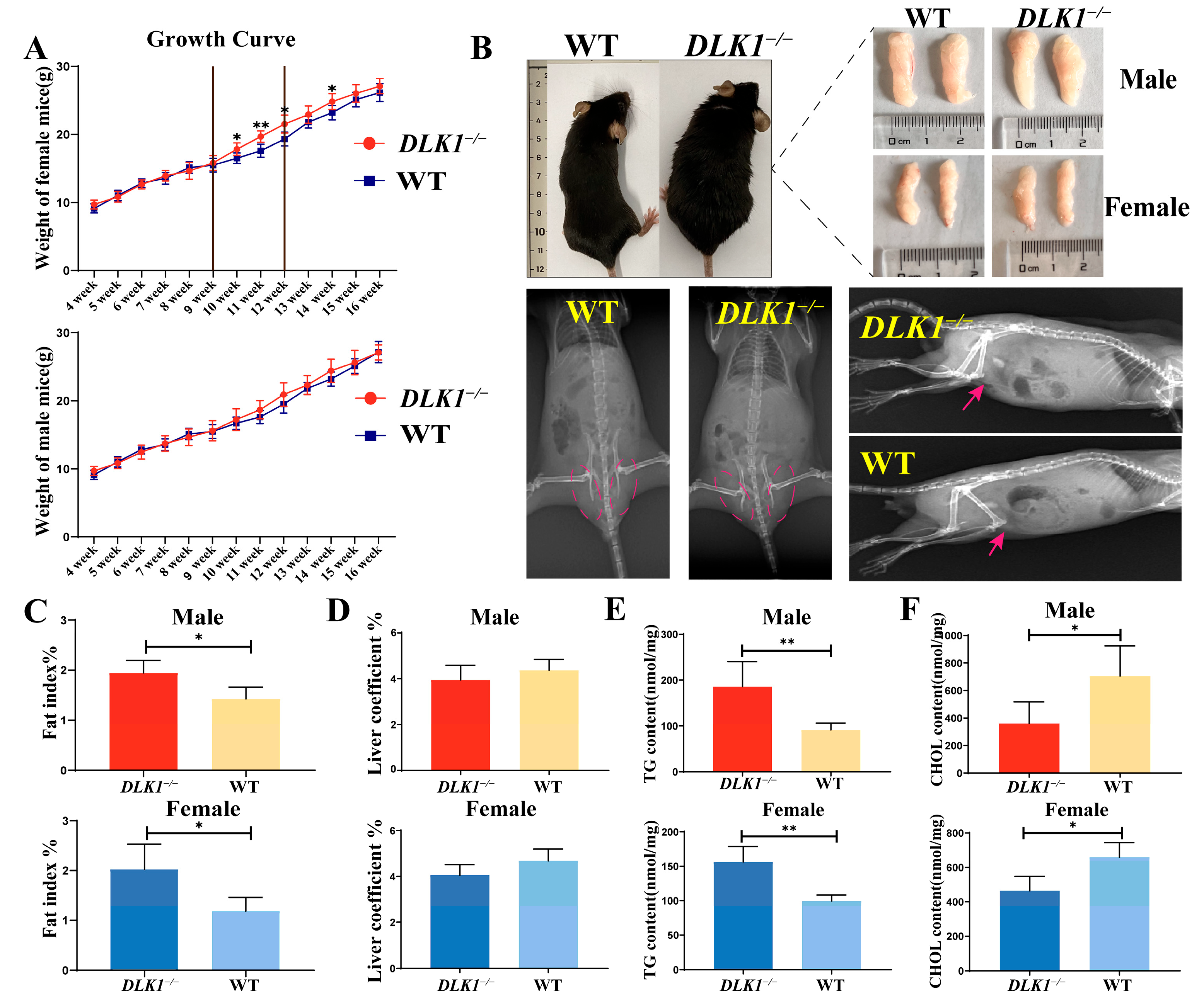
2.2. Effects on Growth and Development Traits in DLK1−/− Mice
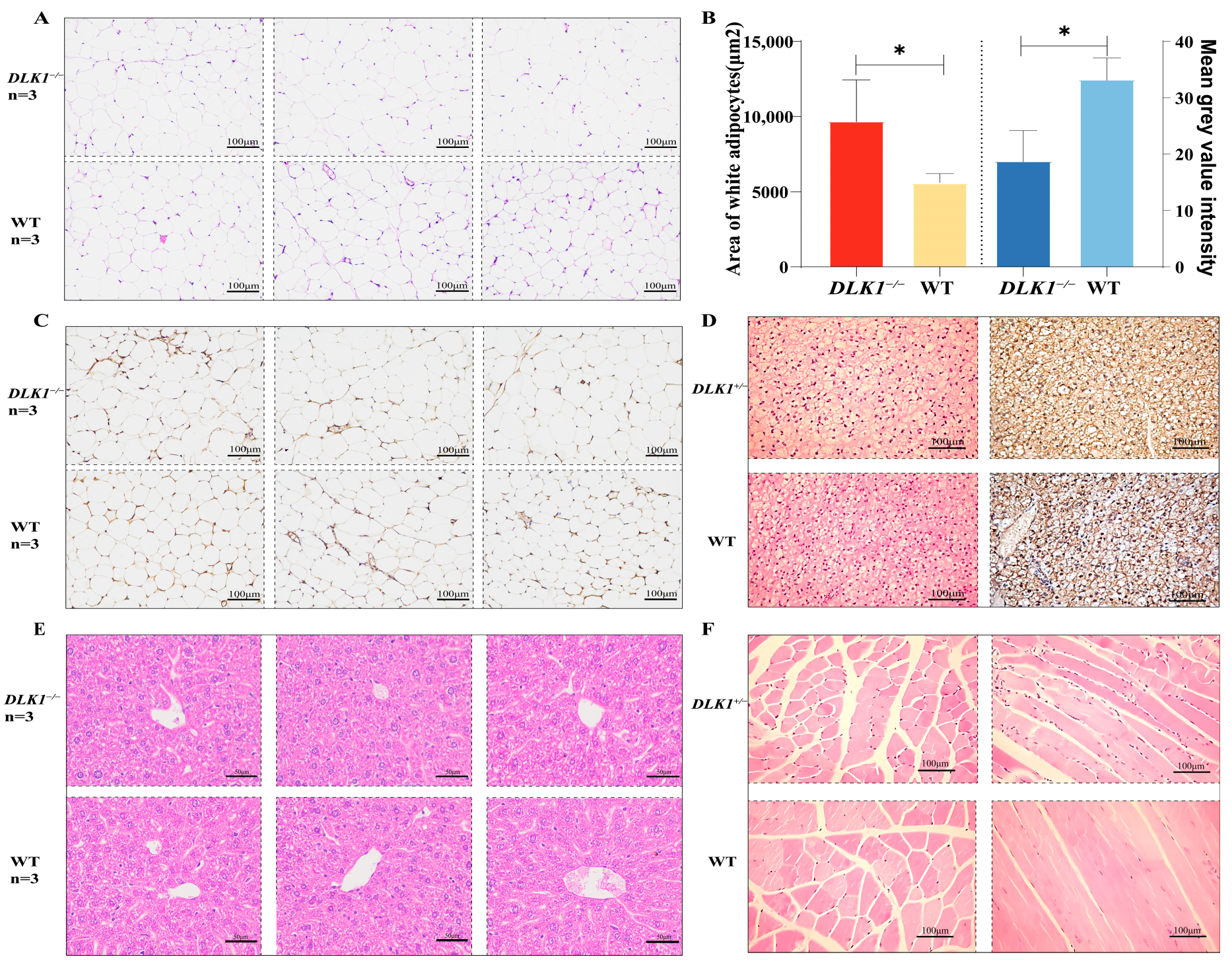
2.3. Examination of Tissue Morphology in DLK1−/− Mice
2.4. Detection of Blood Biochemistry Index in DLK1−/− Mice
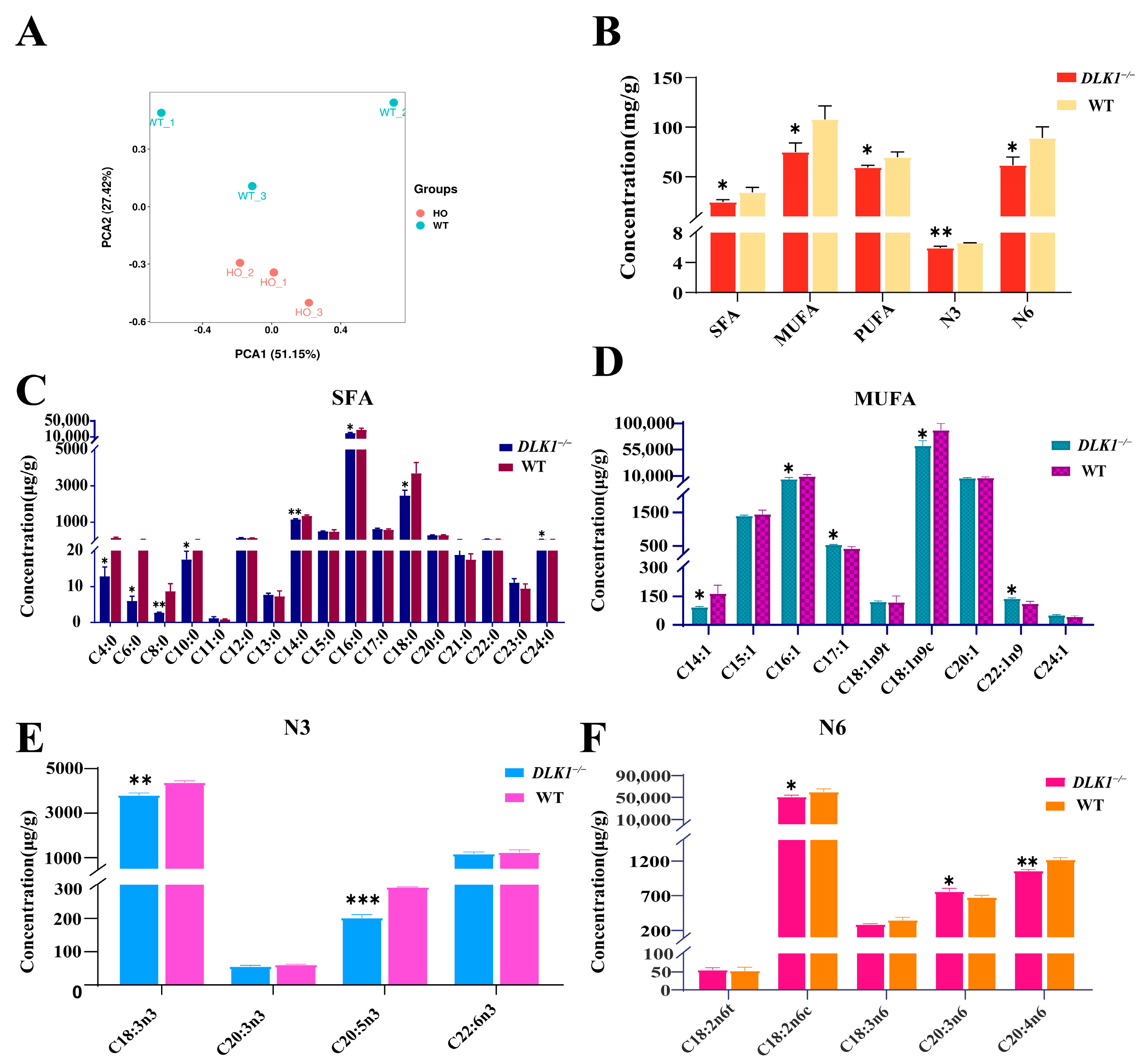
2.5. Effect of Tissue-Specific Knockout of DLK1 on Fatty Acids Content in White Adipose Tissue
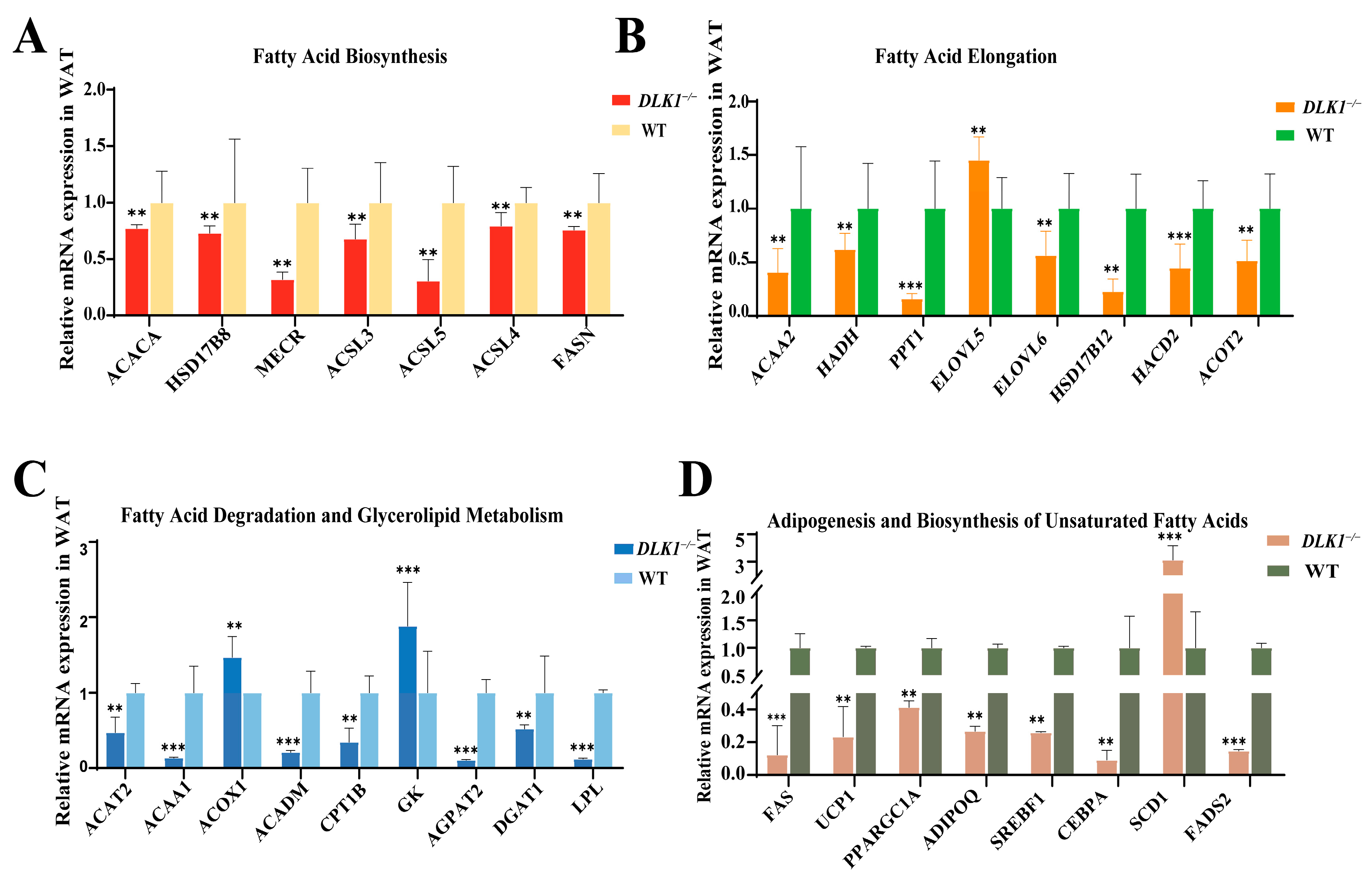
2.6. Differentially Expressed Genes Related to Lipid Metabolism in WAT of DLK1−/− Mice
3. Discussion
4. Materials and Methods
4.1. Preparation of Adipose Tissue-Specific Knockout Mice (DLK1−/− Mice)
4.2. Identification of Mice Genotypes
4.3. Analysis of Triglyceride and Cholesterol Contents in Adipose Mice Tissue
4.4. Tissue HE Staining and Immunohistochemistry
4.5. Assay of Blood Biochemical Indices in Mice
4.6. Analysis of Genes Related to Lipid Metabolism Expression by Quantitative Real-Time PCR
4.7. Fatty Acid Detection in Mouse Adipose Tissue
4.8. Western Blot
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bian, X.; Liu, R.; Meng, Y.; Xing, D.; Xu, D.; Lu, Z. Lipid metabolism and cancer. J. Exp. Med. 2021, 218, 2610–2623. [Google Scholar] [CrossRef] [PubMed]
- Hocquette, J.F.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D.W. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Li, M.; Sun, X.; Plath, M.; Li, C.; Lan, X.; Lei, C.; Huang, Y.; Bai, Y.; Qi, X.; et al. Global Transcriptome Analysis During Adipogenic Differentiation and Involvement of Transthyretin Gene in Adipogenesis in Cattle. Front. Genet. 2018, 9, 463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Bin, Y.; Zhang, J.; Cui, L.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Ren, J.; Huang, L. Genome-wide association studies for fatty acid metabolic traits in five divergent pig populations. Sci. Rep. 2016, 6, 24718. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Virtue, S.; Goie, J.Y.G.; Png, C.W.; Guo, J.; Li, Y.; Jiao, H.; Chua, Y.L.; Campbell, M.; Moreno-Navarrete, J.M.; et al. Regulation of adipogenic differentiation and adipose tissue inflammation by interferon regulatory factor 3. Cell Death Differ. 2021, 28, 3022–3035. [Google Scholar] [CrossRef] [PubMed]
- Smas, C.M.; Sul, H.S. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993, 73, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Takada, S.; Tevendale, M.; Baker, J.; Georgiades, P.; Campbell, E.; Freeman, T.; Johnson, M.H.; Paulsen, M.; Ferguson-Smith, A.C. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr. Biol. 2000, 10, 1135–1138. [Google Scholar] [CrossRef]
- Smas, C.M.; Chen, L.; Zhao, L.; Latasa, M.J.; Sul, H.S. Transcriptional repression of pref-1 by glucocorticoids promotes 3T3-L1 adipocyte differentiation. J. Biol. Chem. 1999, 274, 12632–12641. [Google Scholar] [CrossRef]
- Lu, H.P.; Lin, C.J.; Chen, W.C.; Chang, Y.J.; Lin, S.W.; Wang, H.H.; Chang, C.J. TRIM28 Regulates Dlk1 Expression in Adipogenesis. Int. J. Mol. Sci. 2020, 21, 7245. [Google Scholar] [CrossRef]
- Sul, H.S. Minireview: Pref-1: Role in adipogenesis and mesenchymal cell fate. Mol. Endocrinol. 2009, 23, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Geach, T. Metabolism: DLK1 levels predict fetal growth restriction. Nat. Rev. Endocrinol. 2017, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Charalambous, M.; Ferron, S.R.; da Rocha, S.T.; Murray, A.J.; Rowland, T.; Ito, M.; Schuster-Gossler, K.; Hernandez, A.; Ferguson-Smith, A.C. Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 2012, 15, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Cleaton, M.A.; Dent, C.L.; Howard, M.; Corish, J.A.; Gutteridge, I.; Sovio, U.; Gaccioli, F.; Takahashi, N.; Bauer, S.R.; Charnock-Jones, D.S.; et al. Fetus-derived DLK1 is required for maternal metabolic adaptations to pregnancy and is associated with fetal growth restriction. Nat. Genet. 2016, 48, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.S.; Smas, C.M.; Lee, K.; Villena, J.A.; Kim, K.H.; Yun, E.J.; Sul, H.S. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol. Cell. Biol. 2002, 22, 5585–5592. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, E.; Kuzinski, J.; Komolka, K.; Gotoh, T.; Maak, S. Localization and abundance of early markers of fat cell differentiation in the skeletal muscle of cattle during growth—Are DLK1-positive cells the origin of marbling flecks? Meat Sci. 2015, 100, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Waddell, J.N.; Zhang, P.; Wen, Y.; Gupta, S.K.; Yevtodiyenko, A.; Schmidt, J.V.; Bidwell, C.A.; Kumar, A.; Kuang, S. Dlk1 is necessary for proper skeletal muscle development and regeneration. PLoS ONE 2010, 5, e15055. [Google Scholar] [CrossRef]
- Magee, D.A.; Berry, D.P.; Berkowicz, E.W.; Sikora, K.M.; Howard, D.J.; Mullen, M.P.; Evans, R.D.; Spillane, C.; MacHugh, D.E. Single nucleotide polymorphisms within the bovine DLK1-DIO3 imprinted domain are associated with economically important production traits in cattle. J. Hered. 2011, 102, 94–101. [Google Scholar] [CrossRef]
- Li, J.; Yu, D.; Wang, J.; Li, C.; Wang, Q.; Wang, J.; Du, W.; Zhao, S.; Pang, Y.; Hao, H.; et al. Identification of the porcine IG-DMR and abnormal imprinting of DLK1-DIO3 in cloned pigs. Front. Cell Dev. Biol. 2022, 10, 964045. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, P.; Yu, X.; Mi, J.; Bai, Z.; Zhang, X.; Liu, Y.; Fang, X.; Yang, R.; Zhao, Z. Analysis of the Bovine DLK1 Gene Polymorphism and Its Relation to Lipid Metabolism in Chinese Simmental. Animals 2020, 10, 923. [Google Scholar] [CrossRef]
- Palczewski, G.; Widjaja-Adhi, M.A.; Amengual, J.; Golczak, M.; von Lintig, J. Genetic dissection in a mouse model reveals interactions between carotenoids and lipid metabolism. J. Lipid Res. 2016, 57, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Farrar, J.S.; Lownik, J.C.; Way, G.W.; Rodriguez, M.C.; Celi, F.S.; Martin, R.K. Identification of the transgene insertion site for an adipocyte-specific adiponectin-cre model and characterization of the functional consequences. Adipocyte 2021, 10, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zhu, J. Immunologic applications of conditional gene modification technology in the mouse. Curr. Protoc. Immunol. 2014, 105, 10.34.11–10.34.13. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Sage, M.; Sheppeard, E.A.; Jurecic, V.; Bradley, A. Engineering mouse chromosomes with Cre-loxP: Range, efficiency, and somatic applications. Mol. Cell. Biol. 2000, 20, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Jima, D.D.; Skaar, D.A.; Planchart, A.; Motsinger-Reif, A.; Cevik, S.E.; Park, S.S.; Cowley, M.; Wright, F.; House, J.; Liu, A.; et al. Genomic map of candidate human imprint control regions: The imprintome. Epigenetics 2022, 17, 1920–1943. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a018382. [Google Scholar] [CrossRef] [PubMed]
- Wylie, A.A.; Murphy, S.K.; Orton, T.C.; Jirtle, R.L. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000, 10, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Enterina, J.R.; Enfield, K.S.S.; Anderson, C.; Marshall, E.A.; Ng, K.W.; Lam, W.L. DLK1-DIO3 imprinted locus deregulation in development, respiratory disease, and cancer. Expert Rev. Respir Med. 2017, 11, 749–761. [Google Scholar] [CrossRef]
- Mortensen, S.B.; Jensen, C.H.; Schneider, M.; Thomassen, M.; Kruse, T.A.; Laborda, J.; Sheikh, S.P.; Andersen, D.C. Membrane-tethered delta-like 1 homolog (DLK1) restricts adipose tissue size by inhibiting preadipocyte proliferation. Diabetes 2012, 61, 2814–2822. [Google Scholar] [CrossRef]
- Traustadottir, G.A.; Kosmina, R.; Sheikh, S.P.; Jensen, C.H.; Andersen, D.C. Preadipocytes proliferate and differentiate under the guidance of Delta-like 1 homolog (DLK1). Adipocyte 2013, 2, 272–275. [Google Scholar] [CrossRef]
- Maki, K.C.; Dicklin, M.R.; Kirkpatrick, C.F. Saturated fats and cardiovascular health: Current evidence and controversies. J. Clin. Lipidol. 2021, 15, 765–772. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, C.; Caramujo, M.J. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Torres-Gonzalez, M.; Tripathy, S.; Botolin, D.; Christian, B.; Jump, D.B. Elevated hepatic fatty acid elongase-5 activity affects multiple pathways controlling hepatic lipid and carbohydrate composition. J. Lipid Res. 2008, 49, 1538–1552. [Google Scholar] [CrossRef] [PubMed]
- Dobrzyn, A.; Ntambi, J.M. The role of stearoyl-CoA desaturase in the control of metabolism. Prostagland. Leukot Essent Fat. Acids 2005, 73, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Dales, N.A.; Winther, M.D. Opportunities and challenges in developing stearoyl-coenzyme A desaturase-1 inhibitors as novel therapeutics for human disease. J. Med. Chem. 2014, 57, 5039–5056. [Google Scholar] [CrossRef] [PubMed]
- Dipple, K.M.; Zhang, Y.H.; Huang, B.L.; McCabe, L.L.; Dallongeville, J.; Inokuchi, T.; Kimura, M.; Marx, H.J.; Roederer, G.O.; Shih, V.; et al. Glycerol kinase deficiency: Evidence for complexity in a single gene disorder. Hum. Genet. 2001, 109, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Matschinsky, F.M. Regulation of pancreatic beta-cell glucokinase: From basics to therapeutics. Diabetes 2002, 51 (Suppl. 3), S394–S404. [Google Scholar] [CrossRef]
- Navarro-Tableros, V.; Fiordelisio, T.; Hernández-Cruz, A.; Hiriart, M. Physiological development of insulin secretion, calcium channels, and GLUT2 expression of pancreatic rat beta-cells. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E1018–E1029. [Google Scholar] [CrossRef]
- Kramer, J.; Moeller, E.L.; Hachey, A.; Mansfield, K.G.; Wachtman, L.M. Differential expression of GLUT2 in pancreatic islets and kidneys of New and Old World nonhuman primates. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R786–R793. [Google Scholar] [CrossRef]
- Zhang, L.; Uezumi, A.; Kaji, T.; Tsujikawa, K.; Andersen, D.C.; Jensen, C.H.; Fukada, S.I. Expression and Functional Analyses of Dlk1 in Muscle Stem Cells and Mesenchymal Progenitors during Muscle Regeneration. Int. J. Mol. Sci. 2019, 20, 3269. [Google Scholar] [CrossRef]

| Primers | Sequences (5′–3′) |
|---|---|
| Mmu-DLK1-F3 | CACTGTAGATCACTGTACCAACTG |
| Mmu-DLK1-R3 | CATTTGGTGGTCTTTGTTGAGTCC |
| Mmu-DLK1-R4 | TTAGAATGGAGGGGTCCTACCCTA |
| A-Cre-F | GAACGCACTGATTTCGACCA |
| A-Cre-R | GCTAACCAGCGTTTTCGTTC |
| Primers | Forward Sequences (5′–3′) | Reverse Sequences (5′–3′) |
|---|---|---|
| DLK1 | TGTGACAAGTGTGTAACTGCC | TTCCATTGTTGGCGCAG |
| β-actin | GCACCACACCTTCTACAA | TACGACCAGAGGCATACA |
| ACACA | GCTATGGAAGTCGGCTATGGA | TTGTCAGGAAGAGGCGGATG |
| HSD17B8 | GTCGTGGCTCCATCATCAAC | GCAGTTACCTTGTCCTTCACTT |
| MECR | CGATTGGCTCTCAACTGTGTT | TCATCTGGACTGTGGTTCTTCT |
| ACSL3 | GCGAGAAGGATTCCAAGACTG | TGTGGTGAAGAGTAGCCGATT |
| ACSL5 | AAGTTGGCACAAGGAGAATACA | CCGATCAGGAATGACCGTAAG |
| ACSL4 | GCGTTCCTCCAAGTAGACCAA | GCCTGTCATTCCAGCAATCAA |
| FASN | CCGTGTGACCGCCATCTAT | TGCTGTCGTCTGTAGTCTTGA |
| ACAA2 | GGACTTCTCTGCCACCGATT | ACATTGCCCACGATGACACT |
| HADH | GGAGAACCTGAAGCTGAAGAAC | CTTGTCTGGTGGTGGCATTG |
| PPT1 | GTTCTCACATCTGCGACTTCAT | CTGCCAAGAAGATGCTGTAGTT |
| ELOVL5 | CCTCTGGTGGTACTACTTCTCC | GGTAGCGTGGTGGTAGACAT |
| ELOV6 | ACGAGAACGAAGCCATCCA | ATCAGATGCCGACCACCAA |
| HSD17B12 | GCCTCGTACTCGCTCTTCA | CCATCAGTGCCACCTGTAAC |
| HACD2 | CGGCGTACCTGGTCATCTAC | GGCTCCTGTCTGGAAGAACTT |
| ACOT2 | TGAAGAAGCCGTGAACTACCT | GGAGCCATTGATGACCACAG |
| ACAT2 | TGCTGCTGTGGTCCTTATGA | TATGGCTGGAATTGGTCCTACT |
| ACAA1 | GCTGAGATTGTGCCTGTGAC | GGTAGAGCCTCCATCCTTGAA |
| ACOX1 | GGCGTGGAACTTGACTTCTG | CGGCTCTGTCTTGAATCTTGG |
| ACADM | ATTACCGAAGAGTTGGCGTATG | TCATTGGCTGCTCCGTCAT |
| CPT1B | AAGCACACCAGGCAGTAGC | TCCAGGAGTTGATTCCAGACAG |
| GK | CCTCCTGACAACCGTAGCA | TCTCTTAGCCAGCGGATTACA |
| AGPAT2 | GTGCTCTGCCTGTCCTTCT | CTTCTGTCCGCTGACCTCAA |
| DGAT1 | GCTATCCAGACAACCTGACCTA | GCATCTCAAGAACTCGTCGTAG |
| LPL | ACAAGGTCAGAGCCAAGAGAA | GTTGCTTGCCATCCTCAGTC |
| FAS | CTGGCTCACAGTTAAGAGTTCA | CAGGTTGGCATGGTTGACAG |
| UCP1 | TGGAGGTGTGGCAGTGTTC | TCTGTGGTGGCTATAACTCTGT |
| PPARGC1A | TCGCTGCTCTTGAGAATGGA | TCGTCTGAGTTGGTATCTAGGT |
| ADIPOQ | GCTCTCCTGTTCCTCTTAATCC | ATGCCTGCCATCCAACCT |
| SREBF1 | TCAACAACCAAGACAGTGACTT | GCCAGAGAAGCAGAAGAGAAG |
| CEBPA | ACAAGAACAGCAACGAGTACC | GGTCATTGTCACTGGTCAACT |
| SCD1 | GGTGATGTTCCAGAGGAGGTA | CCAGAGTGTATCGCAAGAAGG |
| FADS2 | TGAAGAAGACTGCTGAGGACAT | GTGCCGAAGTACGAGAGGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, X.; Fang, X.; Mi, J.; Liu, Y.; Liu, R.; Li, G.; Li, Y.; Yang, R. Effects of Adipose Tissue-Specific Knockout of Delta-like Non-Canonical Notch Ligand 1 on Lipid Metabolism in Mice. Int. J. Mol. Sci. 2024, 25, 132. https://doi.org/10.3390/ijms25010132
Lu X, Fang X, Mi J, Liu Y, Liu R, Li G, Li Y, Yang R. Effects of Adipose Tissue-Specific Knockout of Delta-like Non-Canonical Notch Ligand 1 on Lipid Metabolism in Mice. International Journal of Molecular Sciences. 2024; 25(1):132. https://doi.org/10.3390/ijms25010132
Chicago/Turabian StyleLu, Xin, Xibi Fang, Jiaqi Mi, Yue Liu, Ruimin Liu, Guanghui Li, Yue Li, and Runjun Yang. 2024. "Effects of Adipose Tissue-Specific Knockout of Delta-like Non-Canonical Notch Ligand 1 on Lipid Metabolism in Mice" International Journal of Molecular Sciences 25, no. 1: 132. https://doi.org/10.3390/ijms25010132
APA StyleLu, X., Fang, X., Mi, J., Liu, Y., Liu, R., Li, G., Li, Y., & Yang, R. (2024). Effects of Adipose Tissue-Specific Knockout of Delta-like Non-Canonical Notch Ligand 1 on Lipid Metabolism in Mice. International Journal of Molecular Sciences, 25(1), 132. https://doi.org/10.3390/ijms25010132







